Abstract
Molecular motors are instrumental in mRNA localization, which provides spatial and temporal control of protein expression and function. To obtain mechanistic insight into how a class V myosin transports mRNA, we performed single-molecule in vitro assays on messenger ribonucleoprotein (mRNP) complexes that were reconstituted from purified proteins and a localizing mRNA found in budding yeast. mRNA is required to obtain a stable processive transport complex on actin, an elegant mechanism to ensure that only cargo-bound motors are motile. Increasing the number of localizing elements (“zipcodes”) on the mRNA, or configuring the track to resemble actin cables, enhanced run length and event frequency. In multi-zipcode mRNPs, motor separation distance varied during a run, showing the dynamic nature of the transport complex. Building the complexity of single-molecule in vitro assays is necessary to understand how these complexes function within cells
ASH1, the most well-studied localizing mRNA in budding yeast, serves as a paradigm for mRNA transport 1–4. The ASH1 transcript codes for a cell-fate determinant, and is transported and localized to the bud tip by the class V myosin motor Myo4p. The motor associates, via adapter proteins, with mRNA localization elements called “zipcodes”. Class V myosins are uniquely suited to move cellular cargo because they can walk micron long distances on actin filaments without disassociating, a feature called processivity. Type V myosins are generally double-headed to ensure that at least one head remains bound to the track at all times during transport. Myo4p is unique among class V myosins in that it is single-headed and tightly bound to its sole adapter protein She3p, likely forming a hetero-coiled coil that prevents the myosin coiled-coil from self-dimerizing 5. In essence, She3p is a subunit of Myo4p. The tetrameric mRNA binding protein She2p in turn recruits two single-headed Myo4p–She3p motors, hereafter referred to as the “motor complex” (Fig. 1a). Surprisingly, the two myosin motors coupled via She2p walk processively in a hand-over-hand motion, similar to dimeric mammalian myosin Va 6. We showed that the motor complex was processive at low salt (50 mM KCl) 6, but no movement was observed near physiologic ionic strength, implying that the complex had dissociated. This led us to hypothesize that we had not assembled all the components necessary for efficient cargo transport, and that mRNA may be required for full motor activity.
Figure 1.
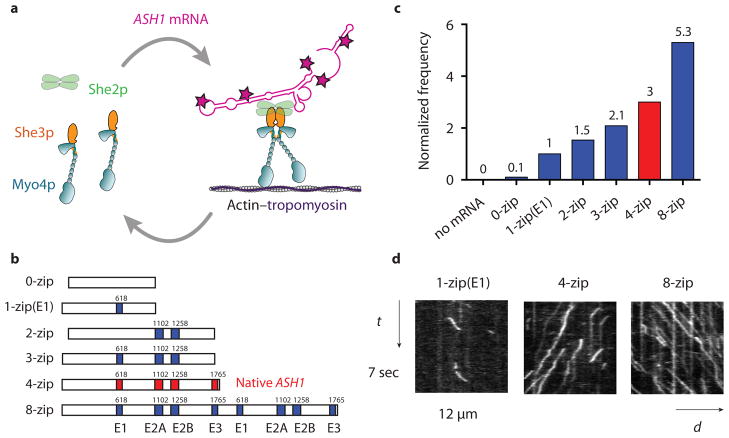
The frequency of mRNP processive runs increases with zipcode number. (a) Diagram showing the She2p and mRNA dependent dimerization of Myo4p–She3p at 140 mM KCl. Motion of the motor-mRNA complex is visualized by incorporating Alexa Fluor 488-5-dUTP (indicated by red stars) into the ASH1 mRNA transcript. (b) Schematic of ASH1 mRNA constructs containing the indicated number of zipcodes. The four zipcode elements in native ASH1 mRNA (E1, E2A, E2B, E3) are indicated in red. The initial base position of each element in the native ASH1 sequence is indicated. The 0-zip construct has no zipcodes. The 1-zip(E1) construct contains only the E1 motif, 2-zip both the E2A and E2B zipcodes, and 3-zip the E1, E2A, and E2B zipcodes, all in their native positions. The 8-zip construct is formed from two concatenated 4-zip sequences. (c) Run frequencies of Myo4p mRNP motility from a representative experiment comparing ASH1 mRNA sequences containing varying numbers of zipcodes. The run frequency (number of runs per μM Myo4p per μm actin per sec) with 1-zip(E1) is normalized to one. The actin track has Tpm1p bound. Motion of the Myo4p motor complex without mRNA is visualized with She2p-YFP. The native ASH1 mRNA run frequency is indicated with a red bar. For ASH1 transcripts containing zipcodes, n ≥ 198, without zipcodes, n=13. (d) Kymographs of Myo4p mRNPs containing ASH1 mRNA with the indicated number of zipcodes. The slope of the trace is the speed of the mRNP. The length of the trace is its run length. The number of traces is related to run frequency. Conditions: 140 mM KCl, pH 7.4, 1 mM MgATP
To understand the molecular basis of mRNA transport, we fully reconstituted an mRNP in vitro. Using single molecule techniques, we showed that mRNA is not a passive cargo, but is instrumental for the stability of the motor complex at physiological salt. Any variation to our in vitro assay that more closely recapitulated conditions found in budding yeast led to more efficient movement of the mRNA transcript. The increased run frequency and run length observed in vitro are features that likely optimize mRNA localization in a cellular context.
RESULTS
ASH1 mRNA triggers assembly of a processive complex
An mRNP complex was reconstituted by adding ASH1 mRNA, synthesized with Alexa Fluor 488 dUTP for visualization, to the motor complex (Fig. 1a, Supplementary Fig. 1). Total internal reflection fluorescence (TIRF) microscopy was used to track mRNP movement on yeast actin–tropomyosin (Tpm1p) filaments. At low ionic strength (25 mM KCl), Myo4p–She3p supported robust motility of transcripts without localizing elements, independent of She2p. Only when the salt was raised to near physiological ionic strength (140 mM KCl) did a control mRNA without zipcodes become non-motile (Fig. 1b–c, Table 1, Supplementary Movie 1), and thus this salt concentration was used for all processivity experiments. Strikingly, when the native four-zipcode ASH1 mRNA was added to the motor complex under these ionic conditions, the assembled mRNP showed long-range continuous motion (Fig. 1d, Table 1, Supplementary Movie 2). Importantly, this result suggests that mRNA triggers the assembly of a processive complex by enhancing the stability of the motor complex, independent of other proteins implicated in promoting correct mRNA localization 7, 8.
Table 1.
Characteristic motion of mRNPs containing various ASH1 mRNA zipcodes
| ASH1 mRNA construct | Normalized Frequency | Run length (μm) | Speed (μm/sec) | n |
|---|---|---|---|---|
| Low concentration (25nM myosin, 0.035nM mRNA) | ||||
| 0-zip | 0.05 | - | - | 13 |
| 4-zip (native sequence) | 3 | 1.1 ± 0.07 | 2.3 ± 0.06 | 211 |
| 8-zip | 5.3 | 1.5 ± 0.11* | 2.2 ± 0.05 | 197 |
| 1-zip(E1) | 1 | 0.93 ± 0.03 | 1.9 ± 0.06 | 302 |
| 1-zip(E2A) | 1.04 | 1.04 ± 0.04 | 1.7 ± 0.05 | 353 |
| 1-zip(E2B) | 1.04 | 1.03 ± 0.02 | 1.9 ± 0.06 | 357 |
| 1-zip(E3) | 1.85 | 1.3 ± 0.04* | 1.6 ± 0.04 | 380 |
| High concentration (250nM myosin, 10nM mRNA) | ||||
| 1-zip(E1) | 1 | 1.4 ± 0.1 | 1.6 ± 0.04 | 189 |
| 4-zip (native sequence) | 2.2 | 2.2 ± 0.15* | 1.4 ± 0.04 | 183 |
| 8-zip | 2.3 | 2.75 ± 0.19* | 1.5 ± 0.04 | 145 |
Speed is calculated from a mean ± s.e.m. The total number of runs analyzed is “n”. Run frequency (number of runs per μM Myo4p per μm actin per sec) with 1-zip(E1) is normalized to one.
Asterisk (*) indicates P < 0.001, using the Kolmogorov-Smirnov Test.
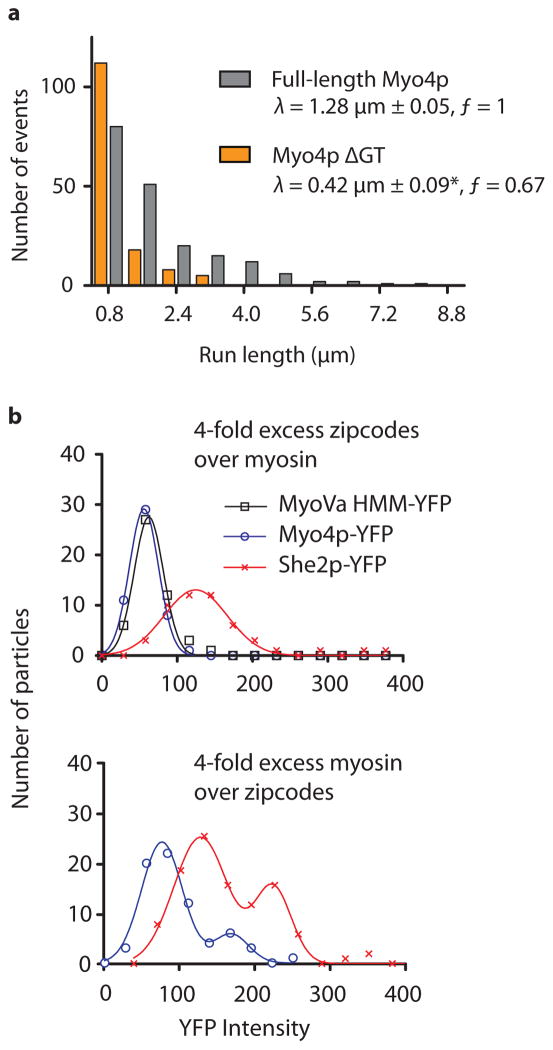
A domain of Myo4p that has not yet been assigned a function is the C-terminal globular tail, and we tested if it also contributes to stabilizing the mRNP. We performed in vitro assays with mRNPs assembled from Myo4p lacking the globular tail, and the single zipcode (E3) mRNA. These mRNPs had significantly reduced run length, and a lower run frequency compared to mRNPs formed with wild type Myo4p (Fig. 2a). Moreover, processive runs were only observed for a short period of time after diluting the mRNP mixture, implying a less stable mRNP.
Figure 2.
The Myo4p globular tail stabilizes the mRNP, and the recruitment of multiple motors to ASH1 is concentration dependent. (a) Characteristic run length (λ) of mRNPs containing the E3 zipcode and either wild type Myo4p (grey) or Myo4p lacking the globular tail (orange). Run length data is binned identically but displaced for presentation purposes. The asterisk (*) indicates that the run length is significantly reduced relative to wild type Myo4p (P<0.001, using the Kolmogorov-Smirnov Test; n ≥ 199). The run frequency (ƒ) (number of runs per μM Myo4p per μm actin per sec) with wild type Myo4p is normalized to one. (b) Histograms of motile mRNP intensities with native ASH1 mRNA. The upper panel shows recruitment of one dimeric Myo4p-YFP (blue) and one tetrameric She2p-YFP (red) at a 1 Myo4p dimer: 4 zipcode mixing ratio. In a given experiment only one protein contained a YFP label. The lower panel shows multiple motor complex recruitment at a 4 Myo4p dimer: 1 zipcode mixing ratio. MyoVa-HMM-YFP (black) serves as a 2-YFP intensity control.
Multiple zipcodes increases mRNP run frequency and length
A feature of localizing mRNAs is the presence of multiple zipcodes, and we wished to determine what advantage this confers. We used ASH1 mRNA, which contains four zipcodes, each capable of recruiting a motor complex for transport and supporting mRNA localization in the cell 9, 10. Three zipcodes (E1, E2A and E2B) are located in the coding region of the transcript, while the fourth element (E3) is located in the 3′ UTR (Fig. 1b) 9–11. Each zipcode has a highly conserved CGA triplet in one loop and a single conserved cytosine in a nearby loop 12, 13. We first tested if each zipcode element of the native ASH1 transcript is equally effective for mRNP transport. mRNPs containing only the E1, E2A or E2B zipcodes were indistinguishable from each other in terms of run length, frequency and speed (Table 1, Supplementary Movie 3). In contrast, mRNPs with the E3 zipcode supported longer run lengths and almost twice the run frequency compared to the other single zipcode transcripts (Table 1). Because Myo4p switches between a monomer and a dimer depending on whether it is in complex with She2p and mRNA, the characteristic run length of Myo4p is not only a function of the intrinsic properties of the motor (“duty ratio”, or fraction of the ATPase cycle spent attached to actin, and strain-dependent coordination between the heads), but also the stability of the motor complex. Assuming that modification of the mRNA cargo does not affect the motor properties of Myo4p, we interpreted the enhanced run lengths and frequencies of the E3 containing mRNP as promoting a more stable mRNP complex than either E1, E2A or E2B.
Because the E1, E2A, and E2B zipcodes showed similar interactions with the motor complex, these elements were used in combination to gain mechanistic insight into how multiple redundant zipcodes promote efficient mRNP transport. The run frequency of mRNPs with one zipcode (E1) was compared to transcripts containing two (E2A and E2B) or three zipcodes (E1, E2A, and E2B) (Fig. 1b). We observed a linear relationship between run frequency and zipcode number (Fig. 1c). This result suggests that ASH1 zipcodes function independently with no long range interactions, and that mRNA transcripts with multiple zipcodes recruit a motor complex more effectively to initiate transport. The lengths of the 2-zip and 3-zip mRNA constructs are comparable (1527 versus 1610 bases), and thus the higher recruitment rate correlates with zipcode number and not mRNA transcript length.
Based on the data described above, the E3 zipcode may be preferentially populated in multi-zipcode mRNAs, at the low nanomolar motor concentrations of the standard single molecule assay. Consistent with this idea, the run frequency with E3 alone was only slightly lower than with the native ASH1 sequence (Table 1). The relationship between zipcode number and run frequency was further tested with an eight zipcode transcript, formed by concatenating two native ASH1 mRNAs (Fig. 1b). mRNPs containing the eight zipcode transcript had approximately twice the run frequency of the four zipcode construct (Fig. 1c–d, Supplementary Movie 4), and a small but significant increase in run length, but no change in speed (Table 1, Supplementary Fig. 2). Recruitment of multiple motors typically increases run length 14, and thus the eight zipcode construct on average has a higher occupancy of bound motors than native ASH1 at low nanomolar protein concentration.
Multiple motors support longer processive runs
To directly quantify the number of She2p tetramers and Myo4p motors associated with the native ASH1 transcript, mRNPs were assembled with a YFP tag on either Myo4p or She2p. The YFP intensity of motile particles was quantified by TIRF microscopy. A dimeric mouse myosin Va-HMM-YFP construct provided a 2-YFP intensity standard. When ASH1 zipcodes were in molar excess to Myo4p motor complexes, two Myo4p–She3p motors and one She2p tetramer were recruited to the transcript. When motor complexes were in excess of ASH1 zipcodes, a second higher intensity peak emerged, indicating binding of an additional motor complex and an additional She2p tetramer (Fig. 2b). Additional zipcode elements thus become populated as motor concentration increases.
To further populate additional zipcodes with motor complexes, excess unlabeled Myo4p motor complex (250 nM) and unlabeled ASH1 mRNA (10 nM) were added to the standard TIRF assay. Under these conditions, run lengths were significantly longer for transcripts with four and eight zipcodes (2.2 μm and 2.75 μm, respectively) compared to ASH1 transcripts containing only the E1 zipcode (1.4 μm), consistent with multiple motor recruitment (Table 1, Supplementary Fig. 3). The run length enhancement was less apparent in the standard TIRF assay (Table 1) because low motor (25 nM) and mRNA (0.35 nM) concentrations failed to promote full occupancy of four or eight zipcode mRNAs with motor complexes.
Electron microscopy of mRNP complexes
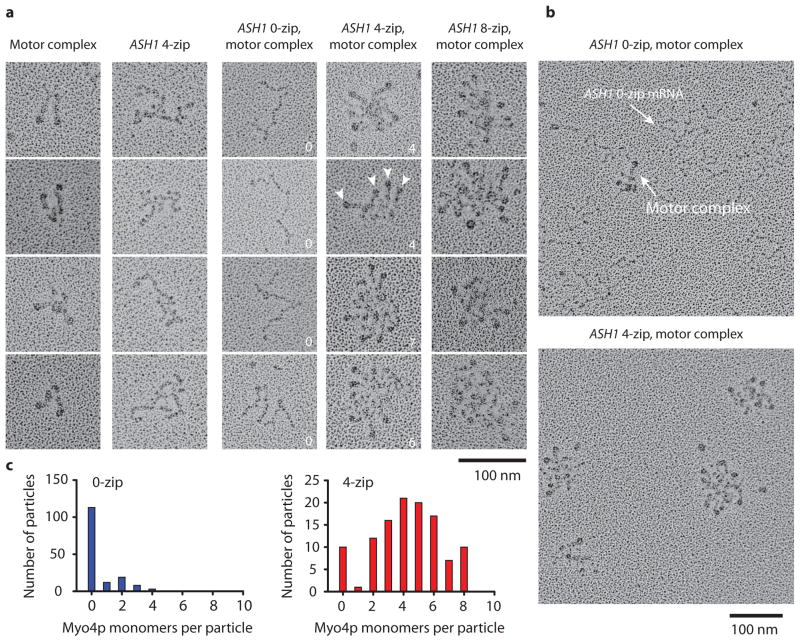
Electron microscopy of metal-shadowed complexes provided structural information about the mRNP. The higher protein concentrations which favor zipcode occupancy were used. As a control, the motor complex was visualized in the absence of mRNA at 25 mM KCl. Similar to our previous study 6, V-shaped structures were observed, consistent with She2p recruiting two single-headed Myo4p motors (Fig. 3a). The images shown are representative of the four types of structures that were observed for the motor complex. Metal-shadowed images of ASH1 mRNA alone showed structures distinctive from Myo4p. At 140 mM KCl, ASH1 mRNA transcripts lacking zipcodes did not bind the motor complex, consistent with the specificity seen at the single molecule level (Fig. 3a, b). When the native four zipcode ASH1 transcript was mixed with the motor complex, the components assembled into mRNPs containing anywhere from zero to eight motor heads, or as many as one motor pair per zipcode (Fig. 3c). mRNPs with eight zipcodes recruited even more motors.
Figure 3.
Metal-shadowed images show variable recruitment of motors. (a) Galleries of images showing (left to right) the recruitment of two Myo4p motors by She2p, the native 4-zip ASH1 transcript alone, and ASH1 transcripts containing 0, 4 or 8 zipcodes in the presence of the Myo4p motor complex(Myo4p–She3p–She2p). Arrowheads indicate Myo4p motors, and the numbers indicate bound heads. (b) Fields of the Myo4p motor complex in the presence of 0-zip or 4-zip ASH1 (c) Histograms showing the number of Myo4p monomers associated with 0-zip or 4-zip ASH1 mRNA.
mRNPs are optimized to walk on actin cables
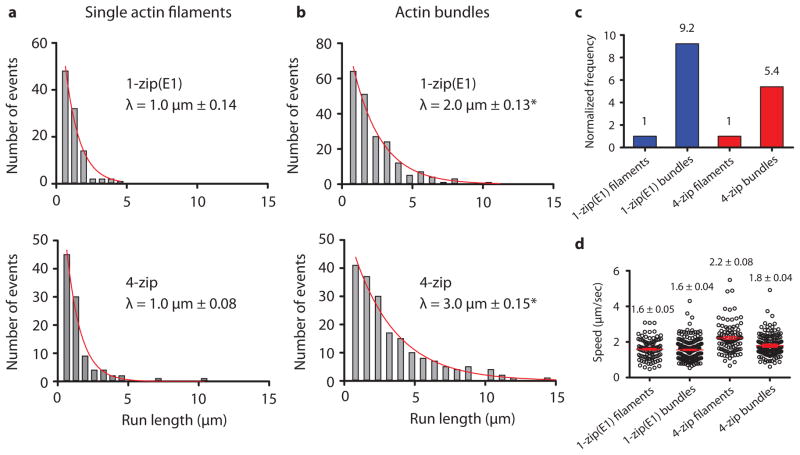
The electron microscopy images highlight the dissimilarity in structure of the Myo4p motor complex in isolation, and the fully assembled mRNP. These images suggest that mRNPs are likely optimized to walk on actin cables, rather than single actin filaments, so that multiple motors do not interfere with each other. We mimicked the actin cables found in vitro with actin–fascin–tropomyosin bundles, composed of parallel actin filaments 15. Motility of mRNPs carrying either the native ASH1 transcript, or the E1 zipcode only, was compared on single yeast actin–tropomyosin filaments versus bundles (Fig. 4a, b). Strikingly, run lengths for mRNPs with the native ASH1 transcript increased from 1 μm to 3 μm. Even the one zipcode mRNP doubled its run length on bundles. Compared to single actin filaments, frequencies on actin bundles were 5-fold higher with the native ASH1 mRNP, and nine-fold higher for the one zipcode mRNPs (Fig. 4c). No significant changes in speed were observed (Fig. 4d).
Figure 4.
Comparison of mRNP movement on single actin filaments versus bundles. Representative run length histograms of Myo4p mRNPs containing 1 zipcode (E1) or 4 zipcodes on (a) single actin filaments or (b) actin bundles. Both tracks have Tpm1p bound. Asterisk (*) indicates run lengths that are significantly different between filaments and bundles with P < 0.001 (Kolmogorov-Smirnov Test; n ≥ 103). (c) Run frequencies of Myo4p mRNP motility containing 1 (E1) or 4 zipcodes on single actin filaments or bundles. The run frequency (number of runs per μM Myo4p per μm actin per sec) with single actin filaments is normalized to one. (d) Speed distributions of Myo4p mRNPs on single actin filaments and bundles. The mean speed is indicated with a red line. Error is in s.e.m; n ≥ 103. Conditions: 140 mM KCl, pH 7.4, 1 mM MgATP
Variable inter-motor spacing during mRNP movement
To obtain more detailed and dynamic information on how multiple motor complexes move on actin filaments, we followed the separation distance between two motor domains on different motor complexes as the mRNP is processively transported on single actin filaments. To simplify the analysis, a two zipcode ASH1 transcript containing only the E1 and E3 zipcodes was used (Fig. 5a). Only one head of each motor complex was labeled with either a red or a green quantum dot (Qdot). The labeled-motor complexes were then assembled into mRNPs with unlabeled transcript. Only motile mRNPs containing a red and a green Qdot, i.e. two motor complexes, were analyzed by TIRF microscopy. Qdot position was tracked with ~6 nm accuracy 16.
Figure 5.
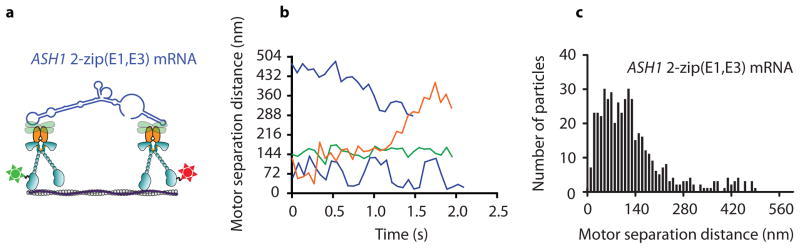
The spacing between two moving motor complexes coupled by mRNA varies. (a) Diagram of the experimental setup for quantifying the distance between two Myo4p motor complexes coupled by a 2-zip ASH1 mRNA construct containing E1 and E3 zipcodes. One head of one motor complex was labeled with a red Q-dot, and one head of a second motor complex was labeled with a green Q-dot. (b) Motor separation distances of four representative mRNP run trajectories as a function of time. (c) Distribution of instantaneous head separation distances for 25 mRNP run trajectories.
The spacing between the coupled motors varied greatly during a run. Fig. 5b shows that motors coupled by a naturally compliant ~140 nm mRNA cargo can get closer together, further apart, remain constant, or oscillate between shorter and longer inter-motor distances as a function of time. Combining measurements from multiple processive runs shows a range of inter-motor separation distances, with 90% of the observed spacings falling in the range of 10 to 250 nm (Fig. 5c).
DISCUSSION
Here we fully reconstituted an mRNP composed of a localizing ASH1 mRNA transcript and the budding yeast class V myosin motor complex (Myo4p–She3p–She2p). Using single molecule techniques, we demonstrated that mRNA is essential to form a processive complex near physiologic strength. Consistent with our direct demonstration of processive movement, in vitro binding assays showed that the She2p–She3p complex had an ~60-fold higher affinity for localizing mRNAs than for control mRNAs 17. In budding yeast, She2p interacts with ASH1 mRNA in the nucleus and then shuttles the complex into the cytoplasm 8, 18. Our observations suggest that at this point each zipcode-bound She2p would recruit a pair of Myo4p–She3p monomers that is capable of initiating processive motion. The requirement for mRNA to stabilize the motor complex provides a regulatory checkpoint such that unbound Myo4p monomers are not motile, and only cargo-bound Myo4p dimers can move processively along actin cables to localize the transcript to the bud tip.
Each of the four zipcode elements in ASH1 is sufficient for supporting transport and localization of a reporter mRNA in budding yeast, but the presence of all four zipcodes increased the quality of cellular localization in the bud 9, 11. By modifying the number of localizing elements in ASH1, we showed that two clear advantages of multiple zipcodes are an enhanced ability to initiate processive movement, and an increased run length. Both of these parameters become optimal when all zipcodes are fully occupied by motors. The single molecule and electron microscopy data indicate that motor recruitment to mRNPs is probabilistic, and multiple zipcodes function to bias the average number of motors associated with the mRNP.
The approximate cellular concentration of Myo4p, based on cell volume (30 μm3 (ref. 19)) and Myo4p copy number (2210 (ref. 20)), is ~100 nM. When motor concentrations similar to this were used, we obtained the highest run frequency and run length in vitro. Electron microscopy showed that the native ASH1 transcript can bind as many as eight Myo4p monomers, implying that ASH1 transcripts in yeast are likely transported by multiple motor complexes. Increased run frequency and length are features that likely promote cellular localization, providing a rationale for why localizing mRNAs tend to have multiple zipcodes 21.
We also showed that a novel function of the Myo4p globular tail is to stabilize the mRNP by providing additional weak interactions with She2p and/or mRNA. In typical dimeric class V myosins, the globular tail interacts with various adapter proteins to specify different cargoes, or it interacts with the myosin motor domain to form an auto-inhibited complex. Biochemical studies with Myo4p indicate that the globular tail is not required for interaction with its adapter protein She3p 5, nor does it contain the conserved residues required for auto-inhibition of the motor 22. In vivo studies, however, demonstrated the importance of the tail for correct mRNA localization 22, 23. Our in vitro results showing reduced run length and frequency, and lower stability of the mRNP formed with Myo4p lacking the globular tail, provides a mechanistic reason for impaired mRNA localization. It has been suggested that the stability of mRNA transport complexes result from a large number of weak interactions, none of which is essential for transport 24. Here we showed potential interactions between all proteins in the motor complex and the mRNA, which leads to the synergistic assembly of the mRNP.
Electron microscopy of an ASH1 transcript with multiple bound motors gave the visual impression that motors would interfere with each other if they walked on single actin filaments. Our results indicated that mRNPs are indeed optimized to walk on yeast actin cables, which provide additional lateral actin binding sites for multi-motor complexes. It should also be noted that all the actin tracks we used had the major tropomyosin isoform Tpm1p bound, to mimic the composition of the actin cables in vivo. Tropomyosin enhanced run frequency compared to bare actin, but was not essential to obtain processive movement. This differs from what we observed for Myo2p, the other class V myosin in budding yeast, where tropomyosin was the switch that was necessary to convert Myo2p into a processive motor 25.
In contrast to the static electron micrographs of the mRNPs, which were fairly uniform in diameter with head spacings no greater than ~100 nm, high resolution single molecule tracking of the separation distance between two motor complexes coupled by ASH1 revealed a highly dynamic view of mRNPs moving on actin. This variable and sometimes long inter-motor spacing is consistent with what we observed for mouse myosin Va motors coupled through a 50 nm DNA scaffold, which supported inter-motor spacings in the range of ~10–225 nm 26. Spacings that exceed the length of the scaffold are likely due to the reach of the motors. Visualizing two motor complexes moving on actin provides the first dynamic view of mRNP transport at the single molecule level.
Implications for mRNA transport in higher organisms
Our results from budding yeast have implications for mRNA transport in higher organisms. Mammalian myoVa, best-known for transport of membranous organelles, has recently been shown to have both mRNA and RNA binding proteins as another class of cargo (reviewed in 27). The similar processive behavior of the yeast motor complex and myosin Va imply that our observations likely apply to both motors. In eukaryotes, microtubule-based motors perform much of the long-range mRNA transport. There, the situation is complicated by having motors that move in opposing directions bound to the mRNA: dynein moving cargo toward the microtubule minus-end, and kinesin driving plus-end directed motion. During Drosophila embryogenesis, both non-localizing and localizing mRNAs undergo bidirectional transport on microtubules. Increasing zipcode number increases the average motor number associated with a localizing transcript in a probabilistic way, which in turn regulates the speed, frequency and duration of minus-end directed runs 28, 29. This variation in motor occupancy was proposed to account for the large variation in dynein driven mRNP motility in the cell. Our results also show variable motor recruitment to ASH1 mRNPs, indicating that this is a widely conserved mechanism to achieve efficient transport.
In conclusion, our study highlights the idea that only by reconstituting biological function from multiple purified components, can one fully understand how molecules that are parts of cellular complexes in vivo work at a mechanistic level.
ONLINE METHODS
Protein constructs
DNA encoding full-length budding yeast MYO4 (1471 amino acids), in pAcSG2, was followed by an eight amino acid FLAG-tag to facilitate purification by affinity chromatography. For experiments tracking motor domain position on actin, an N-terminal biotin tag was added for streptavidin conjugated Qdot (Invitrogen) attachment 6. For experiments quantifying YFP intensity of motile mRNPs, YFP was fused to the N-terminus of MYO4. Myosin constructs without the globular tail (Myo4pΔGT) were truncated at Leu 1024, followed by a FLAG tag. Constructs used to express budding yeast calmodulin (yCaM), Mlc1p, She3p, She2p, Tpm1p and yeast actin were previously described 6. She2p contained four point mutations (C14S, C68S, C106S, and C180S) to prevent aggregation. This construct was functional in a Δ SHE2 background 6. Bacterially expressed Tpm1p constructs included an Ala-Ser before the start codon to mimic acetylation 30. Constructs used for fascin expression were described previously 15.
Protein expression and purification
Myosin constructs were co-expressed with yeast CaM, Mlc1p, and She3p in Sf9 cells using the baculovirus system, and purified by FLAG affinity chromatography as described previously 6. Budding yeast proteins calmodulin (yCaM), Mlc1p, She2p and Tpm1p were expressed and purified as described previously 6. Human fascin purification was described previously 15. Chicken skeletal actin used for myosin Va motility was purified from acetone powder 31 and labeled with rhodamine phalloidin (Invitrogen). S. cerevisiae actin was expressed and purified as described previously 25 and labeled with Alexa Fluor 594 phalloidin.
Actin tracks
All actin tracks have Tpm1p bound (4:1 molar ratio). Yeast actin–tropomyosin bundles were prepared by mixing 0.8 μM Alexa Fluor 594 phalloidin-labeled yeast actin, 2μM Tpm1 and 0.7 μM fascin in motility buffer (25 mM imidazole, pH 7.4, 25 mM KCl, 1 mM EGTA and 4 mM MgCl2) and incubated overnight at 4° C.
ASH1 mRNA Constructs
Sequences encoding the ASH1 transcript were amplified from yeast genomic DNA and placed behind the SP6 promoter in the pSP72 vector (Promega). This sequence was followed by a polyA11 tail for transcript stability. DNA sequences of ASH1 zipcodes and ASH1 constructs used in this study are summarized in Supplementary Table 1 and S2. The ASH1 transcript containing two zipcodes designated 2-zip(E1, E3), used for determining the distance between the two motor domains, was made by mutating specific localization elements in the native ASH1 sequence by site directed mutagenesis of a conserved CGA triplet motif necessary for She2p recognition thereby maintaining native zipcode spacing 13. mRNA was synthesized by linearizing the DNA template with blunt ends at an EcoRV site situated directly after the polyA16 tail and transcribed using a phage SP6 RNA polymerase (RiboMAX system from Promega). ASH1 RNA was labeled by adding a mixture of Alexa488 labeled dUTP (Molecular Probes - Invitrogen) in a 1:10 ratio to unlabeled nucleotides to allow fluorophore incorporation during RNA synthesis.
Total internal reflection fluorescence (TIRF) microscopy
She2p was diluted to 0.6 mg/mL and clarified at 386,000 × g for 15 min. She2p and Myo4p–She3p were premixed at concentrations of 2.8 μM and 5.6 μM respectively in buffer A (25 mM imidazole, pH 7.4, 140 mM KCl, 1 mM EGTA, 4 mM MgCl2 and 1 mM DTT) containing 0.1 mg/mL yCaM and Mlc1p. To this, 20 units of RNasin Plus RNase Inhibitor (Promega) and 0.035 μM of the indicated ASH1 mRNA construct were added and incubated on ice for 30 min.
For experiments in which the distance between two motor pairs was measured, biotin-tagged and untagged full-length Myo4p heterodimers were formed by mixing 0.04 μM tagged Myo4p and 0.04 μM untagged Myo4p in buffer A. Half of this mixture was incubated with 0.2 μM red (655-nm emission) streptavidin Qdots, and the other half with 0.2 μM green (565-nm emission) streptavidin Qdots for 15 min on ice. To both mixtures, clarified She2p was added at 0.1 μM and incubated for 15 min on ice. This results in two populations of dimers: one that has only one head labeled with a red Qdot, and one that has only one head labeled with a green Qdot. Finally, the mixtures were combined 1:1 and unlabeled ASH1 2-zip(E1, E3) mRNA was added to 0.004 μM, giving a myosin dimer:ASH1 zipcode ratio of 2:1. Only motile complexes containing two bound dimeric motors, one with a green head and another with a red head were analyzed.
For experiments measuring YFP intensity of motile partials, She2p-YFP, She2p, and mouse myosin Va 32 HMM-YFP was clarified at 386,000 × g for 15 min. Where indicated, Myo4p-YFP or She2p-YFP was mixed with either She2p or Myo4p in a 1:1 mixing ratio (0.5:1 μM respectively). Unlabeled native ASH1 mRNA was then added to the Myo4p mixture at a 1:4, or 4:1 mixing ratio in buffer D (25 mM imidazole, pH 7.4,140 mM potassium acetate, 1 mM EGTA, 4 mM MgCl2 and 1 mM DTT). Myosin Va HMM-YFP was also prepared in buffer D.
Flow cells containing Alexa Fluor 594 phalloidin-labeled yeast actin–tropomyosin were prepared similar to that described previously 6. The Myo4p–ASH1 mRNA mixture was diluted to obtain final Myo4p concentrations of 14–56 nM and ASH1 mRNA concentrations of 0.2–0.7 nM in buffer A containing 0.1 mg/mL yCaM and Mlc1p, 2 μM Tmp1, 1 mM MgATP and ATP regeneration and oxygen savaging systems as described previously 6. For experiments measuring the distance between two motor pairs, the Myo4p–ASH1–Qdot mixture was diluted to obtain a final Myo4p concentration of 7 nM and an ASH1 mRNA concentration of 0.3 nM. For experiments that included excess Myo4p–She3p and ASH1 mRNA, 250 nM unlabeled motor and 10 nM unlabeled mRNA were added to the mixture. For experiments measuring the YFP intensity of Myo4p–ASH1 and ouse mMyosin Va HMM-YFP, mixtures were diluted in buffer D containing 0.1 mg/mL yCaM and Mlc1p, 2 μM Tmp1, 1 mM MgATP and ATP regeneration and oxygen savaging systems and added to flow cells containing Alexa Fluor 594 phalloidin-labeled yeast actin or rhodamine-phalloidin-labeled chicken skeletal actin respectively.
Data were collected as described previously 6. For experiments measuring mRNA motility, Alexa Fluors 594, 488 and Qdots were excited with a 488 nm argon laser. For YFP intensity measurements, Alexa Fluor 594 labeled actin and YFP was excited with a 532 and 488 nm laser line, respectively. Data were collected at four frames/sec with seven frame averaging. For experiments using labeled mRNA, movement of Alexa Fluor 488-labeled mRNA on actin was measured using ImageJ and the particle tracking plugin, MTrackJ 33. For all processivity assays, frequencies were generated by counting the total number of runs in movies acquired no more than 7 min after dilution of the mRNP mixture. Total number of runs was divided by the total actin length, time and final myosin concentration to generate a run frequency.
For YFP fluorescence intensity measurements, fluorescence intensities of motile mRNPs were measured in ImageJ using the particle tracking plugin, SpotTracker 34. Fluorescence intensities over a 3 ×3 pixel window were recorded and averaged over the entire run and corrected for background intensity. For measuring distances between motor pairs on mRNA, the movement of particles containing red and green Qdots were tracked using SpotTracker and color aligned. The average motor separation distance over an entire run was determined from instantaneous separation distances of runs where the separation distance did not vary significantly over the run.
Electron microscopy
Full-length Myo4p was diluted to 0.32 mg/mL in buffer B (10 mM MOPS, pH 7.0, 140 mM KCl, 1 mM MgCl2, 1 mM EGTA, 2 mM DTT and 80 units RNasin Plus RNase Inhibitor) containing 0.04 mg/mL yCaM and Mlc1p. She2p was diluted to 0.6 mg/mL in buffer B. Both proteins were clarified at 350,000 × g for 15 min, and protein concentration after clarification was determined by a Bradford assay. For samples containing only mRNA, native ASH1 mRNA was diluted to 0.042 μM in buffer C (10 mM MOPS, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 2 mM DTT and 80 units RNasin Plus RNase Inhibitor), after which two volumes of the same buffer containing ~95% glycerol were added. For samples containing the Myo4p–She3p–She2p motor complex, 0.1 mg/mL Myo4p–She3p and 0.02 mg/mL She2p were mixed in buffer C. This equates to a 1:1.2 molar ratio of Myo4p–She3p dimer to She2p tetramer. The mixture was incubated on ice for 30 min and then mixed with two volumes of the same buffer containing ~95% glycerol. For samples containing the full mRNP, Myo4p–She3p and She2p was diluted in buffer B. To the myosin complex, 0.16 μM, 0.16 μM, 0.042 μM and 0.02 μM of ASH1 constructs containing 0, 4 and 8 zipcodes respectively were mixed such that the molar ratio of motors to zipcodes was 2:1, followed by two volumes of the same buffer containing ~95% glycerol were added. 50 μL of the mixture was sprayed onto a freshly split mica surface, metal shadowed with platinum at a stage angle of 7° and carbon-coated using an RMC RFD-9010 Freeze Fracture System. Replicas were imaged at a magnification of 50,000 with a JEOL JEM 1200EXII transmission electron microscope equipped with an AMT XR-60 digital camera.
Supplementary Material
Acknowledgments
The authors thank M. Daniels for EM support and the use of the National Heart, Lung, and Blood Institute Electron Microscopy Core Facility. We also thank G. Kennedy for technical assistance, D. Warshaw for use of the TIRF microscope, E. Krementsova for protein expression, M. Sckolnick, A. Hodges, H. Lu and S. Lowey for helpful discussions. This work was supported by funds from the US National Institutes of Health (GM078097 to K.M.T.).
Footnotes
AUTHOR CONTRIBUTIONS
T.E.S. and K.M.T designed experiments. T.E.S. conducted experiments and analyzed the data. C.S.B. created RNA constructs and cloned and expressed the proteins. M.H. preformed rotary shadowing electron microscopy. T.E.S and K.M.T wrote the manuscript.
References
- 1.Sil A, Herskowitz I. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 2.Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- 3.Long RM, et al. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 4.Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 5.Hodges AR, Krementsova EB, Trybus KM. She3p binds to the rod of yeast myosin V and prevents it from dimerizing, forming a single-headed motor complex. J Biol Chem. 2008;283:6906–6914. doi: 10.1074/jbc.M708865200. [DOI] [PubMed] [Google Scholar]
- 6.Krementsova EB, et al. Two single-headed myosin V motors bound to a tetrameric adapter protein form a processive complex. J Cell Biol. 2011;195:631–641. doi: 10.1083/jcb.201106146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Long RM, et al. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J Cell Biol. 2001;153:307–318. doi: 10.1083/jcb.153.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen Z, Paquin N, Forget A, Chartrand P. Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol Biol Cell. 2009;20:2265–2275. doi: 10.1091/mbc.E08-11-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol. 1999;9:333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- 10.Chartrand P, Meng XH, Huttelmaier S, Donato D, Singer RH. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol Cell. 2002;10:1319–1330. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez I, Buonomo SB, Nasmyth K, von Ahsen U. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr Biol. 1999;9:337–340. doi: 10.1016/s0960-9822(99)80145-6. [DOI] [PubMed] [Google Scholar]
- 12.Jambhekar A, et al. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2005;102:18005–18010. doi: 10.1073/pnas.0509229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olivier C, et al. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol. 2005;25:4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beeg J, et al. Transport of beads by several kinesin motors. Biophys J. 2008;94:532–541. doi: 10.1529/biophysj.106.097881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodges AR, Bookwalter CS, Krementsova EB, Trybus KM. A nonprocessive class V myosin drives cargo processively when a kinesin- related protein is a passenger. Curr Biol. 2009;19:2121–2125. doi: 10.1016/j.cub.2009.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warshaw DM, et al. Differential labeling of myosin V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys J. 2005;88:L30–32. doi: 10.1529/biophysj.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller M, et al. A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol. 2011;9:e1000611. doi: 10.1371/journal.pbio.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du TG, et al. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep. 2008;9:781–787. doi: 10.1038/embor.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jorgensen P, et al. The size of the nucleus increases as yeast cells grow. Mol Biol Cell. 2007;18:3523–3532. doi: 10.1091/mbc.E06-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 21.Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13:625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heuck A, et al. The structure of the Myo4p globular tail and its function in ASH1 mRNA localization. J Cell Biol. 2010;189:497–510. doi: 10.1083/jcb.201002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bookwalter CS, Lord M, Trybus KM. Essential features of the class V myosin from budding yeast for ASH1 mRNA transport. Mol Biol Cell. 2009;20:3414–3421. doi: 10.1091/mbc.E08-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arn EA, Cha BJ, Theurkauf WE, Macdonald PM. Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev Cell. 2003;4:41–51. doi: 10.1016/s1534-5807(02)00397-0. [DOI] [PubMed] [Google Scholar]
- 25.Hodges AR, et al. Tropomyosin is essential for processive movement of a class v Myosin from budding yeast. Curr Biol. 2012;22:1410–1416. doi: 10.1016/j.cub.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu H, et al. Collective dynamics of elastically-coupled myosinV motors. J Biol Chem. 2012 doi: 10.1074/jbc.M112.371393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCaffrey MW, Lindsay AJ. Roles for myosin Va in RNA transport and turnover. Biochem Soc Trans. 2012;40:1416–1420. doi: 10.1042/BST20120172. [DOI] [PubMed] [Google Scholar]
- 28.Bullock SL, Nicol A, Gross SP, Zicha D. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr Biol. 2006;16:1447–1452. doi: 10.1016/j.cub.2006.05.055. [DOI] [PubMed] [Google Scholar]
- 29.Amrute-Nayak M, Bullock SL. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat Cell Biol. 2012 doi: 10.1038/ncb2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maytum R, Geeves MA, Konrad M. Actomyosin regulatory properties of yeast tropomyosin are dependent upon N-terminal modification. Biochemistry. 2000;39:11913–11920. doi: 10.1021/bi000977g. [DOI] [PubMed] [Google Scholar]
- 31.Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85(Pt B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 32.Mercer JA, Seperack PK, Strobel MC, Copeland NG, Jenkins NA. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature. 1991;349:709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- 33.Meijering E, Dzyubachyk O, Smal I. Methods for cell and particle tracking. Methods Enzymol. 2012;504:183–200. doi: 10.1016/B978-0-12-391857-4.00009-4. [DOI] [PubMed] [Google Scholar]
- 34.Sage D, Neumann FR, Hediger F, Gasser SM, Unser M. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans Image Process. 2005;14:1372–1383. doi: 10.1109/tip.2005.852787. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.