Abstract
The polycomb group gene Bmi1 is required for maintenance of adult stem cells in many organs1, 2. Inactivation of Bmi1 leads to impaired stem cell self-renewal due to deregulated gene expression. One critical target of BMI1 is Ink4a/Arf, which encodes the cell cycle inhibitors p16ink4a and p19Arf3. However, deletion of Ink4a/Arf only partially rescues Bmi1 null phenotypes4, indicating that other important targets of BMI1 exist. Here, using the continuously-growing mouse incisor as a model system, we report that Bmi1 is expressed by incisor stem cells and that deletion of Bmi1 resulted in fewer stem cells, perturbed gene expression, and defective enamel production. Transcriptional profiling revealed that Hox expression is normally repressed by BMI1 in the adult, and functional assays demonstrated that BMI1-mediated repression of Hox genes preserves the undifferentiated state of stem cells. As Hox gene upregulation has also been reported in other systems when Bmi1 is inactivated1, 2, 5–7, our findings point to a general mechanism whereby BMI1-mediated repression of Hox genes is required for the maintenance of adult stem cells and for prevention of inappropriate differentiation.
A central goal in stem cell biology is to understand the mechanisms of tissue regeneration and renewal used by diverse organs. In the case of the rodent incisor, continuous growth relies on stem cells that share several characteristics with other regenerating adult systems8–11. These include residence in a discrete niche, slow division kinetics with respect to surrounding cells, and the ability to give rise to differentiated lineages throughout the life of the animal. Label retaining experiments in mice utilizing either BrdU incorporation or genetic labeling with a tetracycline inducible Histone 2B-GFP (H2BGFP) reporter demonstrated that a population of slowly dividing epithelial stem cells was present in a structure called the labial cervical loop (LaCL) at the proximal end of the incisor8, 12. Lineage tracing experiments showed that these cells gave rise to the highly proliferative transit-amplifying (T-A) cells that then differentiated into enamel-secreting ameloblasts11–13. Genetic analyses have shown that development of the incisor stem cells is controlled by TGF-β/BMP and FGF signaling9, 10 and that adult stem cells require active SHH signaling to produce differentiated progeny12. However, it remains relatively unknown how homeostasis is regulated in the adult LaCL. Because Bmi1 plays a key role in adult stem cell homeostasis in a number of mammalian tissues1, 2, 4, 14, we set out to study the role of Bmi1 in the adult incisor.
To investigate Bmi1 expression, we utilized Bmi1GFP reporter mice, in which the second exon of Bmi1 was replaced by GFP, resulting in a null allele15. In Bmi1GFP/+ adult mice, GFP was expressed in cells of the stellate reticulum (SR) and in the outer enamel epithelium (OEE), where the LaCL stem cells reside (Fig. 1a–c). These cells underwent infrequent cell divisions12 and thus retained H2B-GFP or BrdU after extended chase periods (Fig. 1b, 2f), similar to stem cells of the hair follicle bulge16. In addition, in Bmi1GFP/+;Gli1LacZ/+ mice, Bmi1GFP was co-expressed with Gli1LacZ, a marker for adult dental stem cells12 (Supplementary Fig. S1a–d). Together, these data suggested that Bmi1 marks the incisor stem cells.
Figure 1. Bmi1-expressing cells in the dental epithelium are stem cells.
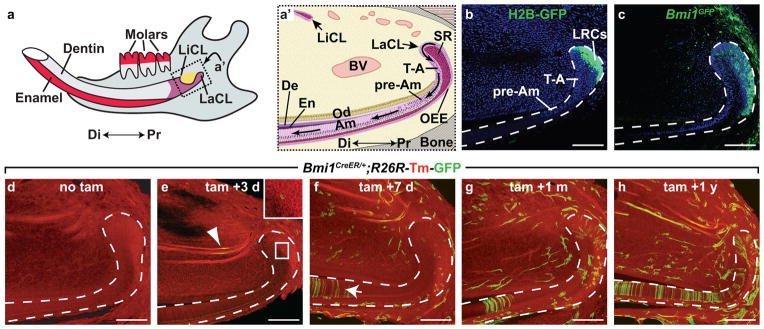
(a) Schematic diagram of an adult mandible. The incisor is a long tooth that grows under the molars. Enamel is produced by ameloblasts, which are present only on the labial surface. Dentin, produced by odontoblasts, is deposited on both the labial and lingual surfaces. Di, distal. LiCL, lingual cervical loop. LaCL, labial cervical loop. Pr, proximal. (a′) Schematic representation of the cell types associated with the dental epithelium and stem cell niche. Arrows in labial epithelium represent direction of movement of cells as they differentiate. Am, ameloblasts. BV, blood vessel. De, dentin. En, enamel. Od, odontoblasts. OEE, outer enamel epithelium. pre-AM, pre-ameloblasts. SR, stellate reticulum. T-A, transit amplifying. (b) K5tTA;H2BGFP mice treated for 2 months with doxycycline reveal label retention in the SR and OEE of the labial cervical loop. LRC, label retaining cells. (c) Bmi1GFP expression is localized to the OEE and SR of the cervical loop. (d–h) Bmi1CreER;R26R-Tm-GFP mice were induced at 6 weeks with tamoxifen and chased for the indicated time period. Dotted lines in b–h outline the dental epithelium. Scale bar = 100 μm.
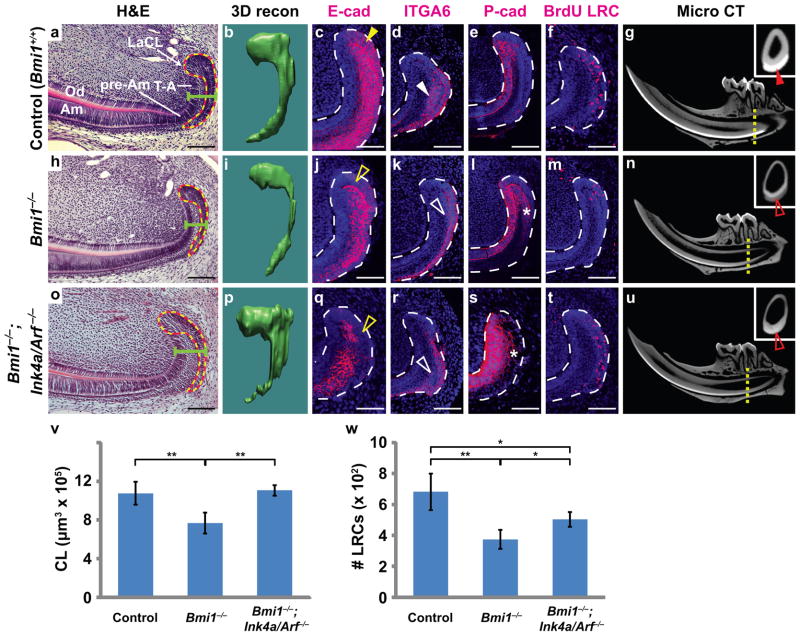
Figure 2. Deletion of Bmi1 affects adult LaCL through both Ink4a/Arf-dependent and independent mechanisms.
(a,h,o) Haematoxylin and eosin staining comparing LaCLs from 5-month-old control (Bmi1+/+), Bmi1−/−, and Bmi1−/−;Ink4a/Arf−/− mice. Dotted lines outline region traced on coronal sections for 3-dimensional (3D) renderings. Green bar demarcates width of cervical loop. (b,i,p) 3D renderings enable reconstruction (recon) of the control, Bmi1−/− and Bmi1−/−;Ink4a/Arf−/− (triple mutant) LaCLs. (c,j,q) E-cadherin staining of the LaCL in control, Bmi1, and triple mutants. E-cadherin expression is downregulated in both the single and triple mutants (open yellow arrowheads) when compared to the control (yellow arrowhead). (d,k,r) Expression of ITGA6 detected by immunostaining is decreased in the Bmi1−/− and Bmi1−/−;Ink4a/Arf−/− LaCLs (open white arrowheads) compared to the control (white arrowhead). (e,l,s) P-cadherin expression in the LaCL is expanded in both the Bmi1 and Bmi1−/−;Ink4a/Arf−/− LaCLs (asterisks). (f,m,t) Representative sections of LaCLs from control, Bmi1−/−, and Bmi1−/−;Ink4a/Arf−/− jaws. Animals were pulsed with BrdU post-natally and aged for 1.5 months for identification of label-retaining cells (LRCs). (g,n,u) MicroCT scans showing mandibles from 5-month-old control, Bmi1−/− and Bmi1−/−;Ink4a/Arf−/− mice. Enamel is thinner and less mineralized in both single and triple mutants when compared to the control. Insets are coronal sections through the distal root of the second molar (yellow dotted lines) and show that enamel is less mineralized in mutants (open red arrowheads) than in the control (red arrowhead). (v) Quantification of volume of the LaCL stem cell compartment in control, Bmi1−/−, and Bmi1−/−;Ink4a/Arf−/− (n = 4 mice for each genotype). (w) Quantification of BrdU LRCs by sectioning and staining through the entire LaCL (n = 5 control, 3 Bmi1−/−, and 4 Bmi1−/−;Ink4a/Arf−/− mice). Error bars indicate means ± s.d. * is p < 0.05, and ** is p < 0.001. Scale bar = 100 μm for a,h,o, and 75 μm for c–f, j–m, and q–t. Source data of statistical analyses are shown in Supplementary Table S2 and S3.
To determine whether Bmi1-expressing cells are indeed stem cells that can give rise to differentiated cell types over a long period of time, we performed inducible genetic lineage tracing. This technique permanently labels a cell and its progeny and definitively identifies adult stem cells in vivo17. To this end, we genetically labeled Bmi1-expressing cells and traced their descendants using an inducible Bmi1CreER strain18 crossed to a Cre-responsive reporter line (R26R-Tm-GFP)19. Induction of Cre activity by tamoxifen injection results in permanent expression of GFP in Bmi1-expressing cells and their progeny. While we did not detect GFP labeling in uninjected Bmi1CreER/+;R26R-Tm-GFP mice (Fig. 1d), we observed a few labeled cells in the LaCL and surrounding mesenchyme 3 days after tamoxifen injection (Fig. 1e, insert). GFP was also observed in blood vessels (Fig. 1e, arrowhead), consistent with a previous report of Bmi1 expression in endothelium20. By 7 days after tamoxifen injection, increased numbers of GFP-positive cells were observed in the LaCL and mesenchyme (Fig. 1f). Importantly, as cells in the pre-ameloblast region move ~350 μm per day21, 22, the appearance of GFP-positive pre-ameloblasts in the dental epithelium (Fig. 1f, arrow) several days after tamoxifen injection established the production of differentiated cells from Bmi1-positive progenitors. Longer chases revealed the accumulation of labeled cells in the LaCL, indicating that LaCL stem cells had undergone self-renewal (Fig. 1g,h). The appearance of groups of labeled ameloblasts suggested that each cluster represented the progeny of a single stem cell whose descendants underwent several rounds of replication. Together, the expression and lineage tracing data demonstrated that Bmi1-expressing cells in the LaCL are stem cells.
Next, to identify the function of Bmi1 in the mouse incisor, we analyzed the architecture of the dental epithelium in Bmi1 null animals (Bmi1GFP/GFP, hereafter referred to as Bmi1−/−) at 5 months of age by performing haematoxylin and eosin staining. Bmi1−/− animals had LaCLs that were consistently thinner in both the sagittal and coronal planes, with a substantial decrease in the amount of SR compared to Bmi1+/+ siblings (Fig. 2a,h). To obtain a clearer picture of the morphological defect in Bmi1−/− LaCLs, we prepared 3D reconstructions of the LaCL from Bmi1+/+ and Bmi1−/− animals. The 3D reconstructions demonstrated a significant loss of tissue in the LaCL, specifically in the SR on the labial side (Fig. 2b,i, and Supplementary Fig. S2). On average, Bmi1−/− LaCLs showed a 28% decrease in total volume compared to controls (Fig. 2v).
We then asked if the expression profile was altered in Bmi1−/− LaCLs by analyzing markers of dental epithelium. E-cadherin is normally expressed in stem cells residing in the SR and OEE and downregulated in the T-A zone21 (Fig. 2c). Conversely, P-cadherin is absent in the stem cell region but upregulated in the T-A region21 (Fig. 2e). In Bmi1−/− LaCLs, the domain of E-cadherin expression was smaller (Fig. 2j), and P-cadherin staining was expanded posteriorly into the OEE (Fig. 2l, asterisk), indicating that cells in this region assumed a more differentiated character. We next examined the expression of integrin alpha 6 (ITGA6), a marker of hematopoietic23, neural23, tracheal24, and epidermal25 stem cells that is also expressed in the dental epithelial stem cells26. Similar to E-cadherin, ITGA6 is normally expressed in the SR and OEE of the LaCL (Fig. 2d), and its domain of expression was dramatically smaller in the LaCL of Bmi1−/− animals (Fig. 2k). Together, the expression pattern of these markers indicated that a loss of Bmi1 activity caused a reduction in the size of the stem cell-containing region in the incisor.
Because Bmi1 is known to regulate stem cell self-renewal1, 2, 4, we asked whether the reduction of cells in the Bmi1−/− LaCLs was due at least in part to a reduced population of stem cells, using BrdU label retention as a marker of these cells8, 12. We administered BrdU to perinatal Bmi1+/+ and Bmi1−/− pups and analyzed the number of label-retaining cells (LRCs) in 6-week-old adult mice (Fig. 2f,m). On average, Bmi1−/− LaCLs had 45% fewer LRCs compared to Bmi1+/+ controls (Fig. 2w), indicating that Bmi1 regulates stem cell number in the incisor. Similarly low levels of apoptosis were observed in the mutants and controls (Supplementary Fig. S1e,f).
Finally, as Bmi1-expressing stem cells give rise to enamel-secreting ameloblasts, we compared the mineralized enamel in Bmi1+/+ and Bmi1−/− lower incisors using microtomography scans (microCT). In 5-month-old Bmi1+/+ adults, mature enamel was present in a swath that extended from a region proximal to the molars to the distal tip of the tooth (Fig. 2g). In contrast, mature Bmi1−/− enamel receded to a position between the proximal and distal roots of the first molar, indicating defective enamel formation by Bmi1−/− progeny (Fig. 2n).
In other contexts, BMI1 and other polycomb group proteins preserve stem cell self-renewal through repression of the Ink4a/Arf locus, which encodes two tumor suppressor proteins that negatively regulate the cell cycle1, 3, 4. As Ink4a/Arf was also upregulated in the Bmi1 null incisor epithelium (Fig. 3a), it was possible that BMI1 functions similarly in mouse incisor stem cells by repressing Ink4a/Arf expression. To test this possibility and to dissect phenotypes associated with Ink4a/Arf upregulation, we bred Bmi1−/−;Ink4a/Arf−/− triple mutants. We first observed that the additional loss of Ink4a/Arf was able to restore LaCL volume (Fig. 2o,p,v). Ink4a/Arf deletion also partially rescued LRC number (Fig. 2t,w), consistent with the notion that Bmi1 is required to maintain an adequate population of stem cells for homeostasis by repressing Ink4a/Arf expression. However, the incompleteness of the rescue pointed to the existence of additional BMI1 targets that may contribute to the mutant phenotype. Similarly, in other organs, deletion of Ink4a/Arf in Bmi1 null animals was insufficient to completely rescue either the morphological defects4 or the maintenance of downstream lineage specification14. Indeed, the marker expression pattern of the Bmi1−/−;Ink4a/Arf−/− mutant LaCL resembled that of the Bmi1 null incisor, with diminished E-cadherin and ITGA6 expression in the SR and OEE, accompanied by an expanded P-cadherin domain (Fig. 2q–s), suggesting that Bmi1 maintains incisor stem cell identity through mechanisms that do not involve Ink4a/Arf. Importantly, despite the rescue in LaCL size, Bmi1−/−;Ink4a/Arf−/− mice exhibited defects in enamel deposition (Fig. 2u), a phenotype that is likely due to abnormal gene expression in stem cells and their progeny.
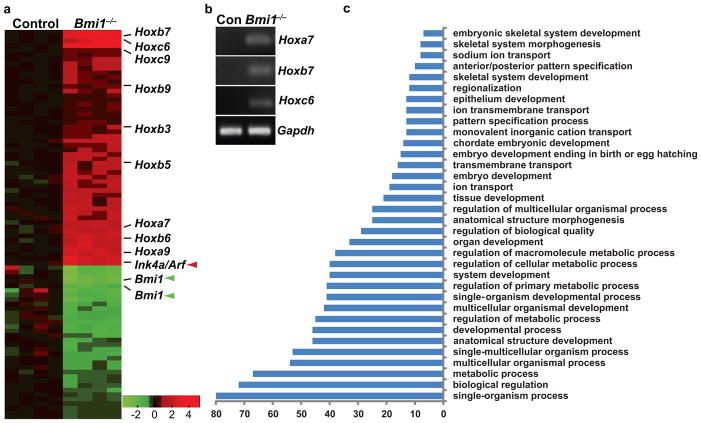
Figure 3. Bmi1 suppresses expression of Ink4a/Arf and Hox genes.
(a) Microarray analysis on Bmi1+/+ (control or con) and Bmi1−/− dental epithelia shows that inactivation of Bmi1 leads to de-regulation of several Hox genes, in addition to Ink4a/Arf (red arrowhead). Loss of Bmi1 expression is indicated by green arrowheads. (b) RT-PCR analysis showing upregulation of Hoxa7, b7, and c6 in Bmi1−/− LaCLs. (c) Gene ontology analysis reveals upregulation of genes normally involved in developmental processes and cell differentiation.
To identify additional BMI1 targets, we performed microarray analysis on dental epithelia from control and Bmi1 null incisors. In addition to Ink4a/Arf, many other genes were upregulated in the mutants, including 9 Hox genes from 3 different Hox clusters (Fig. 3a). To validate the upregulation of Hox genes in Bmi1 null dental epithelia, we performed RT-PCR on Bmi1+/+ and Bmi1−/− LaCLs. Whereas Hoxa7, b7, and c6 transcripts were undetectable in control LaCLs, their expression was readily detected in Bmi1−/− LaCLs (Fig. 3b). Similarly, Hoxc9 and a9 expression was dramatically increased in the Bmi1−/− LaCLs. Thus, BMI1 suppressed the expression of several Hox genes in the LaCL, suggesting that de-repressed Hox transcription may lead to premature differentiation. Gene ontology analysis further confirmed this notion, as genes associated with differentiation were upregulated in the mutants (Fig. 3c and Supplementary Table S1). Specifically, genes important for enamel formation that are normally expressed in maturing ameloblasts, such as amelotin (Amtn) and kallikrein-related peptidase (Klk4)27, 28, were prematurely upregulated in the LaCL (Supplementary Table S1).
It was next important to determine if BMI1-mediated repression of Hox genes was functionally required for the regulation of adult incisor stem cells. We utilized a tissue culture system21, 29 to enable concurrent manipulation of Hox expression levels in incisor stem cells. LaCL epithelia from Bmi1+/+ or Bmi1−/− incisors were dissociated to create single cell suspensions. 10 days after plating control cells, three morphological types of colonies appeared, with the majority composed of small and tightly packed cells that continued to express E-cadherin, suggesting that they maintained some of the incisor stem cell characteristics (Supplementary Fig. S3a–g). In contrast, dissociated LaCL cells from Bmi1−/− incisors grew very poorly and produced far fewer colonies (Supplementary Fig. S3h), consistent with upregulation of Ink4a/Arf and the self-renewal defects described above, but no change in apoptosis was observed in cultured Bmi1−/− cells (Supplementary Fig. S1g,h). Finally, sorting of cells from Bmi1GFP/+ animals showed that cells with higher GFP expression (GFPHI) produced significantly more colonies than cells with low GFP (GFPLO) (Supplementary Fig. S3i,j). Together, these data supported the functional results and confirmed that Bmi1 is required to maintain dental epithelial stem cells.
We then tested whether Ink4a/Arf deletion could rescue the colony-forming defect associated with loss of Bmi1 activity by culturing LaCL cells from Bmi1−/−;Ink4a/Arf−/− mice. In contrast to the Bmi1 mutants, the dissociated Bmi1−/−;Ink4a/Arf−/− LaCL cells readily formed colonies that were comparable in numbers to the control (Supplementary Fig. S3h), however the Bmi1−/−;Ink4a/Arf−/− colonies contained larger cells (Fig. 4a,b). We reasoned that whereas Bmi1−/− cells normally cannot self-renew due to elevated Ink4a/Arf expression, deletion of the Ink4a/Arf locus enabled these cells to proliferate. The altered morphology in Bmi1−/−;Ink4a/Arf−/− cells suggested that BMI1 additionally regulates a developmental program that is independent of Ink4a/Arf expression. Consistent with this notion, the transcriptomes of Bmi1−/−;Ink4a/Arf−/− colonies were dramatically different from wild-type controls and very similar to Bmi1−/− colonies (Supplementary Fig. S4). Thus, BMI1 must additionally control cell morphology and differentiation by regulating the expression of other genes.
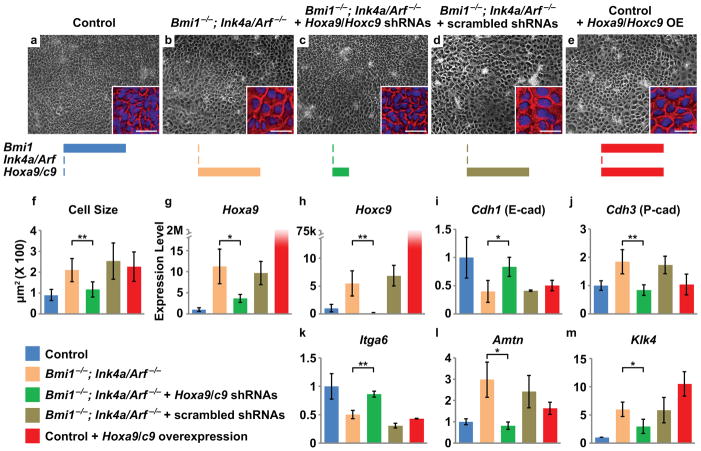
Figure 4. Hox gene upregulation contributes to the Bmi1 loss of function phenotype.
(a) Stem cell colonies derived from control LaCLs are composed of small, rounded cells. (b) Deletion of Ink4a/Arf enables colony formation by Bmi1−/− cells, but cell size is increased. (c) Knockdown of Hoxa9 and Hoxc9 rescues the morphological defects in Bmi1−/−;Ink4a/Arf−/− colonies. (d) Scrambled shRNA does not rescue the phenotype. (e) Overexpression of Hoxa9 and Hoxc9 in control cells phenocopies the morphology of Bmi1−/−;Ink4a/Arf−/− colonies. Insets in a–e are enlarged images of representative cells with pseudo-colored cell boundary (red) and DAPI nuclear staining (blue). Schematics represent the level of expression for each gene. (f) Quantification of cell size under different conditions (n = 3 independent experiments with 100 cells scored for each experiment). (g–m) Relative expression level by qPCR of Hoxa9, Hoxc9, Cdh1, Cdh3, Itga6, Amtn, and Klk4 in cells cultured under different conditions (n = 3 independent experiments). Error bars indicate means ± s.d. * is p < 0.05, and ** is p < 0.001. Scale bar = 100 μm for a–e and 30 μm for the insets. Source data of statistical analyses are shown in Supplementary Table S4 and S5.
To test whether upregulation of Hox genes was an important contributor to the mutant phenotype in Bmi1 null incisors, we used lentiviral transduction to introduce shRNAs against Hoxa9 and Hoxc9. These genes were the most upregulated Hox family members in both Bmi1−/− and Bmi1−/−;Ink4a/Arf−/− mutants as well as in colonies derived from these animals (Fig. 3a, Fig. 4 g,h). qPCR results demonstrated that the shRNAs effectively knocked down Hoxa9 and Hoxc9, whereas control scrambled sequences did not (Fig. 4 g,h). Furthermore, the doubly-infected colonies exhibited morphology and cell size similar to controls (Fig. 4 a,c,f). The rescue by Hoxa9/c9 shRNAs coincided with upregulation of Cdh1 (encoding E-cadherin) and Itga6 and with downregulation of Cdh3 (encoding P-cadherin), Amtn, and Klk4 (Fig. 4 i–m, Supplementary Fig. S5c–g), although single knockdown of either Hox gene alone did not show a clear rescue (Supplementary Fig. S5h,i), most likely due to functional redundancy. Thus, reducing Hoxa9/c9 expression reconstituted the incisor epithelial stem cell expression signature, which indicated that deregulated expression of Hoxa9 and Hoxc9 was at least partially responsible for the changes in stem cell morphology and gene expression in Bmi1−/− cells.
Repression of Hox genes in the adult dental stem cells is therefore an important function of Bmi1, and this finding led us to predict that overexpression of Hox genes in the LaCL would phenocopy the Bmi1−/− mutants. To that end, we first introduced exogenous Hoxa9/c9 by lentiviral transduction into cultured control LaCL cells (Fig. 4e,g,h). This overexpression caused enlarged cell size, downregulated expression of Cdh1 and Itga6, and upregulated Amtn and Klk4 expression, all of which are characteristic of the Bmi1−/− mutants (Fig. 4f,i–m). However, Cdh3 expression was not altered, suggesting that additional genes and/or factors were required for its upregulation. To further test whether Hox genes function similarly in vivo, we generated Gli1CreER/+;R26Hoxc9/YFP mice, in which ectopic Hoxc9 expression and a YFP Cre-reporter were induced specifically in the LaCL stem cells by tamoxifen injection (Fig. 5 a,f). Overexpression of Hoxc9 led to downregulation of E-cadherin and ITGA6 in the SR and OEE and to expanded P-cadherin expression (Fig. 5b–d,g–i), matching the gene expression phenotype exhibited by the Bmi1−/− mutants, although LaCL size was not affected (Fig. 5e,j,k).
Figure 5. Overexpression of Hoxc9 in LaCLs phenocopies Bmi1 mutants.
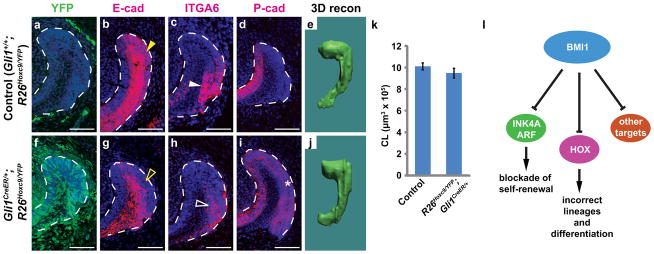
(a,f) Hoxc9 and YFP Cre-reporter were overexpressed using a Gli1CreER driver in the LaCL by tamoxifen induction, but not in the absence of Cre. YFP expression is shown here 10 days after induction. (b,g) E-cadherin is expressed in SR and OEE in the control LaCL (yellow arrowhead) but downregulated in the mutant (open yellow arrowhead). (c,h) ITGA6 is similarly downregulated in the mutant (compare solid and open white arrowheads). (d,i) P-cadherin expression is restricted in the T-A region in the control but expanded in the mutant (asterisk). All LaCLs are outlined by white dashed lines. Scale bar = 75 μm. (e,j,k) 3D reconstruction (recon) of LaCLs shows no difference in LaCL size between control and Gli1CreER/+;R26Hoxc9/YFP (n = 3 animals for each genotype). Error bars indicate means ± s.d. Source data of statistical analysis are shown in Supplementary Table S2. (i) Model for function of BMI1 in incisor stem cells.
Together, these data demonstrate that Bmi1-positive cells in the LaCL are adult stem cells and that Bmi1 function is required to maintain these cells through two distinct mechanisms (Fig. 5l). The first is via repression of Ink4a/Arf expression to permit stem cell self-renewal, a well-documented function of Bmi1 in other systems1, 3, 4. However, the Bmi1−/− phenotype was not fully rescued by the deletion of Ink4a/Arf, and thus, prevention of arrested cell division is not the sole function of BMI1. We found that the second function of BMI1 is to suppress the expression of Hox genes to prevent inappropriate cell differentiation. A similar observation was made in the Drosophila testis, where mutated polycomb genes resulted in Abdominal-B upregulation and abnormal cyst stem cell differentiation30, hinting at an evolutionarily conserved role for Bmi1 in regulating stem cell differentiation. It is notable that upregulation of Hox genes has also been documented in other Bmi1−/− adult stem cells1, 2, 5–7, and it will be important to consider the role of repression of Hox genes and other targets by BMI1 in those systems. For example, in haematopoietic stem cells, Ebf1 and Pax514 regulate lineage specification, and their upregulation in Bmi1 mutants causes a lineage shift during haematopoiesis. In the Bmi1 null incisor, we observed upregulation of genes associated with maturing ameloblasts in addition to upregulation of Hox genes, which is likely the cause of the abnormal enamel deposition by Bmi1−/− ameloblasts. Thus, we propose that Bmi1 has two distinct functions: (1) maintain adequate stem cell self-renewal by suppressing Ink4a/Arf and (2) prevent inappropriate differentiation by inhibiting the expression of Hox genes and other genes important for cellular maturation (Fig. 5i).
Methods
Mouse lines and injections
Mice carrying the Bmi1GFP 15, Bmi1CreER 18, Gli1CreER 31, Gli1lacZ 32, K5tTA33, R26R-Tm-GFP 19, and H2B-GFP16 alleles or transgenes were maintained and genotyped as previously described. For R26Hoxc9/+, chicken (Gallus gallus) Hoxc9 was cloned and inserted into the ROSA locus as described 34, 35. PCR genotyping for the R26Hoxc9 allele was performed using the following primers: R26wt_FW: 5′-AAAGTCGCTCTGAGTTGTTAT-3′, R26wt_REV: 5′-GGAGCGGGAGAAATGGATATG-3′, and R26loxC9_REV: 5′-GTTATGTAACGCGGAACTCCA-3′. Gli1CreER;R26Hoxc9/YFP mice were subsequently generated by mating Gli1CreER/+;R26YFP/+ male to R26Hoxc9/Hoxc9 females. For lineage-tracing studies, 5 mg Tamoxifen dissolved in corn oil was injected IP into adult mice between 8 and 12 weeks of age at 5 mg per 25 g body weight. For label retention studies, postnatal day 2 animals were injected with 200 μg BrdU for 3 consecutive days and sacrificed after 6 weeks. At least three mice were examined at each time point for all experiments. Unless specified in the main text, 6-week-old mice were used. Both male and female mice were included in this study.
Lentiviral production and colony forming assay
All shRNAs were designed using pSicoOligomaker 1.5 and cloned into the pSicoR-GFP vector (Tyler Jacks lab protocols, http://web.mit.edu/jacks-lab/protocols_table.html). Two sets of shRNAs against Hoxa9 and Hoxc9 were designed. The antisense sequences are: Hoxa9 (1st set): TTAATGCCATAAGGCCGGC, Hoxa9 scrambled (1st set): TACTATCGGTAACGGACGC, Hoxa9 (2nd set): TAAACAGAAACTCCTTCTC, Hoxa9 scrambled (2nd set): AGTCTACAACTATTCACAC, Hoxc9 (1st set): ATTGAAGAGAAACTCCTTC, Hoxc9 scrambled (1st set): ACATGAATTAAGTTCCGAC, Hoxc9 (2nd set): ATAGACCACAGACGACTGC, Hoxc9 scrambled (2nd set): ACGTACGAGAAACAGCTCC. Lentiviral packaging was performed using a 3rd generation packaging system (Addgene) according to established protocols. The in vitro culture system was modeled after a system for culture of hair follicle stem cells36–38 and has been previously published29. Briefly, whole incisors, including the entire dental epithelium, were isolated from mandibles and incubated in collagenase (Worthington) for 4 hours on ice. Labial cervical loops were mechanically isolated and dissociated in 100 μl Accumax (Sigma) at 37°C for 1 hour using gentle pippetting. Cells were counted and plated at a density of 15,000 cells per well on a 6 well plate. Colonies were then either harvested for RNA extraction or counted and imaged on a Leica inverted microscope after 10 days. All experiments were performed at least three times. Colony forming efficiency was expressed as a ratio of the number of small-cell containing colonies divided by the total number of plated cells. Cell sizes were measured using ImageJ (n = 3 independent experiments, and 100 cells were scored for each experiment).
Flow cytometry
LaCLs from Bmi1GFP/+ and Bmi1+/+ mice were isolated and dissociated as described above. Single cell suspensions were sorted for GFP expression using a FACSAria 2 cell sorter (BD Biosciences) at the Eli and Edythe Broad Center of Regeneration Medicine and Stem Cell Research at UCSF, Cell Analysis core. Two separate experiments were performed.
3D reconstruction of cervical loops
The epithelium was outlined from ~40 consecutive 7 μm thick coronal sections at the most proximal end of the mouse incisor (n = 4 animals per genotype). BioVis software (http://www.biovis3d.com) was used to generate 3-dimensional (3D) reconstructions from this data and to calculate the volume of the structures.
Microarray analysis and qPCR
Total RNA from dissected whole LaCLs or colonies was extracted using the Ambion Mirvana RNA Isolation kit. Sample preparation, labeling, and array hybridizations were performed according to standard protocols from the UCSF Shared Microarray Core Facilities and Agilent Technologies (http://www.arrays.ucsf.edu and http://www.agilent.com). Total RNA quality was assessed using a Pico Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). The datasets were normalized using the quantile normalization method39. No background subtraction was performed, and the median feature pixel intensity was used as the raw signal before normalization. A one-way ANOVA linear model was fit to the comparison to estimate the mean M values and calculate moderated t-statistic, B statistic, false discovery rate, and p-value for each gene for the comparison of interest. All procedures were carried out using functions in the R package limma in Bioconductor. Gene ontology analysis was performed using the Term Enrichment tool in AmiGO (http://amigo.geneontology.org). For qPCR analysis, cDNA was generated using the Advantage RT for PCR kit (Clontech), and Taqman assays (Ambion) were used to perform qPCR. Primers for Hoxc9, Amtn, Klk4, and L19 were ordered from IDT PrimeTime. Primer sequences for Hoxa9, Cdh1, Cdh3, and Itga6 are:Hoxa9: forward- GAATGAGAGCGGCGGAGAC, reverse- GAGCGAGCATGTAGCCAGTTG, Cdh1: forward- AATGAAGCCCCCATCTTTAT, reverse- GAGATGGACAGAGAAGACGC, Cdh3: forward-CCGCATCTTAAGGAGACGAA, reverse- AAATCTTGGTGCCTCTGTCC, Itga6: forward- GGAGCCTCTTCGGCTTCTC, reverse- AGTGCTTCTGCCCGAGGT. All samples were normalized to L19. CT values were extracted, and relative gene expression levels were calculated using the ΔΔCT method. All experiments were performed at least three times.
Histology and Immunofluorescence
Jaws were dissected from perfusion-fixed animals, post-fixed in 4% PFA overnight, decalcified in RNase-free 0.5 M EDTA for 16 days, and processed for paraffin embedding. 7 μm sections were prepared and stained with Hematoxylin and Eosin using standard methods. For all images shown, representative samples were chosen after sectioning through the entire jaw, in order to avoid plane of section artifacts. Brightfield images were obtained using a Leica DFC 500 camera with a Leica DM 5000B microscope. For immunostaining, paraffin sections were rehydrated, incubated in 1mM EDTA just below boiling temperature for 30 min for antigen retrieval, washed in distilled H2O, and treated for 20 min with 3% H2O2 in PBS. Primary antibodies against GFP (1:5000, Torrey Pines, TP401), E-cadherin (1:1000, Invitrogen,131900), P-cadherin (1:1000, Invitrogen,132000Z), BrdU (1:500, Abcam, ab6326), and ITGA6 (1:1000, Santa Cruz, sc-10730) were used. Washes in PBS (3 × 20 min) and PBS-T (1 × 5 min) were followed by incubation with Alexa fluor 488 and 555 secondary antibodies (1:500, Invitrogen) or biotinylated anti-rat secondary antibody (Vector BA-4001) followed by signal amplification (Perkin Elmer). Sections were counterstained with DAPI (Vector Laboratories) and mounted in 1% DABCO in glycerol. Images were acquired using a Leica-TCS SP5 confocal microscope. BrdU staining was quantified using ImageJ. TUNEL staining was performed according to the manufacturer’s protocol (Roche 12156792910). All experiments were performed at least three times.
MicroCT analysis
Hemimandibles (n = 3 per genotype) were imaged under wet conditions using a Micro XCT (Xradia, Pleasanton, CA) at a 2X magnification using a tungsten anode setting of 90keV and 66 microamps. Virtual sections from reconstructed tomographs were used to study the X-ray attenuation of incisor enamel across groups under identical experimental parameters and at similar sectioning planes.
Statistical analysis
For statistical analyses, mean values with standard deviation (s.d.) are shown in most graphs. All experiments were performed independently at least three times, and the exact n numbers are listed in the figure legends. P values were obtained from student t-tests with paired samples. P<0.05 was determined to be significant for all experiments. Actual P values are shown in each figure or figure legend.
Accession Codes
Gene Expression Omnibus: GSE46001
Supplementary Material
Acknowledgments
We thank members of the Klein laboratory for helpful advice, Fred Michon for discussion, D.-K. Tran and S. Alto for technical assistance, X.-P. Wang for help with the culture system, Andrea Barczak and the UCSF Microarray Core Facilities for help with experiments and analysis, and Irving Weissman for mice. This work was supported by R01-DE021420 (NIH/NIDCR) and a CIRM New Faculty Award II, both to O. D. Klein.
Footnotes
Author Contributions
B.B., J.K.H., N.B.S., H.J., E.S., R.-P.H., A.G., J.S.D and O.D.K. designed and performed experiments. B.B., J.K.H, and O.D.K. wrote the manuscript. All authors discussed results, analyzed data and edited the manuscript.
References
- 1.Molofsky AV, et al. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park IK, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 3.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 4.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zacharek SJ, et al. Lung stem cell self-renewal relies on BMI1-dependent control of expression at imprinted loci. Cell Stem Cell. 2011;9:272–281. doi: 10.1016/j.stem.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruggeman SW, Hulsman D, van Lohuizen M. Bmi1 deficient neural stem cells have increased integrin dependent adhesion to self-secreted matrix. Biochim Biophys Acta. 2009;1790:351–360. doi: 10.1016/j.bbagen.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Fasano CA, et al. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Harada H, et al. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. The Journal of cell biology. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang XP, et al. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 2007;5:e159. doi: 10.1371/journal.pbio.0050159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein OD, et al. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377–385. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith CE, Warshawsky H. Cellular renewal in the enamel organ and the odontoblast layer of the rat incisor as followed by radioautography using 3H-thymidine. Anat Rec. 1975;183:523–561. doi: 10.1002/ar.1091830405. [DOI] [PubMed] [Google Scholar]
- 12.Seidel K, et al. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development. 2010;137:3753–3761. doi: 10.1242/dev.056358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juuri E, et al. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Developmental cell. 2012;23:317–328. doi: 10.1016/j.devcel.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oguro H, et al. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell. 2010;6:279–286. doi: 10.1016/j.stem.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 15.Hosen N, et al. Bmi-1-green fluorescent protein-knock-in mice reveal the dynamic regulation of bmi-1 expression in normal and leukemic hematopoietic cells. Stem Cells. 2007;25:1635–1644. doi: 10.1634/stemcells.2006-0229. [DOI] [PubMed] [Google Scholar]
- 16.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joyner AL, Zervas M. Genetic inducible fate mapping in mouse: establishing genetic lineages and defining genetic neuroanatomy in the nervous system. Dev Dyn. 2006;235:2376–2385. doi: 10.1002/dvdy.20884. [DOI] [PubMed] [Google Scholar]
- 18.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 20.Sangiorgi E, Capecchi MR. Bmi1 lineage tracing identifies a self-renewing pancreatic acinar cell subpopulation capable of maintaining pancreatic organ homeostasis. Proc Natl Acad Sci U S A. 2009;106:7101–7106. doi: 10.1073/pnas.0902508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li CY, et al. E-cadherin regulates the behavior and fate of epithelial stem cells and their progeny in the mouse incisor. Dev Biol. 2012;366:357–366. doi: 10.1016/j.ydbio.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang WS, Tonna EA. Autoradiographic Analysis of Labeling Indices and Migration Rates of Cellular Component of Mouse Incisors Using Tritiated Thymidine (H3tdr) J Dent Res. 1965;44:42–53. doi: 10.1177/00220345650440012901. [DOI] [PubMed] [Google Scholar]
- 23.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 24.Rock JR, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tani H, Morris RJ, Kaur P. Enrichment for murine keratinocyte stem cells based on cell surface phenotype. Proc Natl Acad Sci U S A. 2000;97:10960–10965. doi: 10.1073/pnas.97.20.10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiong J, Mrozik K, Gronthos S, Bartold PM. Epithelial Cell Rests of Malassez Contain Unique Stem Cell Populations Capable of Undergoing Epithelial-Mesenchymal Transition. Stem Cells Dev. 2012 doi: 10.1089/scd.2011.0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simmer JP, Richardson AS, Smith CE, Hu Y, Hu JC. Expression of kallikrein-related peptidase 4 in dental and non-dental tissues. European journal of oral sciences. 2011;119 (Suppl 1):226–233. doi: 10.1111/j.1600-0722.2011.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwasaki K, et al. Amelotin--a Novel Secreted, Ameloblast-specific Protein. Journal of dental research. 2005;84:1127–1132. doi: 10.1177/154405910508401207. [DOI] [PubMed] [Google Scholar]
- 29.Chavez MG, et al. Characterization of dental epithelial stem cells from the mouse incisor with two-dimensional and three-dimensional platforms. Tissue engineering Part C, Methods. 2013;19:15–24. doi: 10.1089/ten.tec.2012.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morillo Prado JR, Chen X, Fuller MT. Polycomb group genes Psc and Su(z)2 maintain somatic stem cell identity and activity in Drosophila. PloS one. 2012;7:e52892. doi: 10.1371/journal.pone.0052892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahn S, Joyner AL. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 32.Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- 33.Diamond I, Owolabi T, Marco M, Lam C, Glick A. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J Invest Dermatol. 2000;115:788–794. doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]
- 34.Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, Fishell G. Characterization of Nkx6-2-derived neocortical interneuron lineages. Cerebral cortex. 2009;19 (Suppl 1):i1–10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung H, et al. Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron. 2010;67:781–796. doi: 10.1016/j.neuron.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rochat A, Kobayashi K, Barrandon Y. Location of stem cells of human hair follicles by clonal analysis. Cell. 1994;76:1063–1073. doi: 10.1016/0092-8674(94)90383-2. [DOI] [PubMed] [Google Scholar]
- 37.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci U S A. 1987;84:2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.