Abstract
Peroxisome proliferator-activated receptor α (PPARα) belongs to the family of ligand-dependent nuclear transcription factors that regulate energy metabolism. Although there exists remarkable overlap in the activities of PPARα across species, studies utilizing exogenous PPARα ligands suggest species differences in binding, activation, and physiological effects. While unsaturated long-chain fatty acids (LCFA) and their thioesters (long-chain fatty acyl-CoA; LCFA-CoA) function as ligands for recombinant mouse PPARα (mPPARα), no such studies have been conducted with full-length human PPARα (hPPARα). The objective of the current study was to determine whether LCFA and LCFA-CoA constitute high-affinity endogenous ligands for hPPARα or whether there exist species differences for ligand specificity and affinity. Both hPPARα and mPPARα bound with high affinity to LCFA-CoA; however, differences were noted in LCFA affinities. A fluorescent LCFA analog was bound strongly only by mPPARα, and naturally occurring saturated LCFA was bound more strongly by hPPARα than mPPARα. Similarly, unsaturated LCFA induced transactivation of both hPPARα and mPPARα, whereas saturated LCFA induced transactivation only in hPPARα-expressing cells. These data identified LCFA and LCFA-CoA as endogenous ligands of hPPARα, demonstrated species differences in binding specificity and activity, and may help delineate the role of PPARα as a nutrient sensor in metabolic regulation.
Keywords: PPAR, transcription factor, endogenous ligand, species differences, fatty acid, long chain fatty acyl-CoA
Whole-body energy homeostasis is regulated in part by nutrient-sensing members of the nuclear hormone receptor superfamily of ligand-dependent transcription factors, such as the peroxisome proliferator-activated receptor α (PPARα). Like other nuclear hormone receptors, the PPARα protein is comprised of several distinct domains, including a highly conserved DNA-binding domain (DBD) and a less conserved C-terminal ligand-binding domain (LBD). In highly metabolic tissues such as liver and heart, PPARα heterodimerizes with the retinoid X receptor α (RXRα), and this heterodimer potently activates genes involved in fatty acid oxidation (1–3). At a cellular level, PPARα regulates fatty acid metabolism, glucose metabolism, inflammation, differentiation, and proliferation (4–6).
Although a multitude of exogenous ligands have been shown to activate both human and mouse PPARα (1, 7–9), the identity of high-affinity endogenous ligands has been more elusive. Studies utilizing recombinant PPARα proteins have largely focused on the ligand binding domain of mouse PPARα (mPPARα). These studies suggest that long-chain fatty acids (LCFA) and their activated metabolites (long-chain acyl-CoA, LCFA-CoA) may function as endogenous PPARα ligands (10–13). Such ligand binding has been shown to induce PPARα conformational changes and increase transactivation, consistent with expectations for an endogenous ligand of a nuclear receptor.
While LCFA and LCFA-CoA have been studied as putative ligands for mPPARα, no such studies have been conducted with the full-length mPPARα or human PPARα (hPPARα). Although there exists remarkable overlap in the activities of PPARα across species, human and mouse PPARα proteins promote transcription to a different extent in response to certain hypolipidemic agents and pthalate monoesters (9, 14, 15), suggesting species differences may exist. Administration of PPARα agonists (e.g., Wy-14,643) to rodents results in peroxisome proliferation and hepatic cancer; these effects are not observed in humans (16). Even though human and mouse PPARα proteins share 91% identity (17), the observed physiological responses to exogenous activators suggest that minor sequence differences may be important to PPARα function.
The objective of the current study was to elucidate whether LCFA and/or LCFA-CoA constitute high-affinity endogenous ligands for full-length hPPARα and to determine whether species differences affect ligand specificity. Since elevated LCFA are associated with metabolic, endocrine, and cardiovascular complications, these data are important for understanding the molecular role of dietary nutrients in PPARα-mediated energy homeostasis. As putative ligands of PPARα, LCFA and/or LCFA-CoA may control their own metabolism by binding PPARα and inducing PPARα-regulated genes important for fatty acid uptake, transport, and oxidation. Thus, dysregulated LCFA could alter the transcriptional activity of PPARα, leading to hyper- or hypoactivation of these genes and further contributing to the metabolic imbalance.
MATERIALS AND METHODS
Chemicals
Fluorescent fatty acid (BODIPY-C16) was purchased from Molecular Probes, Inc. (Eugene, OR). Eicosapentaenoyl-CoA (EPA-CoA), docosapentaenoyl-CoA (DPA-CoA), docosahexaenoyl-CoA (DHA-CoA), and BODIPY C16-CoA were synthesized and purified by HPLC as previously described (12, 18) and found to be >99% unhydrolyzed. All other putative ligands were from Sigma-Aldrich (St. Louis, MO).
Purification of recombinant PPARα protein
Full-length hPPARα (amino acids 1–468) and full-length mPPARα (amino acids 1–468) were used for all experiments presented herein. An N-terminal polyhistidine tag (6xHis) was added to the GST open reading frame in the pGEX-6P vector (Amersham Biosciences, Piscataway, NJ) by overlap PCR, resulting in 6xHis and GST tags upstream of the PreScission Protease and multiple cloning sites. The hPPARα coding sequence was amplified from cDNA derived from HepG2 cells with the following primers: 5′-c ggatcc ATGGTGGACACGGAAAGCCC-3′ and 5′-c gtcgac CTATCAGTACATGTCCCTGTAG-3′. In these and subsequent primers, lowercase represents nucleotides outside of the PPARα open-reading frame with restriction sites underlined. The mPPARα coding sequence was amplified from cDNA derived from mouse liver with the following primers: 5′- cggatccATGGTGGACACAGAGAGCCC-3′ and 5′-gaagcttcactcgagCTATCAGTACATGTCTCTG-3′. Each PCR product was cloned into the pGEM-T easy vector (Promega Corporation, Madison, WI) and subsequently transferred into the Bam HI / Sal I sites or the Bam HI / Xho I sites of the pGEX-6P derivative to produce 6xHis-GST-hPPARα and 6xHis-GST-mPPARα, respectively. These 6xHis-GST-PPARα fusions were expressed in RosettaTM2 cells (Novagen, Gibbstown, NJ), and each soluble protein fraction was applied to a glutathione cartridge (Bio-Rad Laboratories, Hercules, CA) per the manufacturer's instructions. Washes and on-column digestion with PreScission Protease (GE Healthcare, Pittsburgh, PA) were conducted as recommended, producing full-length, untagged hPPARα and mPPARα. Eluted proteins were concentrated, dialyzed, and tested for purity by SDS-PAGE with Coomassie blue staining and immunoblotting as previously described (12, 13). Protein concentrations were estimated by Bradford Assay (Bio-Rad Laboratories) and by absorbance spectroscopy using the molar extinction coefficient for the protein.
Direct fluorescent ligand-binding assays
Fluorescent ligand (BODIPY C16 or BODIPY C16-CoA) binding measurements were performed as described earlier (12, 19). Briefly, 0.1 μM hPPARα or mPPARα was titrated with increasing concentrations of fluorescent ligand. This concentration of PPARα protein was chosen because it gave the maximal signal-to-noise ratio, while allowing saturable binding of most of the examined ligands to be reached at concentrations below their critical micellular concentrations (data not shown). Fluorescence emission spectra (excitation, 465 nm; emission, 490–550 nm) were obtained at 24°C with a PC1 photon counting spectrofluorometer (ISS Inc., Champaign, IL) corrected for background (protein only and fluorescent ligand only), and maximal intensities were used to calculate the dissociation constant (Kd) and number of binding sites (n) (12).
Displacement of bound fluorescent BODIPY C16-CoA by nonfluorescent ligands
Based on the binding affinities obtained with the direct fluorescent ligand-binding assays for BODIPY C16-CoA, 0.1 μM PPARα was mixed with BODIPY C16-CoA at the concentration where maximal fluorescence intensity first occurred (75 nM for hPPARα and 130 nM for mPPARα). The maximal fluorescence intensity was measured, and the effect of increasing concentrations of naturally occurring ligands was measured as a decrease in fluorescence (19). Emission spectra were obtained and corrected for background as described above for BODIPY. Changes in fluorescence intensity were used to calculate the dissociation constant (Kd), inhibition constant (Ki), and the number of binding sites as described (12, 19).
Quenching of PPARα aromatic amino acid residues by nonfluorescent ligands
The direct binding of hPPARα or mPPARα to nonfluorescent ligands was determined by quenching of intrinsic PPARα aromatic amino acid fluorescence as described (12, 13). Briefly, hPPARα or mPPARα (0.1 μM) was titrated with increasing concentrations of ligand. Emission spectra at 300–400 nm were obtained at 24°C upon excitation at 280 nm with a PC1 photon counting spectrofluorometer (ISS Inc., Champaign, IL). Data were corrected for background and inner filter effects, and maximal intensities were used to calculate the dissociation constant (Kd) and number of binding sites (n) (12).
Secondary structure determination: effect of ligand binding on PPARα circular dichroism
Circular dichroism (CD) spectra of hPPARα or mPPARα (0.6 μM in 600 µM HEPES at pH 8.0, 24 μM dithiothreitol, 6 μM EDTA, 6 mM KCl, and 0.6% glycerol) were taken in the presence and absence of LCFA and LCFA-CoA (0.6 μM) with a J-815 spectropolarimeter (Jasco Inc., Easton, MD) as previously described (12, 13). Spectra was recorded at 260–187 nm with a bandwidth of 2.0 nm, sensitivity of 10 millidegrees, scan rate of 50 nm/min, and a time constant of 1 s. Ten scans were averaged for percentage compositions of α-helices, β-strands, turns, and unordered structures with the CONTIN/LL program of the software package CDPro (12, 13, 20).
Mammalian expression plasmids
hPPARα was amplified from 6xHis-GST-hPPARα using the following primers: 5′ catcggatccaccATGGTGGACACGGAAAGCCCA-3′ and 5′-cgtcgacCTATCAGTACATGTCCCTGTAG-3′. mPPARα was amplified from 6xHis-GST-mPPARα using the following primers: 5′-cggatccaccATGGTGGACACAGAGAGCCC-3′ and ctcctcgagTCAGTACATGTCTCTGTAGA-3′. The PCR products were cloned into the pGEM-T easy vector. A Bam HI / end-filled Sal I fragment for hPPARα and a Bam HI / end-filled Xho I mouse PPARα fragment were subcloned into the Bam HI / end-filled Bgl II multiple-cloning site of pSG5 (Stratagene, La Jolla, CA) to produce pSG5-hPPARα and pSG5-mPPARα, respectively. The human retinoid X receptor α (hRXRα) coding sequence was amplified from HepG2 cDNA using the following primers: 5′-catcgaattccaccATGGACACCAAACATTTCCTGCCGCT-3′ and 5′-ctcgagCTAAGTCATTTGGGTGCGGCGCCTCC-3′. The mRXRα coding sequence was amplified from cDNA derived from mouse liver with the following primers: 5′-cgaattc caccATGGACACCAAACATTTCCTGCCGCT-3′ and 5′-actcgagCTAGGTGGCTTGATGTGGT-3′. The PCR products were cloned into the pGEM-T easy vector, and Eco RI / end-filled Xho I fragments for each gene were subsequently transferred into the multiple-cloning site of pSG5 (Eco RI / end-filled Bgl II) to produce pSG5-hRXRα and pSG5-mRXRα. The reporter construct, PPRE×3 TK LUC was a kind gift of Dr. Bruce Spiegelman (Harvard Medical School, Boston, MA) (Addgene plasmid # 1015) and contained three copies of the acyl-CoA oxidase (ACOX) peroxisome proliferator response element (PPRE) (21).
Cell culture and transactivation assay
COS-7 cells (ATCC, Manassas, VA) were grown in DMEM supplemented with 10% fetal bovine serum (Invitrogen, Grand Island, NY) at 37°C with 5% CO2 in a humidified chamber. Cells were seeded onto 24-well culture plates and transfected with Lipofectamine™ 2000 (Invitrogen, Grand Island, NY) and 0.4 μg of each full-length mammalian expression vector (pSG5-hPPARα, pSG5-hRXRα, pSG5-mPPARα, pSG5-mRXRα) or empty plasmid (pSG5), 0.4 μg of the PPRE×3 TK LUC reporter construct, and 0.04 μg of the internal transfection control plasmid pRL-CMV (Promega Corp., Madison, WI) as previously described (12, 19). Following transfection incubation, medium was replaced with serum-free medium for 2 h, ligands (1 μM) were added, and the cells were grown for an additional 20 h. Fatty acids were added as a complex with BSA (BSA) as described (22). Firefly luciferase activity, normalized to Renilla luciferase (for transfection efficiency), was determined with the dual luciferase reporter assays system (Promega) and measured with a SAFIRE2 microtiter plate reader (Tecan Systems, Inc. San Jose, CA). Clofibrate-treated samples overexpressing both PPARα and RXRα were arbitrarily set to 1.
Statistical analysis
Data were analyzed by SigmaPlot™ (Systat Software, San Jose, CA), and a one-way ANOVA was used to evaluate overall significance. A Fisher least-significant difference (LSD) posthoc test was used to identify individual group differences. The results are presented as means ± SEM. The confidence limit of P < 0.05 was considered statistically significant.
RESULTS
Full-length hPPARα and mPPARα protein purification
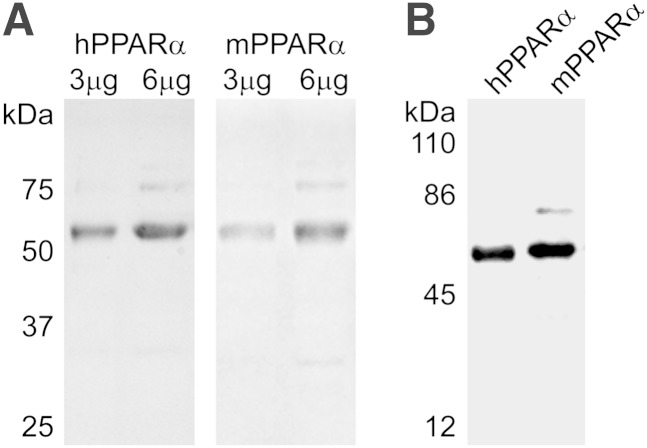
Based on recent demonstrations that truncation of a nuclear transcription factor can significantly affect ligand-binding affinity, specificity, and consequently, receptor activity (23, 24), full-length hPPARα and mPPARα were used for all experiments. SDS-PAGE and Coomassie blue staining indicated predominant bands of 52 kDa corresponding to the expected size of full-length hPPARα and mPPARα, for which densitometry indicated greater than 85% purity (Fig. 1A). Western blotting confirmed that the predominant protein bands were PPARα (Fig. 1B).
Fig. 1.
(A) SDS-PAGE and Coomassie blue staining of 3 μg and 6 μg purified recombinant hPPARα (left) and mPPARα (right) showing relative purity of the protein. The prominent band at 52 kDa is full-length, untagged recombinant PPARα. (B) Western blot of 1 μg purified recombinant hPPARα (left) and mPPARα (right) confirming the 52 kDa band is untagged, full-length PPARα.
Binding of fluorescent fatty acid and fatty acyl-CoA to PPARα
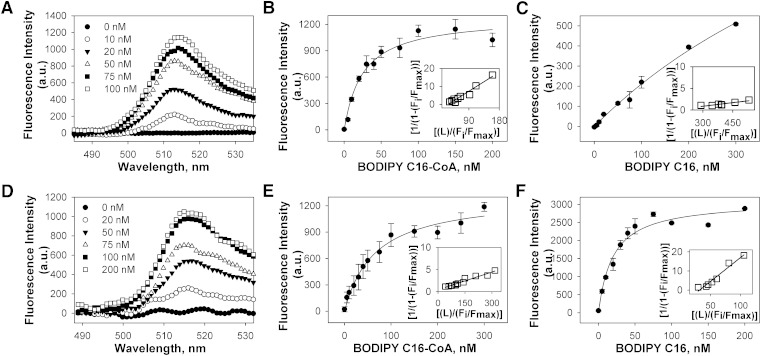
The sensitivity of the BODIPY fluorophore to environmental hydrophobicity is useful for determining whether binding represents a direct molecular interaction within the hydrophobic ligand-binding pocket of PPARα. In aqueous buffer without protein, BODIPY fluorescence was low for each of the examined fluorophores. Titration of hPPARα with BODIPY C16-CoA resulted in increased fluorescence with an emission maximum near 515 nm (Fig. 2A). This increased fluorescence was saturable near 100 nM (Fig. 2B, circles), indicating high-affinity binding (Kd = 25 ± 4 nM). These data transformed into a linear double reciprocal plot (Fig. 2B, inset), consistent with a single binding site (R2 > 0.95). In contrast, a smaller, nonsaturable increase in fluorescence was seen upon titration of hPPARα with BODIPY C16 fatty acid (Fig. 2C), indicating only weak or nonspecific binding. Titration of mPPARα with BODIPY C16-CoA resulted in a similar increase in BODIPY C16-CoA fluorescence (Fig. 2D) as noted for hPPARα, with the exception that slightly higher BODIPY C16-CoA concentrations were required to reach saturation (Fig. 2E). This resulted in a lower binding affinity (Kd = 65 ± 9 nM), but it was still consistent with a single binding site (Fig. 2E). While hPPARα binding to BODIPY C16 fatty acid was nonsaturable, mPPARα binding to BODIPY C16 fatty acid resulted in strong fluorescence changes with saturation near 50 nM (Fig. 2F), suggesting high-affinity binding (Kd = 19 ± 4 nM). Although these data were consistent with previous data suggesting that a truncated mPPARα protein can bind to both BODIPY C16 fatty acid derivative and BODIPY C16-CoA with high affinity (19), these data also suggested that species differences exist in ligand-binding specificity.
Fig. 2.
(A) Corrected fluorescence emission spectra of 0.1 μM hPPARα titrated with 0 (filled circles), 10 (open circles), 20 (filled triangles), 50 (open triangles), 75 (filled squares), and 100 nM (open squares) of BODIPY C16-CoA upon excitation at 465 nm, demonstrating increased fluorescence intensity upon binding to hPPARα. Plot of hPPARα maximal fluorescence emission as a function of BODIPY C16:0-CoA (B) and BODIPY C16:0 FA (C). (D) Corrected fluorescence emission spectra of 0.1 μM mPPARα titrated with 0 (filled circles), 20 (open circles), 50 (filled triangles), 75 (open triangles), 100 (filled squares), and 200 nM (open squares) of BODIPY C16-CoA upon excitation at 465 nm, demonstrating increased fluorescence intensity upon binding to mPPARα. Plot of mPPARα maximal fluorescence emission as a function of BODIPY C16:0-CoA (E) and BODIPY C16:0 FA (F). Insets represent linear plots of the binding curve from each panel. All values represent the mean ± SE, n ≥ 3.
Binding of endogenous LCFA and LCFA-CoA to hPPARα: displacement of bound BODIPY C16-CoA
To determine the ligand specificity of hPPARα for naturally occurring, endogenous fatty acids, LCFA and LCFA-CoA were examined for their ability to displace BODIPY C16-CoA from the hPPARα ligand-binding pocket, which was observed as decreased BODIPY fluorescence. With the exception of lauric acid and lauryl-CoA, titration with fatty acids and fatty acyl-CoA resulted in significantly decreased BODIPY fluorescence (supplementary Fig. I). Quantitative analyses of these data suggested strong affinity binding (Ki = 10–40 nM, Table 1). By comparison, the synthetic PPARα agonist clofibrate showed slightly weaker affinity (Ki = 48 nM), while the synthetic PPARγ agonist rosiglitazone showed no displacement (Table 1). These data revealed that both LCFA and LCFA-CoA are capable of displacing a fluorescent fatty acyl-CoA, suggesting that both LCFA and LCFA-CoA could be endogenous ligands of hPPARα. These data are in contrast with displacement studies conducted with a truncated form of mPPARα, which showed that only unsaturated LCFA, but not saturated LCFA, could displace a bound fluorescent fatty acid (11), and suggest that important differences may exist between hPPARα and mPPARα.
TABLE 1.
Affinity of hPPARα for nonfluorescent ligands determined by quenching of hPPARα aromatic amino acid fluorescence and by displacement of hPPARα-bound BODIPY C16-CoA
| Ligand | Chain Length: Double Bonds (Position) | Kd (nM) Fatty Acid | Kd (nM) Fatty Acyl-CoA | Ki (nM) Fatty Acid | Ki (nM) Fatty Acyl-CoA |
| Lauric acid/CoA | C12:0 | ND | ND | ND | ND |
| Palmitic acid/CoA | C16:0 | 22 ± 3 | 11 ± 1 | 16 ± 2 | 10 ± 2 |
| Palmitoleic acid/CoA | C16:1 (n-7) | 16 ± 2 | 29 ± 4 | 26 ± 6 | 46 ± 8 |
| Stearic acid/CoA | C18:0 | 14 ± 2 | 16 ± 2 | 13 ± 3 | 15 ± 2 |
| Oleic acid/CoA | C18:1 (n-9) | 19 ± 3 | 13 ± 1 | 13 ± 2 | 16 ± 3 |
| Linoleic acid/CoA | C18:2 (n-6) | 12 ± 1 | 12 ± 2 | 26 ± 6 | 40 ± 8 |
| Arachidonic acid/CoA | C20:4 (n-6) | 24 ± 5 | 23 ± 3 | 24 ± 3 | 17 ± 2 |
| EPA/CoA | C20:5 (n-3) | 34 ± 4 | 16 ± 2 | 38 ± 5 | 26 ± 5 |
| DPA/CoA | C22:5 (n-3) | 13 ± 2 | 18 ± 4 | 10 ± 2 | 30 ± 6 |
| DHA/CoA | C22:6 (n-3) | 30 ± 5 | 14 ± 1 | 18 ± 3 | 28 ± 5 |
| Clofibrate | 58 ± 6 | 48 ± 6 | |||
| Rosiglitazone | ND | ND |
Values represent the mean ± SE (n ≥ 3). ND, not determinable.
Binding of endogenous LCFA and LCFA-CoA to mPPARα: displacement of bound BODIPY C16-CoA
To compare the ability of naturally occurring LCFA and LCFA-CoA to displace BODIPY C16-CoA from the binding pocket of mPPARα (versus hPPARα), we first mixed mPPARα and BODIPY C16-CoA at the same molar ratio used for the hPPARα displacement assays. However, very little displacement was noted for any ligand and only at high LCFA concentrations (data not shown). Since the BODIPY C16-CoA binding affinity for mPPARα is much weaker than for hPPARα, a higher concentration of BODIPY C16-CoA is needed to reach saturation and ensure BODIPY C16-CoA-bound mPPARα. Thus, these experiments were repeated with a saturating concentration of BODIPY C16-CoA, and displacement was observed as a decrease in BODIPY fluorescence. With the exception of lauric acid and lauryl-CoA, titration with fatty acids and fatty acyl-CoA resulted in significantly decreased BODIPY fluorescence (supplementary Fig. II). Quantitative analyses of these data suggested that, with the exception of the saturated LCFA (palmitic acid, Ki = 135 nM and stearic acid, Ki = 134 nM), most LCFA and LCFA-CoA demonstrated strong affinity binding (Ki = 13–38 nM, Table 2) for mPPARα. The mPPARα showed similar displacement and affinity for the synthetic PPARα agonist clofibrate (Ki = 46 nM, Table 2) compared hPPARα (Table 1), and the synthetic PPARγ agonist rosiglitazone showed no displacement (Table 2). These data show that LCFA and LCFA-CoA are both capable of displacing a fluorescent fatty acyl-CoA, suggesting that both LCFA and LCFA-CoA could be endogenous ligands of mPPARα. When compared with binding data from hPPARα (Table 1), these data also suggest differences in the ligand-binding specificity between hPPARα and mPPARα, particularly for saturated LCFA.
TABLE 2.
Affinity of mPPARα for nonfluorescent ligands determined by quenching of mPPARα aromatic amino acid fluorescence and by displacement of mPPARα-bound BODIPY C16-CoA
| Ligand | Chain Length: Double Bonds (Position) | Kd (nM) Fatty Acid | Kd (nM) Fatty Acyl-CoA | Ki (nM) Fatty Acid | Ki (nM) Fatty Acyl-CoA |
| Lauric acid/CoA | C12:0 | ND | ND | ND | ND |
| Palmitic acid/CoA | C16:0 | 92 ± 13 | 14 ± 2 | 135 ± 13 | 23 ± 4 |
| Palmitoleic acid/CoA | C16:1 (n-7) | 32 ± 3 | 24 ± 5 | 35 ± 3 | 31 ± 4 |
| Stearic acid/CoA | C18:0 | 81 ± 15 | 28 ± 5 | 134 ± 30 | 37 ± 5 |
| Oleic acid/CoA | C18:1 (n-9) | 22 ± 5 | 37 ± 5 | 37 ± 4 | 38 ± 6 |
| EPA/CoA | C20:5 (n-3) | 24 ± 6 | 17 ± 3 | 33 ± 5 | 21 ± 3 |
| DHA/CoA | C22:6 (n-3) | 31 ± 2 | 24 ± 2 | 34 ± 3 | 13 ± 3 |
| Clofibrate | 39 ± 6 | 46 ± 3 | |||
| Rosiglitazone | ND | ND |
Values represent the mean ± SE (n ≥ 3). ND, not determinable.
Binding of endogenous LCFA and LCFA-CoA to hPPARα: quenching of intrinsic aromatic amino acid fluorescence
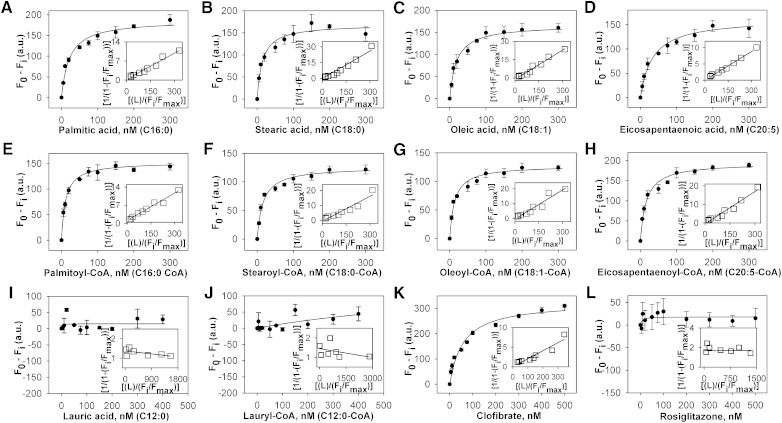
Since previous data has suggested that fluorescent fatty acid analogs are not always bound the same as endogenous fatty acids due to bulky side chains altering the energy-minimized state of the molecule (12, 19), the binding of LCFA and LCFA-CoA to hPPARα was also measured directly by spectroscopically monitoring the quenching of hPPARα aromatic amino acid emission. Titration of hPPARα with the saturated LCFA palmitic acid (Fig. 3A) and stearic acid (Fig. 3B) yielded sharp saturation curves with maximal fluorescence changes at 100 nM, and both transformed into linear reciprocal plots (insets), indicating high-affinity binding at a single binding site (R2 > 0.9). Similar results were obtained for all examined LCFA and LCFA-CoA (Fig. 3C–H), with single-site binding affinities in the 10–30 nM range (Table 1), similar to affinities determined by displacement assays. Titration with lauric acid (Fig. 3I) and lauryl-CoA (Fig. 3J) did not significantly alter hPPARα fluorescence, and no binding was detected (Table 1). The PPARα agonist clofibrate strongly quenched hPPARα fluorescence (Fig. 3K) but displayed weaker affinity than the LCFA (Table 1), while the PPARγ agonist rosiglitazone showed no binding (Fig. 3L), further confirming that hPPARα bound saturated, monounsaturated, and polyunsaturated LCFA and LCFA-CoA with high affinity.
Fig. 3.
Interaction of naturally occurring fatty acids and fatty acyl-CoA with hPPARα. Direct binding assay based on quenching of hPPARα aromatic amino acid fluorescence emission when titrated with the following ligands: (A) palmitic acid, (B) stearic acid, (C) oleic acid, (D) EPA, (E) palmitoyl-CoA, (F) stearoyl-CoA, (G) oleoyl-CoA, (H) EPA-CoA, (I) lauric acid, (J) lauryl-CoA, (K) clofibrate, and (L) rosiglitazone. Data are presented as the change in fluorescence intensity (F0−Fi) plotted as a function of ligand concentration. Insets represent linear plots of the binding curve from each panel. All values represent mean ± SE, n ≥ 3.
Binding of endogenous LCFA and LCFA-CoA to mPPARα: quenching of intrinsic aromatic amino acid fluorescence
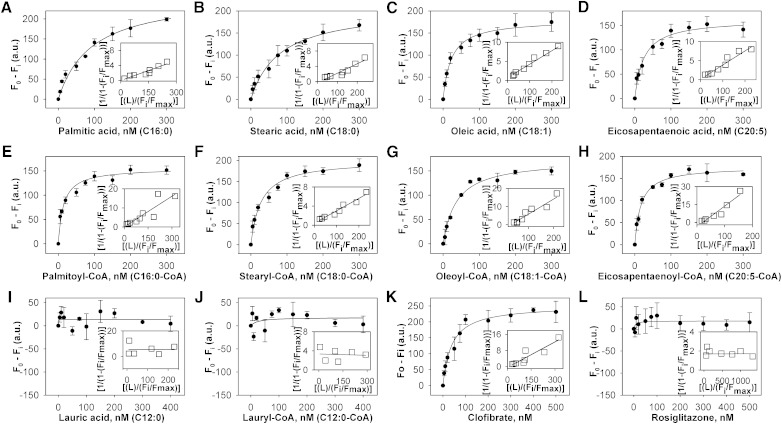
Binding of full-length mPPARα to LCFA and LCFA-CoA was also measured by spectroscopically monitoring the quenching of mPPARα aromatic amino acid emission. Although titration with the saturated LCFA palmitic acid (Fig. 4A) and stearic acid (Fig. 4B) resulted in decreased mPPARα fluorescence, the slopes of these curves were much shallower than that of hPPARα with palmitic acid (Fig. 3A) or stearic acid (Fig. 3B), with the change in fluorescence intensity plateauing off at approximately 300 nM. Transformation of these data into double reciprocal plots yielded single lines (Fig. 4A, B, insets), indicating single binding sites for both. However, multiple replicates yielded much weaker binding affinities for mPPARα (Kd = 92 nM for palmitic acid and 81 nM for stearic acid, Table 2) than for hPPARα (Table 1). Titration of mPPARα with the other examined LCFA and LCFA-CoA yielded sharp saturation curves with the maximal change in fluorescence intensity noted at approximately 100 nM (Fig. 4C–H), indicating high-affinity binding (Kd = 14-37 nM, Table 2). These data transformed into linear reciprocal plots (insets), indicating binding at a single binding site (R2 > 0.9). Similar to hPPARα, no significant mPPARα binding was noted for lauric acid (Fig. 4I), lauryl-CoA (Fig. 4J), or rosiglitazone (Fig. 4L), while clofibrate binding resulted in the strongest fluorescence changes (Fig. 4K). Although the weak binding of palmitic acid and stearic acid to full-length mPPARα was consistent with previous data using mPPARαΔAB (11–13), it was significantly different from the binding of hPPARα with the same ligand (Table 1). On the other hand, while mPPARαΔAB demonstrated weak binding toward polyunsaturated fatty acids (PUFA), such as eicosapentanoic acid (EPA) and docosahexaenoic acid (DHA), our data employing full-length mPPARα and hPPARα demonstrated high-affinity binding for both these PUFA (Figs. 3D and 4D, Tables 1 and 2). These findings suggest two important conclusions: species-dependent differences exist in the ligand-binding specificity and affinity between human and mouse PPARα, and the N-terminal domain of PPARα plays an unexpected, but important, role in the ligand-binding function of the protein.
Fig. 4.
Interaction of naturally occurring fatty acids and fatty acyl-CoA with mPPARα. Direct binding assay based on quenching of mPPARα aromatic amino acid fluorescence emission when titrated with the following ligands: (A) palmitic acid, (B) stearic acid, (C) oleic acid, (D) EPA, (E) palmitoyl-CoA, (F) stearoyl-CoA, (G) oleoyl-CoA, (H) EPA-CoA, (I) lauric acid, (J) lauryl-CoA, (K) clofibrate, and (L) rosiglitazone. Data are presented as the change in fluorescence intensity (F0−Fi) plotted as a function of ligand concentration. Insets represent linear plots of the binding curve from each panel. All values represent mean ± SE, n ≥ 3.
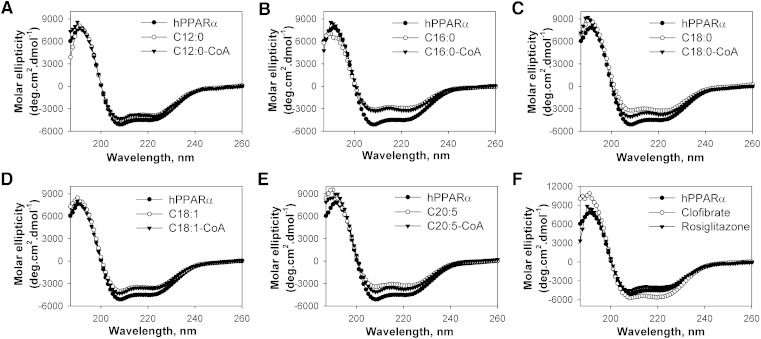
Effect of endogenous fatty acids and fatty acyl-CoA on hPPARα secondary structure
Ligand-activated receptors, such as PPARα, undergo conformational changes upon ligand binding, which allows for altered cofactor interactions (12, 25, 26). Circular dichroism was used to examine whether the binding of LCFA or LCFA-CoA altered the hPPARα secondary structure. The far UV CD spectrum of hPPARα suggested the presence of substantial α-helical content, exhibiting a large positive peak at 192 nm and two negative peaks at 207 and 222 nm (Fig. 5, filled circles). Quantitative analyses of the CD spectra confirmed that hPPARα was composed of approximately 32% α-helix, 18% β-sheets, 21% β-turns, and 29% unordered structures (Table 3).
Fig. 5.
Far UV CD spectra of hPPARα in the absence (filled circles) and presence of added ligand: (A) lauric acid (open circles) or lauryl-CoA (filled triangles); (B) palmitic acid (open circles) or palmitoyl-CoA (filled triangles); (C) stearic acid (open circles) or stearoyl-CoA (filled triangles); (D) oleic acid (open circles) or oleoyl-CoA (filled triangles); (E) EPA (open circles) or EPA-CoA (filled triangles); and (F) clofibrate (open circles) or rosiglitzone (filled triangles). Each spectrum represents an average of 10 scans for a given representative spectrum from at least three replicates.
TABLE 3.
Effect of ligands on the relative proportion of hPPARα secondary structure determined by CD
| Average | Total H ± SE | Total S ± SE | Trn ± SE | Unrd ± SE |
| hPPARα | 32 ± 1 | 19 ± 1 | 21.3 ± 0.3 | 29.3 ± 0.5 |
| hPPARα + lauric acid | 30 ± 1 | 20 ± 2 | 21.8 ± 0.4 | 28.7 ± 0.3 |
| hPPARα + lauryl-CoA | 31 ± 3 | 18.2 ± 0.2 | 20 ± 1 | 29 ± 1 |
| hPPARα + palmitic acid | 16 ± 3** | 32 ± 2** | 21.7 ± 0.4 | 30 ± 1 |
| hPPARα + palmitoyl-CoA | 13 ± 3** | 34 ± 2** | 22.5 ± 0.2 | 30 ± 1 |
| hPPARα + palmitoleic acid | 22 ± 4* | 28 ± 3* | 21 ± 1 | 28 ± 1 |
| hPPARα + palmitoleoyl-CoA | 24 ± 5# | 27 ± 3* | 21 ± 1 | 29 ± 1 |
| hPPARα + stearic acid | 14 ± 3** | 33 ± 2** | 22.0 ± 0.2 | 31 ± 2 |
| hPPARα + stearyl-CoA | 24 ± 4# | 27 ± 2* | 21 ± 1 | 29 ± 1 |
| hPPARα + oleic acid | 18 ± 2** | 31 ± 2** | 22 ± 1 | 29 ± 1 |
| hPPARα + oleoyl-CoA | 26 ± 3 | 25 ± 2# | 21 ± 1 | 28.3 ± 0.3 |
| hPPARα + linoleic acid | 27 ± 6 | 28 ± 2* | 19 ± 2* | 26 ± 3 |
| hPPARα + linoleoyl-CoA | 24 ± 3# | 26 ± 2* | 21 ± 1 | 28.8 ± 0.1 |
| hPPARα + arachidonic acid | 19 ± 1* | 30 ± 1** | 21.8 ± 0.3 | 28.9 ± 0.1 |
| hPPARα + arachidonoyl-CoA | 30 ± 1 | 23.4 ± 0.4 | 19.4 ± 0.5# | 26.9 ± 0.4 |
| hPPARα + EPA | 14 ± 7** | 24 ± 6 | 23 ± 2 | 33 ± 5 |
| hPPARα + EPA-CoA | 21 ± 1* | 29 ± 1* | 21.6 ± 0.3 | 29 ± 1 |
| hPPARα + DPA | 17 ± 4** | 32 ± 3** | 21.9 ± 0.1 | 30 ± 1 |
| hPPARα + DPA-CoA | 20 ± 1* | 30 ± 1** | 21 ± 1 | 29.6 ± 0.2 |
| hPPARα + DHA | 12 ± 3** | 38 ± 4** | 21 ± 1 | 30 ± 1 |
| hPPARα + DHA-CoA | 20 ± 2* | 29 ± 2* | 22 ± 1 | 28.9 ± 0.2 |
| hPPARα + clofibrate | 33 ± 1 | 15 ± 1* | 22 ± 1 | 30 ± 1 |
| hPPARα + rosiglitazone | 29 ± 1 | 22 ± 2 | 20 ± 1 | 28 ± 1 |
Structure abbreviations: H, total helices (sum of regular α-helices and distorted α-helices); S, total sheets (sum of regular β-sheets and distorted β-sheets); Trn, β-turns; Unrd, unordered. Asterisks represent significant differences between hPPARα only and hPPARα in the presence of added ligand. *P < 0.05, **P < 0.001, #P = 0.07.
Since most of the examined ligands were shown to bind at a single binding site, ligand effects were measured at a molar concentration equivalent to that of hPPARα. The addition of high-affinity LCFA and LCFA-CoA ligands resulted in alterations in molar ellipticity at 192, 207, and 222 nm (Fig. 5B–E), demonstrating hPPARα conformational changes. Although both increases and decreases of the 192 nm peak were noted, most of the examined LCFA and LCFA-CoA resulted in less negative peaks at 207 and 222 nm (Fig. 5B–E), suggestive of decreased α-helical content. Quantitative analyses confirmed that most high-affinity LCFA and LCFA-CoA ligands significantly decreased the estimated fraction of α-helical content and concomitantly increased the estimated fraction of β-sheets (Table 3). However, lauric acid and its CoA thioester, which showed no binding, resulted in only minor, nonsignificant changes to the hPPARα secondary structure (Fig. 5A, Table 3). Contrary to previously published mPPARα data (12, 13), the strongest conformational changes were noted with palmitic acid, stearic acid, EPA, and DHA (Fig. 5, Table 3). These changes in spectra and percentage composition were stronger than those observed with the addition of clofibrate (Fig. 5F, open circles, Table 3), and no changes were observed with the addition of rosiglitazone (Fig. 5F, filled triangles, Table 3), consistent with the decreased affinity of hPPARα for these compounds.
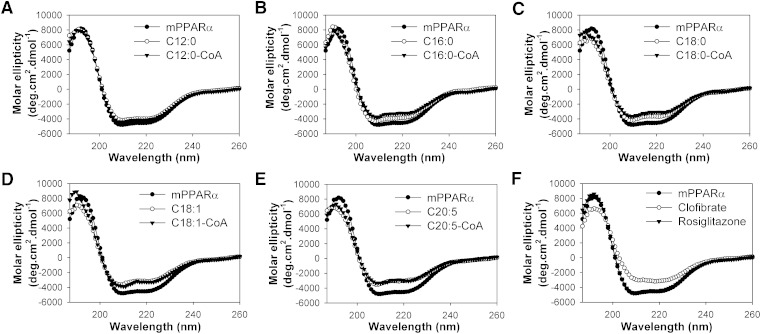
Effect of endogenous fatty acids and fatty acyl-CoA on mPPARα secondary structure
Consistent with hPPARα, the far UV CD spectrum of mPPARα suggested the presence of substantial α-helical content, exhibiting a large positive peak at 192 nm and two negative peaks at 207 and 222 nm (Fig. 6, filled circles). Quantitative analyses of the CD spectra confirmed that mPPARα was composed of approximately 30% α-helix, 19% β-sheets, 22% β-turns, and 29% unordered structures (Table 4), similar to hPPARα (Table 3). With the exception of lauric acid and lauryl-CoA (Fig. 6A), the addition of fatty acids (Fig. 6B–E, open circles) and fatty acyl-CoA (Fig. 6B–E, filled triangles) resulted in mPPARα conformational changes consistent with decreased molar ellipticity at 192 nm and increased molar ellipticity at 207 and 222 nm. Addition of clofibrate resulted in the strongest changes to the mPPARα spectrum, but consistent with binding data, no changes were seen with the addition of rosiglitazone (Fig. 6F). Quantitative analyses of multiple replicates indicated that LCFA and LCFA-CoA significantly decreased the mPPARα estimated α-helical content and concomitantly increased the estimated percentage of β-sheets (Table 4), a trend similar to that seen with hPPARα. However, for several ligands, the magnitude of the change was different between the two proteins. While palmitic acid and stearic acid resulted in some of the strongest changes to the hPPARα structure, addition of these same ligands resulted in some of the weakest changes seen to the mPPARα structure. Moreover, clofibrate had the strongest effect on mPPARα secondary structure and a very small effect on hPPARα secondary structure. The changes in CD spectra and estimated percentage composition were consistent with the affinity of mPPARα for each ligand. These data further suggest that species differences in ligand specificity and affinity exist between mouse and human PPARα.
Fig. 6.
Far UV CD spectra of mPPARα in the absence (filled circles) and presence of added ligand: (A) lauric acid (open circles) or lauryl-CoA (filled triangles); (B) palmitic acid (open circles) or palmitoyl-CoA (filled triangles); (C) stearic acid (open circles) or stearoyl-CoA (filled triangles); (D) oleic acid (open circles) or oleoyl-CoA (filled triangles); (E) EPA (open circles) or EPA-CoA (filled triangles); and (F) clofibrate (open circles) or rosiglitzone (filled triangles). Each spectrum represents an average of 10 scans for a given representative spectrum from at least three replicates.
TABLE 4.
Effect of ligands on the relative proportion of mPPARα secondary structure determined by CD
| Average | Total H ± SE | Total S ± SE | Trn ± SE | Unrd ± SE |
| mPPARα | 30 ± 1 | 19 ± 2 | 22 ± 1 | 29 ± 1 |
| mPPARα + lauric acid | 29 ± 1 | 20 ± 1 | 22 ± 1 | 28.8 ± 0.1 |
| mPPARα + lauryl-CoA | 27 ± 3 | 23 ± 3 | 22.1 ± 0.1 | 28.9 ± 0.1 |
| mPPARα + palmitic acid | 23 ± 3* | 23 ± 2 | 21 ± 2 | 30 ± 2 |
| mPPARα + palmitoyl-CoA | 16 ± 1** | 32 ± 1** | 23 ± 1 | 29.2 ± 0.2 |
| mPPARα + palmitoleic acid | 14 ± 1** | 29 ± 1* | 23 ± 1 | 34 ± 5 |
| mPPARα + palmitoleoyl-CoA | 19 ± 1* | 34 ± 5** | 21 ± 1 | 28 ± 1 |
| mPPARα + stearic acid | 21.8 ± 0.5* | 28 ± 0.5* | 21.2 ± 0.1 | 28.6 ± 0.2 |
| mPPARα + stearyl-CoA | 21 ± 2* | 30 ± 4* | 21 ± 1 | 29.7 ± 0.3 |
| mPPARα + oleic acid | 10 ± 4** | 36 ± 3** | 23 ± 2 | 31 ± 1 |
| mPPARα + oleoyl-CoA | 22 ± 4* | 28 ± 2* | 20 ± 1 | 29 ± 1 |
| mPPARα + linoleic acid | 21 ± 1* | 30 ± 1* | 22 ± 1 | 28.5 ± 0.3 |
| mPPARα + linoleoyl-CoA | 17 ± 2** | 33 ± 2** | 22.0 ± 0.5 | 28.7 ± 0.1 |
| mPPARα + arachidonic acid | 18 ± 1** | 31 ± 1* | 22.5 ± 0.5 | 28.7 ± 0.2 |
| mPPARα + arachidonoyl-CoA | 22 ± 3* | 28 ± 3* | 21.7 ± 0.1 | 28 ± 1 |
| mPPARα + EPA | 15 ± 2** | 31 ± 3* | 21 ± 1 | 30 ± 1 |
| mPPARα + EPA-CoA | 22.5 ± 1.5* | 28 ± 2* | 20.1 ± 0.3 | 30 ± 1 |
| mPPARα + DPA | 20 ± 1* | 29 ± 1* | 22 ± 1 | 29.1 ± 0.3 |
| mPPARα + DPA-CoA | 16 ± 3** | 34 ± 3** | 22.1 ± 0.2 | 27.9 ± 0.5 |
| mPPARα + DHA | 16 ± 5** | 30 ± 4* | 21 ± 1 | 30 ± 2 |
| mPPARα + DHA-CoA | 9.5 ± 0.5** | 37 ± 1** | 21.9 ± 0.2 | 31.8 ± 0.2 |
| mPPARα + clofibrate | 13 ± 3** | 34 ± 3** | 22.4 ± 0.1 | 31 ± 1 |
| mPPARα + rosiglitazone | 27 ± 2 | 24 ± 3 | 25.5 ± 3.5 | 23 ± 2 |
Structure abbreviations: H, total helices (sum of regular α-helices and distorted α-helices); S, total sheets (sum of regular β-sheets and distorted β-sheets); Trn, β-turns; Unrd, unordered. Asterisks represent significant differences between mPPARα only and mPPARα in the presence of added ligand. *P < 0.05, **P < 0.001.
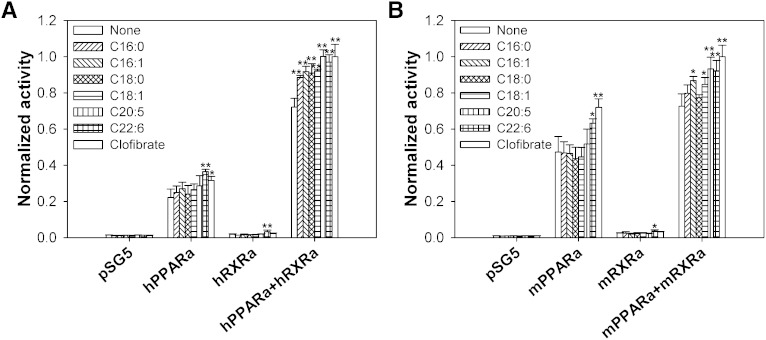
Effect of fatty acids and fatty acyl-CoA on transactivation of PPARα-RXRα heterodimers
Since PPARα heterodimerizes with RXRα to induce transactivation (27), COS-7 cells were cotransfected with pSG5 empty vector, PPARα alone, RXRα alone, or PPARα with RXRα, and then analyzed for transactivation of an acyl-CoA oxidase PPRE-luciferase reporter construct in the absence or presence of ligands (Fig. 7). Transactivation was measured as percentage firefly luciferase activity normalized to Renilla luciferase (internal control). In cells overexpressing only hPPARα (Fig. 7A) or mPPARα (Fig. 7B), DHA and clofibrate significantly increased transactivation. Although normalized activity was extremely low in hRXRα-overexpressing (Fig. 7A) and mRXRα-overexpressing (Fig. 7B) cells, DHA significantly increased transactivation in both, suggesting that this ligand (or its metabolite) is a strong activator of endogenous PPARα. While cells overexpressing hPPARα and hRXRα (Fig. 7A) or mPPARα and mRXRα (Fig. 7B) both showed increased activity, even in the absence of ligand, differences were noted in their ligand-induced effects. For cells overexpressing hPPARα and hRXRα, addition of palmitic acid, palmitoleic acid, stearic acid, oleic acid, EPA, and DHA resulted in similar effects on transactivation as did the PPARα agonist clofibrate (Fig. 7A). These data further validated LCFA or their metabolites as endogenous ligands of hPPARα needed to induce PPARα activity. However, addition of only the examined unsaturated LCFA and clofibrate significantly increased activity levels in COS-7 cells overexpressing mPPARα and mRXRα (Fig. 7B). The addition of the palmitic acid and stearic acid resulted in no significant changes in activity (Fig. 7B), consistent with the weak binding affinity of mPPARα for these ligands. In addition to suggesting that LCFA and LCFA-CoA represent high-affinity ligands for mPPARα, these data also suggested that differences in binding affinity for saturated LCFA could significantly affect the activity of PPARα.
Fig. 7.
PPARα ligands alter PPARα transactivation. COS-7 cells transfected with pSG5 empty vector, PPARα, RXRα, and both PPARα and RXRα were analyzed for transactivation of the acyl-CoA oxidase-PPRE-luciferase reporter construct in the presence of vehicle (open bars), 1 µM palmitic acid (diagonally upward bars), 1 µM palmitoleic acid (diagonally downward bars), 1 µM stearic acid (cross-hatched bars), 1 µM oleic acid (horizontal lined bars), 1 µM EPA (vertically lined bars), 1 µM DHA (hatched bars), and 1 µM clofibrate (open bars). For comparison between human and mouse effects, COS-7 cells were transfected with human versions of these proteins (A) or mouse versions of these proteins (B). The y axis represents values for firefly luciferase activity that have been normalized to Renilla luciferase (internal control), where PPARα- and RXRα-overexpressing cells in the presence of 1 μM clofibrate were arbitrarily set to 1. The bar graph represents the mean values (n ≥ 3) ± SE. *P < 0.05, **P < 0.01.
DISCUSSION
Although lipids have been shown to be endogenous ligands of PPARα from several species, including mouse, studies with hPPARα have focused on exogenous ligands. Since an increasing number of studies suggest species differences exist for ligand specificity and affinity (9, 14–16), this study focused on LCFA and/or LCFA-CoA as putative endogenous ligands of hPPARα. These data are the first to demonstrate full-length hPPARα binding to LCFA and LCFA-CoA at physiologically relevant concentrations. Human PPARα displayed high-affinity binding for saturated, monounsaturated, and polyunsaturated LCFA and LCFA-CoA (Kd = 11–40 nM), consistent with previously reported nuclear concentrations (3–68 nM) of LCFA and LCFA-CoA (28, 29). These high-affinity ligands significantly altered the secondary structure of hPPARα, while ligands that did not bind hPPARα (lauric acid, lauryl-CoA, and rosiglitazone) did not demonstrate any significant change in the structure of the protein. LCFA that bound to hPPARα in vitro transactivated the ACOX PPRE-luciferase reporter in a PPARα-dependent manner in COS-7 cells, further suggesting that LCFA and LCFA-CoA are endogenous ligands of hPPARα. These data are consistent with experiments using peroxisomal ACOX and/or PPARα knockout mice, which also suggest that LCFA and their thioester derivatives serve as natural ligands for PPARα in vivo (30–32).
Apart from identifying LCFA and LCFA-CoA as physiologically relevant endogenous ligands for hPPARα, these data highlight important species differences with respect to ligand specificity and affinity. While affinities for LCFA-CoA and unsaturated LCFA were similar between full-length human and murine PPARα, mPPARα only weakly bound the saturated palmitic acid and stearic acid, yet hPPARα strongly bound both. Similarly, some of the strongest changes in hPPARα secondary structure occurred with the addition of saturated and polyunsaturated LCFA, whereas saturated LCFA had only minor effects on mPPARα secondary structure. Consistent with these data, COS-7 cells overexpressing mPPARα and mRXRα treated with these saturated LCFA did not transactivate the ACOX-PPRE-luciferase reporter at the examined concentrations, while unsaturated LCFA did. Taken together, these data suggested that the human and mouse PPARα proteins bind and respond differently to specific ligands.
Given the high evolutionary rate exhibited by PPARα (33), it is not surprising to see such differences between hPPARα and mPPARα. In addition, strong physiological differences exist between human and rodent PPARα activation. Long-term administration of PPARα agonists are associated with hepatic carcinomas in rodents, but “humanized” PPARα mice are resistant to PPARα agonist-induced hepatocellular adenomas and carcinomas (16, 34). The potency and efficacy of many hypolipidemic agents and phthalate monoesters on the activation of human and mouse PPARα are also different (9, 14, 15). As previous microarray experiments have demonstrated a strong divergence between PPARα-regulated genes in mouse and human hepatocytes (15), it is likely that a combination of ligand-binding differences and target gene differences are responsible for the overall physiological variations. Other factors, including differences in ligand uptake and ligand metabolism between cell types, may account for some of these differences as well. However, this same study showed a high conservation in PPARα regulation of genes involved in lipid metabolism (15), suggesting that differences in these processes must be due to another mechanism - not just variation in target genes. Since a single mutation in the mouse PPARα ligand-binding domain (E282G) results in altered activity but displays similar DNA binding capacity, protein levels, and protein localization (35), it suggests that individual amino acid differences in the ligand-binding domain can affect activity through ligand binding. Such differences in specificity of mouse and human PPARα for specific nutrients could reflect an adaptation to different physiological and/or nutritional patterns of the species.
Additionally, these data suggest that differences exist in the binding affinity of full-length versus truncated PPARα. Data presented herein indicate that both full-length hPPARα and mPPARα bound polyunsaturated LCFA with strong affinity. This data challenges previously published data indicating that mouse PPARα does not bind saturated LCFA in the physiological range and only weakly interacts with PUFA (11–13). While such differences may exist due to variations in protein preparation, ligand-binding techniques, or changes in the protein's secondary structure, it should be noted that the previously published data were generated using a truncated mouse PPARα protein that lacked the N-terminus (mPPARαΔAB). Therefore, it is possible that the N-terminal domain of PPARα influences ligand binding. This hypothesis is supported in the case of PPARγ, where it was shown that mutation of specific residues within the N-terminal A/B domain affects the binding affinity of a synthetic PPARγ agonist (36).
In summary, LCFA and LCFA-CoA function as endogenous hPPARα ligands, binding with high affinity, altering PPARα secondary structure, and affecting transactivation. Although LCFA-CoA similarly bound both hPPARα and mPPARα, several ligands (including fluorescent LCFA/LCFA-CoA analogs, saturated LCFA, PUFA, and clofibrate) resulted in significant species differences. These data suggest that even though there is overlap in the endogenous ligands for mouse and human PPARα, significant species differences exist, and these differences may affect downstream gene regulation. These findings corroborate the importance of PPARα in allosteric regulation of fatty acid metabolism, where PPARα acts as a sensor to monitor the levels of fatty acids and their metabolites, then transcriptionally activates enzymes involved in their metabolism.
Supplementary Material
Acknowledgments
The authors thank Ms. Genesis Hines for assistance with binding assays, and Ms. Alagammai Kaliappan and Ms. Andrea Davis for technical expertise and helpful conversations.
Footnotes
Abbreviations:
- ACOX
- acyl-CoA oxidase
- CD
- circular dichroism
- DBD
- DNA-binding domain
- DHA
- docosahexanoic acid
- DPA
- docosapentanoic acid
- EPA
- eicosapentanoic acid
- hPPARα
- human PPARα
- LBD
- ligand-binding domain
- LCFA-CoA
- long-chain fatty acyl-CoA
- mPPARα
- mouse PPARα
- mPPARαΔAB
- truncated form of mPPARα lacking the N-terminal A/B region
- PPARα
- peroxisome proliferator-activated receptor α
- PPRE
- peroxisome-proliferator response element
- RXRα
- retinoid X receptor α
This work was supported by National Institutes of Health Grant DK-77573 and by funds from the Boonshoft School of Medicine and the College of Science and Mathematics, Wright State University.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of seven figures and four tables.
REFERENCES
- 1.Kliewer S. A., Umesono K., Noon D. J., Heyman R. A., Evans R. M. 1992. Convergence of 9-cis-retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 358: 771–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottlicher M., Widmark E., Li Q., Gustafsson J. A. 1992. Fatty acids activate a chimera of the clofibric acid-activated receptor and the glucocorticoid receptor. Proc. Natl. Acad. Sci. USA. 89: 4653–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reddy J. K., Hashimoto T. 2001. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 21: 193–230. [DOI] [PubMed] [Google Scholar]
- 4.Mangelsdorf D. J., Evans R. M. 1995. The RXR heterodimers and orphan receptors. Cell. 83: 841–850. [DOI] [PubMed] [Google Scholar]
- 5.Desvergne B., Michalik L., Wahli W. 2004. Be fit or be sick: peroxisome proliferator-activated receptors are down the road. Mol. Endocrinol. 18: 1321–1332. [DOI] [PubMed] [Google Scholar]
- 6.Frederiksen K. S., Wulf E. M., Wassermann K., Sauerberg P., Fleckner J. 2003. Identification of hepatic transcriptional changes in insulin-resistant rats treated with peroxisome proliferator activated receptor-alpha agonists. J. Mol. Endocrinol. 30: 317–329. [DOI] [PubMed] [Google Scholar]
- 7.Dreyer C., Krey G., Keller H., Givel F., Helftenbein G., Wahli W. 1992. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 68: 879–887. [DOI] [PubMed] [Google Scholar]
- 8.Maloney E. K., Waxman D. J. 1999. Trans-activation of PPARalpha and PPARgamma by structurally diverse environmnetal chemicals. Toxicol. Appl. Pharmacol. 161: 209–218. [DOI] [PubMed] [Google Scholar]
- 9.Bility M. T., Thompson J. T., McKee R. H., David R. M., Butala J. H., Vanden Heuvel J. P., Peters J. M. 2004. Activation of mouse and human peroxisome proliferator-activated receptors (PPARs) by phthalate monoesters. Toxicol. Sci. 82: 170–182. [DOI] [PubMed] [Google Scholar]
- 10.Ellinghaus P., Wolfrum C., Assmann G., Spener F., Seedorf U. 1999. Phytanic acid activates the peroxisome proliferator-activated receptor alpha (PPARalpha) in sterol carrier protein-2-/sterol carrier protein x-deficient mice. J. Biol. Chem. 274: 2766–2772. [DOI] [PubMed] [Google Scholar]
- 11.Lin Q., Ruuska S. E., Shaw N. S., Dong D., Noy N. 1999. Ligand selectivity of the peroxisome proliferator-activated receptor a. Biochemistry. 38: 185–190. [DOI] [PubMed] [Google Scholar]
- 12.Hostetler H. A., Petrescu A. D., Kier A. B., Schroeder F. 2005. Peroxisome proliferator activated receptor alpha (PPARalpha) interacts with high affinity and is conformationally responsive to endogenous ligands. J. Biol. Chem. 280: 18667–18682. [DOI] [PubMed] [Google Scholar]
- 13.Hostetler H. A., Kier A. B., Schroeder F. 2006. Very-long-chain and branched-chain fatty acyl CoAs are high affinity ligands for the peroxisome proliferator-activated receptor alpha (PPARalpha). Biochemistry. 45: 7669–7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller H., Devchand P. R., Perroud M., Wahli W. 1997. PPAR alpha structure-function relationships derived from species-specific differences in responsiveness to hypolipidemic agents. Biol. Chem. 378: 651–655. [DOI] [PubMed] [Google Scholar]
- 15.Rakhshandehroo M., Hooiveld G., Muller M., Kersten S. 2009. Comparative analysis of gene regulation by the transcription factor PPARalpha between mouse and human. PLoS ONE. 4: e6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonzalez F. J., Shah Y. M. 2008. PPARα: mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology. 246: 2–8. [DOI] [PubMed] [Google Scholar]
- 17.Sher T., Yi H-F., McBride O. W., Gonzalez F. J. 1993. cDNA cloning, chromosomal mapping, and functional characterization of the human peroxisome proliferator activated receptor. Biochemistry. 32: 5598–5604. [DOI] [PubMed] [Google Scholar]
- 18.Hubbell T., Behnke W. D., Woodford J. K., Schroeder F. 1994. Recombinant liver fatty acid binding protein interactions with fatty acyl-coenzyme A. Biochemistry. 33: 3327–3334. [DOI] [PubMed] [Google Scholar]
- 19.Hostetler H. A., Huang H., Kier A. B., Schroeder F. 2008. Glucose directly links to lipid metabolism through high-affinity interactionwith peroxisome proliferator activated receptor-alpha. J. Biol. Chem. 283: 2246–2254. [DOI] [PubMed] [Google Scholar]
- 20.Sreerama N., Woody R. 2000. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and DCSSTR methods with an expanded reference set. Anal. Biochem. 287: 252–260. [DOI] [PubMed] [Google Scholar]
- 21.Kim J. B., Wright H. M., Wright M., Spiegelman B. M. 1998. Add1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc. Natl. Acad. Sci. USA. 95: 4333–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spector A. A. 1969. Influence of pH of the medium on free fatty acid utilization by isolated mammalian cells. J. Lipid Res. 10: 207–215. [PubMed] [Google Scholar]
- 23.Petrescu A. D., Huang H., Hertz R., Bar-Tana J., Schroeder F., Kier A. B. 2005. Role of regulatory F-domain in hepatocyte nuclear factor-4alpha ligand specificity. J. Biol. Chem. 280: 16714–16727. [DOI] [PubMed] [Google Scholar]
- 24.Berbaum J., Harrison R. K. 2005. Comparison of full-length versus ligand binding domain constructs in cell-free and cell-based peroxisome proliferator-activated receptor alpha assays. Anal. Biochem. 339: 121–128. [DOI] [PubMed] [Google Scholar]
- 25.Francis G. A., Fayard E., Picard F., Auwerx J. 2003. Nuclear receptors and the control of metabolism. Annu. Rev. Physiol. 65: 261–311. [DOI] [PubMed] [Google Scholar]
- 26.Escher P., Wahli W. 2000. Peroxisome proliferator activated receptors: insights into multiple cellular functions. Mutat. Res. 448: 121–138. [DOI] [PubMed] [Google Scholar]
- 27.Kersten S., Desvergne B., Wahli W. 2000. Roles of PPARs in health and disease. Nature. 405: 421–424. [DOI] [PubMed] [Google Scholar]
- 28.Huang H., Starodub O., McIntosh A., Kier A. B., Schroeder F. 2002. Liver fatty acid binding protein targets fatty acids to the nucleus: real-time confocal and multiphoton fluorescence imaging in living cells. J. Biol. Chem. 277: 29139–29151. [DOI] [PubMed] [Google Scholar]
- 29.Huang H., Starodub O., McIntosh A., Atshaves B. P., Woldegiorgis G., Kier A. B., Schroeder F. 2004. Liver fatty acid binding protein colocalizes with peroxisome proliferator receptor alpha and enhances ligand distribution to nuclei of living cells. Biochemistry. 43: 2484–2500. [DOI] [PubMed] [Google Scholar]
- 30.Lee S. S. T., Pineau T., Drago J., Lee E. J., Owens J. W., Kroetz D. L., Fernandez-Salguero P. M., Wesphal H., Gonzalez F. J. 1995. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotrophic effects of peroxisome proliferators. Mol. Cell. Biol. 15: 3012–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoyama T., Peters J. M., Iritani N., Nakajima T., Furihata K., Hashimoto T., Gonzalez F. J. 1998. Altered constitutive expression of fatty acid metabolizing enzymes in mice lacking PPARalpha. J. Biol. Chem. 273: 5678–5684. [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto T., Fujita T., Usuda N., Cook W., Qi C., Peters J. M., Gonzalez F. J., Yeldandi A. V., Rao M. S., Reddy J. K. 1999. Peroxisomal and mitochondrial fatty acid beta-oxidation in mice nullizygous for both PPARalpha and peroxisomal fatty acyl CoA oxidase. J. Biol. Chem. 274: 19228–19236. [DOI] [PubMed] [Google Scholar]
- 33.Laudet V. 1997. Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. J. Mol. Endocrinol. 19: 207–226. [DOI] [PubMed] [Google Scholar]
- 34.Morimura K., Cheung C., Ward J. M., Reddy J. K., Gonzalez F. J. 2006. Differential susceptibility of mice humanized for peroxisome proliferator-activated receptor alpha to Wy-14, 643-induced liver tumorigenesis. Carcinogenesis. 27: 1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu M-H., Palmer C. N. A., Griffin K. J., Johnson E. F. 1995. A single amino acid change in the mouse peroxisome proliferator activated receptor alters transcriptional response to peroxisome proliferators. Mol. Pharmacol. 48: 559–567. [PubMed] [Google Scholar]
- 36.Shao D., Rangwala S. M., Bailey S. T., Krakow S. L., Reginato M. J., Lazar M. A. 1998. Interdomain communication regulating ligand binding by PPAR-gamma. Nature. 396: 377–380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.