Abstract
Proprotein convertase subtilisin-kexin-9 (PCSK9) inhibition markedly augments the LDL lowering action of statins. The combination is being evaluated for long-term effects on atherosclerotic disease outcomes. However, effects of combined treatment on hepatic cholesterol and bile acid metabolism have not yet been reported. To study this, PCSK9-Y119X mutant (knockout) and wild-type mice were treated with or without atorvastatin for 12 weeks. Atorvastatin progressively lowered plasma LDL in each group, but no differences in liver cholesterol, cholesterol ester, or total bile acid concentrations, or in plasma total bile acid levels were seen. In contrast, atorvastatin increased fecal total bile acids (∼2-fold, P < 0.01) and cholesterol concentrations (∼3-fold, P < 0.01) versus controls for both PCSK9-Y119X and wild-type mice. All 14 individual bile acids resolved by LC-MS, including primary, secondary, and conjugated species, reflected similar increases. Expression of key liver bile acid synthesis genes CYP7A1 and CYP8B1 were ∼2.5-fold higher with atorvastatin in both strains, but mRNA for liver bile acid export and reuptake transporters and conjugating enzymes were not unaffected. The data suggest that hepatocyte cholesterol and bile acid homeostasis is maintained with combined PCSK9 and HMG-CoA reductase inhibition through efficient liver enzymatic conversion of LDL-derived cholesterol into bile acids and excretion of both, with undisturbed enterohepatic recycling.
Keywords: proprotein convertase subtilisin-kexin-9 inhibitors, 3-hydroxy-3-methylglutaryl-coenzyme A reductase inhibitors, statins, CYP7A1, bile acid transporter, cholestasis, low desnsity lipoprotein receptor, N-ethyl N-nitrosourea mutagenesis
Proprotein convertase subtilisin-kexin-9 (PCSK9) activity has a profound effect on cholesterol metabolism in humans and animal models, as revealed through genetic studies (1–4), knockout and transgenic mice (5–7), and recent studies with PCSK9-inhibiting monoclonal antibodies, Adnectins, and antisense oligonucleotides (8–10). Evaluation of PCSK9 inhibitors in combination with statins is important to understand the full potential of this new class of low density lipoprotein cholesterol (LDL-C) lowering agents. Both statins and PCSK9 inhibitors lower circulating LDL-C levels by increasing LDL receptor (LDLR) activity primarily in liver. The hepatic production and enterohepatic circulation of bile acids is important for regulation of whole body cholesterol metabolism, recycling of bile acids, and absorption of nutrients. However the effects of PCSK9 suppression, in the presence and absence of statins, on bile acid metabolism have not been previously reported. Greater LDL clearance increases liver cholesterol exposure, potentially leading to cholesterol accumulation and/or increased conversion to bile acids through liver metabolic pathways, which could affect gastrointestinal exposure to bile acids. Therefore the present study was conducted to analyze the pharmacological effects of statins combined with PCSK9 suppression on bile acid and cholesterol balance and gene regulation in a mouse model.
Some PCSK9 biologic inhibitors exhibit species-dependent target binding, and potential immune responses to humanized antibodies in animal models can confound chronic pharmacological modeling in animals. Therefore we used a genetic model of PCSK9 suppression, the PCSK9-Y119X mutant (knockout) mouse model, to evaluate the long-term effects of combination inhibition. The mouse tyrosine-119 mutation (Y119X) in the PCSK9 coding sequence results in loss of PCSK9 expression and reduced plasma LDL, analogous to the PCSK9-Y142X loss-of-function mutation in humans. The loss of PCSK9 expression in this model leads to increased liver LDLR protein and activity, and decreased circulating LDL and total cholesterol. Using this mutant mouse model compared with wild-type mice, the effects of atorvastatin treatment (vs. statin-free controls) were studied over 12 weeks. Forty-eight hour fecal samples were collected and comprehensive analyses of fecal bile acids and cholesterol were conducted using LC-MS. At the end of the study, liver cholesterol content was assayed and liver mRNA concentrations for 16 key sterol and bile acid pathway genes were conducted by RT-PCR. Significant changes in sterol and bile acid metabolism were observed, with metabolic regulation driven by increased expression of a very few liver genes accommodating the increased flux of cholesterol in liver and the increased production of bile acids following stimulated cholesterol uptake with combined statin/PCSK9 suppression.
METHODS
Mouse model
Mice carrying a nonsense mutation (point mutation) in the PCSK9 coding sequence at tyrosine 119 (Y119X) were generated through N-ethyl N-nitrosourea (ENU) mutagenesis (11). Frozen embryos produced from matings of G1 male mice heterozygous for the Y119X allele of PCSK9 and wild-type female mice were used to establish a breeding colony of the mutant mice at Bristol-Myers Squibb. The Y119X positive progeny were extensively backcrossed with C57BL/6J mice through more than eight generations using speed congenics to derive the Y119X mice with >99.8% homogenous genetic background. The mice were fertile and bred as homozygous matings to maintain the colony. Control C57BL/6J mice, age- and weight-matched to the mutant mice, were obtained from The Jackson Laboratory.
Study design
For the study, each group comprised 10 homozygous mutant or wild-type mice (6 males and 4 females for each treatment group) aged 3 months at the start. Mice were placed on a diet containing 0.05% atorvastatin, or normal chow for 12 weeks total baseline plasma cholesterol and LDL-C values were obtained and mice were randomized into control diet and atorvastatin supplemented diet (0.05% by weight). The use of 0.05% dietary atorvastatin has previously been used in mouse studies of cholesterol and bile acid metabolism (12). In our past work, this resulted in atorvastatin plasma exposure equivalent to ∼10–15 mg/kg/day by oral gavage. Animals had continuous access to diet for the duration of the study. Blood plasma samples collected in ethylenediamine tetraacetic acid were taken at 9:00–10:00 AM in the morning (light phase) without fasting, every 2 weeks for 8 weeks. At week 9, 48 h accumulated fecal samples were collected for each mouse for bile acid and cholesterol analyses. The study was terminated at week 12 and liver samples were collected for assay of free and esterified cholesterol, and for isolation of RNA.
Plasma chemistry
EDTA plasma samples from treated and control mice were assayed for total cholesterol, LDL cholesterol (direct LDL), and total bile acids by standard plasma chemistry laboratory enzymatic methods using an Olympus automated analyzer (model AU680). In addition, interim plasma samples at 6 weeks were assayed for alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels to assess potential liver toxicity. The plasma chemistry analyses used enzymatic reagent kits: Cholesterol Assay and Direct LDL Assay, Roche Diagnostics; Total Bile Acids Assay, Bio Quant Laboratories.
Liver cholesterol and total bile acids
Homogenates of 0.3–0.5 g samples of livers were prepared in PBS buffer. Aliquots were assayed for free and esterified cholesterol using enzymatic methods according to Wako free cholesterol and Infinity esterified cholesterol reagent kits. Total bile acids were assayed using the enzymatic recycling method from Bio Quant Laboratories.
LC-MS analysis of bile acids and cholesterol
The method for liquid chromatography-mass spectrometry (LC-MS) quantitation of fecal cholesterol and bile acids from mice was as follows. The MS responses and LC retention time characteristics were confirmed during method development using commercially available standards. Mouse fecal samples were taken over a 48 h collection period at week 9 of the study and mixed before drying overnight (50°C). Aliquots (0.10 g) of dried samples received 0.7 ml water and were vortexed to a paste, and 1.4 ml of 1 N NaOH in ethanol was added followed by vortexing and heating at 85°C for 2 h. Aliquots then received addition of 0.7 ml of water, 80 μg D6- cholesterol (deuterated cholesterol standard) and 4.5 ug of D4-cholic acid (deuterated deoxycholic acid standard) in 25 μl working solution to all samples. After vortexing, 3.0 ml of petroleum ether was added with vortexing. Samples were centrifuged at 500 g for 5 min, and for cholesterol 0.5 ml of the upper phase was transferred to a 96-well assay block; for bile acids 0.6 ml of the lower clear layer was transferred to the 96-well block. For cholesterol LC-MS analysis, the dried samples were reconstituted in 200 ul of methanol/well in the 96-well plate, vortexed for 2 min, and centrifuged for 10 min. Supernatant (120 ul) was transferred to new 96-well plates for LC-MS analysis. The LC-MS analysis was performed on a Thermo Acella uHPLC system interfaced with a Thermo Exactive mass spectrometer. The uHPLC column used was a Waters BEH C8 2.1, 1.7 u, 50 mm and the detection was performed in APCI positive ion mode at 25 K resolution and data collection between 200 and 600 Da. For bile acid LC-MS analysis, the dried samples were reconstituted in 200 ul of methanol in the 96-well plate. The plate was vortexed for 2 min and centrifuged for 10 min. Supernatant (120 ul) was transferred to a new 96-well plate for LC-MS analysis. The LC-MS analysis was performed on a Thermo Acella uHPLC system interfaced with a Thermo Exactive mass spectrometer. The uHPLC column used was a Waters BEH C18 2.1, 1.7 u, 150 mm and the detection was performed in ESI negative ion mode at 25 K resolution and data collection between 200 and 1,000 Da.
Liver mRNA extraction and RT-PCR assay
At 12 weeks in the mouse study, whole liver was processed for RNA isolation as follows. Tissue samples were immediately placed in RNAlater reagent and kept at 4°C for 24 h, then removed and placed at −80°C in cassettes. To prepare RNA, ∼30 mg tissue samples were added to Biopur tubes with stainless steel bead on dry ice, followed by addition of 1.1 ml TRIzol and lysing by TissueLyser at 30 Hz for 3 min. Samples then received 0.4 ml chloroform, were mixed and incubated for 5 min at 20°C, and centrifuged for 25 min at 12,000 g at 4°C. Supernatants (0.35 ml) from each sample were extracted in QIAcubes, and the extracted mRNA samples were stored at −80°C in 96-well plates with 1 μl of Protector RNase Inhibitor added.
For real-time qPCR measurements, total RNA was quantitated on a NanoDrop ND-1000 UV-Vis spectrophotometer, and RNA quality was assessed on an Agilent 2100 BioAnalyzer. Aliquots of 1.0 ug of RNA from each sample were converted to cDNA using the Applied Biosystems (ABI) High-Capacity cDNA Archive Kit. Quantitative real-time PCR was conducted in 384-well reaction plates on an Applied Biosystems Prism 7900HT sequence detector. The mRNA levels for specific genes were normalized to 18S rRNA expression (ABI primer-probe set Hs99999901_s1). Standard curves for each mRNA as well as 18S rRNA were generated by serially diluting cDNA from saline treated animals. All measurements were performed in duplicate and mRNA was averaged within treatment groups (n = 10 mice per group), and unpaired two-tailed t-tests were performed to evaluate statistical difference; P ≤ 0.05 was considered significant.
RESULTS
Phenotypic characterization
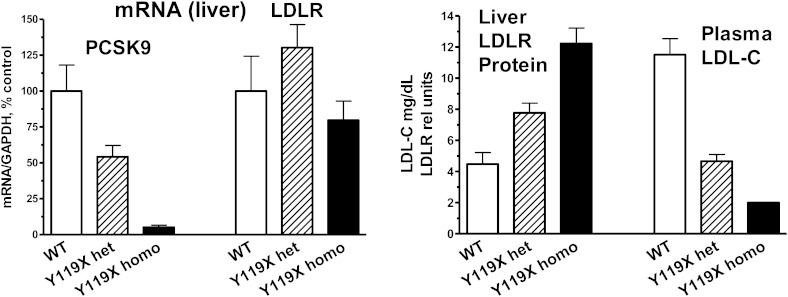
The Y119X point mutation in the PCSK9 coding sequence lies in the prodomain of the protein, analogous to the human Y142X loss-of-function mutation (2). In initial studies of the mutant mice, very low levels of PCSK9 mRNA for homozygous Y119X mice and ∼50% lower PCSK9 mRNA for the heterozygotes were measured by RT-PCR compared with wild type, suggesting nonsense mediated decay of the point-mutant PCSK9 mRNA. LDLR mRNA levels in liver were not significantly different between the mutant and wild type (Fig. 1, left panel). PCSK9 protein was undetectable in liver or plasma of the Y119X homozygous mice (not shown). In initial studies, wild-type (C57BL/6J) mice, heterozygous Y119X, and homozygous Y119X mutant mice were studied at ∼6 months of age, maintained on a normal chow diet. Liver LDLR protein levels were increased ∼2-fold in the heterozygous and 3-fold in the homozygous mutants compared with wild type, and plasma LDL-C was ∼50% lower in the heterozygous and ∼80% lower in the Y119X homozygous mice compared with wild-type controls (Fig. 1, right panel). For these endpoints, an intermediate phenotype in heterozygous mutant mice was observed. These findings show that the Y119X mutation acts as a loss-of-function mutation, and that loss of PCSK9 function leads to increased LDLR protein in liver and marked decreases in circulating LDL-C levels as seen in humans with the Y142X mutation.
Fig. 1.
Biochemical phenotype of PCSK9-Y119X mice. Liver mRNA for PCSK9 and LDLR in PCSK9-Y119X heterozygous and homozygous mutant versus wild-type (WT) mice (left panel). PCSK9-Y119X homozygous and heterozygous mice exhibit increased liver LDLR protein and reduced circulating LDL-C levels versus WT (right panel). Mean values (n = 6 male mice) and 95% confidence interval are shown.
Statin treatment effects on plasma lipids
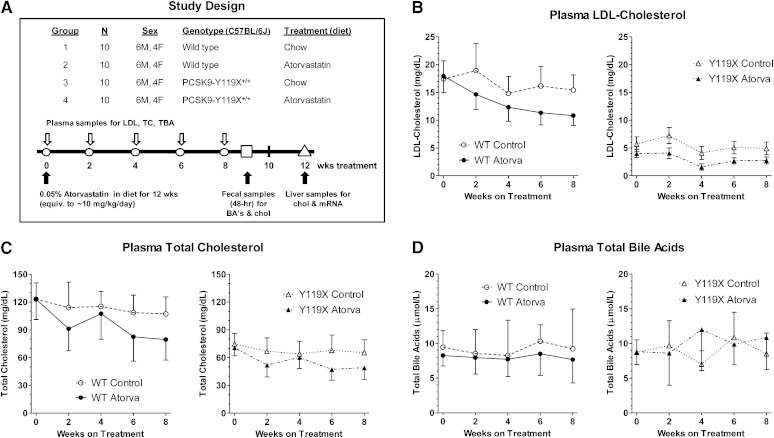
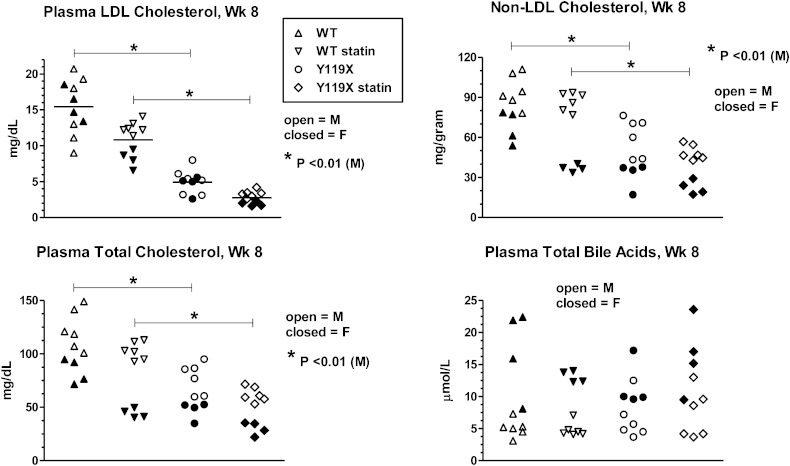
In the study design, age-matched cohorts of 20 C57BL/6J (wild-type) and 20 PCSK9-Y119X homozygous mice were randomized into groups of 10 mice each (6 males, 4 females) for atorvastatin treatment and sampling over time (Fig. 2A). Plasma LDL-C (Fig. 2B) and total cholesterol (Fig. 2C) tended to decrease progressively and approached plateau values by 8 weeks. Plasma total bile acids remained relatively constant in all groups (Fig. 2D). At 8 weeks treatment, atorvastatin significantly reduced LDL-C in homozygous PCSK9-Y119X mice and in wild-type mice (Fig. 3), reaching levels near ∼3 mg/dl in the mutant mice. Non-LDL cholesterol and total cholesterol reflected similar atorvastatin effects although female mice showed greater apparent changes than males, whereas plasma total bile acids were not different between any groups despite wider variability between animals (Fig. 3), averaging ∼8–10 μmoles/l for all time points. Furthermore plasma ALT and AST were not different in any group (assayed at week 6, not shown). These findings suggest that bile acid enterohepatic recirculation and hepatocyte integrity were not significantly disturbed with the combination of PCSK9 and HMG-CoA reductase inhibition in mice despite strong effects on LDL and total cholesterol.
Fig. 2.
Time course of plasma lipid responses to atorvastatin for wild-type (WT) versus PCSK9-Y119X mice. A: Study design. B: Plasma LDL-C levels at biweekly intervals over 8 weeks treatment. C: Plasma total cholesterol at intervals over 8 weeks. D: Plasma total bile acids at intervals over 8 weeks. Mean values [n = 10 mice; 6 males (M), 4 females (F)] and 95% confidence intervals are shown.
Fig. 3.
Plasma LDL-C, HDL-C, total cholesterol, and total bile acids after 8 weeks atorvastatin. Values for each lipid measured (indicated in titles for each panel) are shown as scatterplots of individual animal data for male mice (M) (closed symbols, n = 6 per group) and female mice (F) (open symbols, n = 4 per group). Bars with asterisk (*) indicate significance for means of male group data analyzed by one-way ANOVA with Bonferroni test for multiple comparisons.
Liver cholesterol and total bile acids concentrations
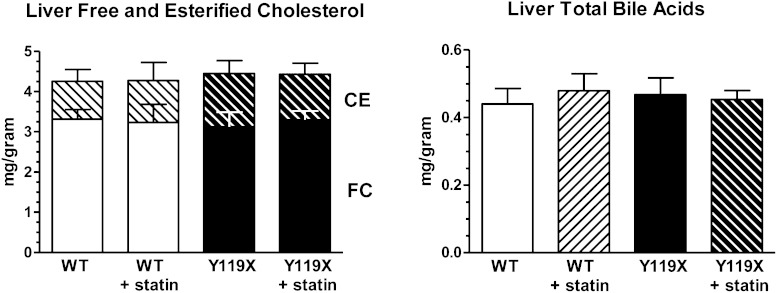
We next examined whether the changes in plasma cholesterol were associated with any differences in cholesterol accumulation in liver tissue. At 12 weeks treatment hepatic cholesterol and total bile acid concentrations were assayed by enzymatic method. No differences in liver free cholesterol or esterified cholesterol mean concentrations were seen between any of the four study groups (Fig. 4, left panel). Male and female mice exhibited similar values. Approximately 1/4 of total liver cholesterol was esterified in each group, and total levels observed were consistent with previous studies in our laboratories. Similarly, liver total bile acid concentrations measured enzymatically were essentially identical for all four groups in the study (Fig. 4, right panel) and similar levels were found in males and females. The levels of liver cholesterol relative to total bile acids in liver were ∼10:1 for all mice, and the liver total bile acid concentrations were similar to those reported previously for normal mouse liver extracts using LC-MS/MS methodology (13). Overall the data indicate that livers accumulated neither cholesterol nor bile acids despite the increased LDLR-mediated hepatic uptake of plasma cholesterol stimulated by the combined mechanism of PCSK9 and HMG-CoA reductase inhibition. Liver cholesterol homeostasis was maintained throughout the study.
Fig. 4.
Liver free and esterified cholesterol and total bile acid concentrations at 12 weeks. Bars indicate mean liver tissue concentrations of free cholesterol (lower bars) and esterified cholesterol (upper hatched bars) by enzymatic assay (left panel). Liver tissue concentrations of total bile acids determined by enzymatic assay. Combined male and female data are plotted as bars for clarity, as no sex dependency was observed in these data (right panel). For both panels, mean values (n = 10 per group; mg analyte per gram liver wet weight) and 95% confidence intervals are shown. WT, wild type.
Fecal cholesterol
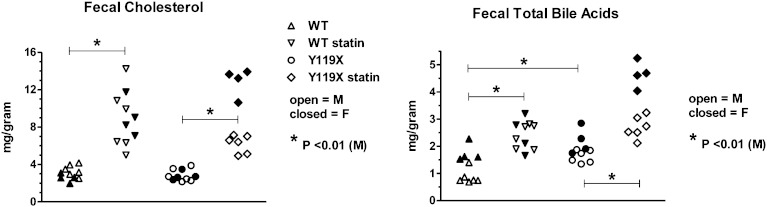
Forty-eight hour accumulated fecal samples from each mouse were taken during the 9th week of the study. The fecal samples were analyzed for free cholesterol by LC-MS, and for total bile acids by both enzymatic method, as well as individual bile acids determined by LC-MS. Without statin treatment, fecal cholesterol concentrations at 9 weeks were the same for wild-type and PCSK9-mutant mice, and males and females exhibited similar levels (Fig. 5). With atorvastatin treatment, the fecal cholesterol concentrations were approximately 2- to 4-fold higher than statin-free controls for both wild-type and PCSK9-Y119X mice (Fig. 5, left panel; significance shown for male mice). Female PCSK9-Y119X mice appeared to be more sensitive than males to the atorvastatin-induced increase in fecal cholesterol concentrations. Because liver cholesterol concentrations were not affected by atorvastatin, the data indicate that the statin-induced increase in flux of cholesterol from plasma into liver was excreted via bile into the intestinal tract for elimination from the body. The finding that PCSK9-Y119X mice (without statin) had lower plasma LDL-C but did not accumulate cholesterol or cholesterol esters in the liver, and had identical fecal cholesterol levels as wild-type mice, suggested that genetic suppression of PCSK9 may influence hepatic conversion of cholesterol into bile acids in mice.
Fig. 5.
Fecal cholesterol and total bile acids at 9 weeks treatment. Free cholesterol concentration determined by LC-MS from 48 h fecal samples of wild-type (WT) and PCSK9-Y119X homozygous mice (left panel). Fecal total bile acids determined by enzymatic assay method (right panel). Values are shown as scatterplots of individual animal data for male mice (M) (closed symbols, n = 6 per group) and female mice (F) (open symbols, n = 4 per group). Bars with asterisk (*) indicate significance for means of male group data analyzed by one-way ANOVA with Bonferroni test for multiple comparisons.
Fecal bile acids
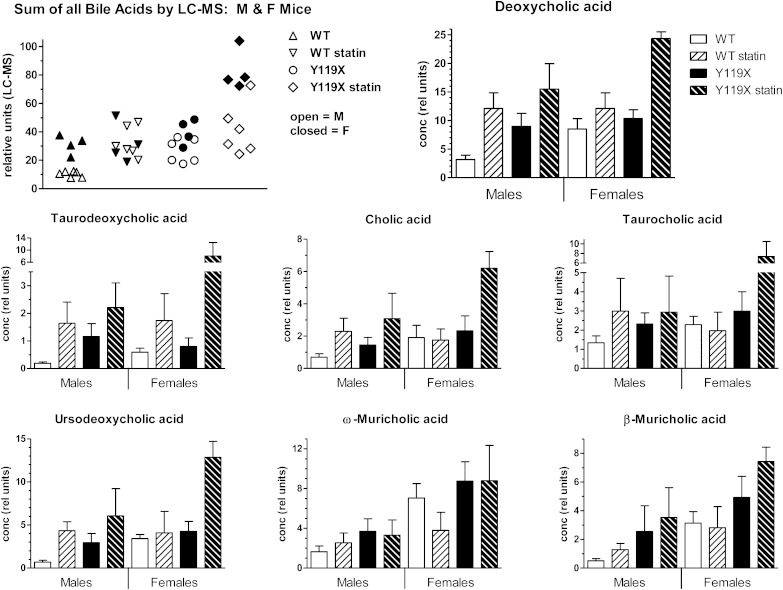
Total bile acids measured enzymatically in 48 h fecal samples taken at week 9 showed significant effects of both statin treatment and genotype. Samples from PCSK9-Y119X mice had ∼1.5- to 2-fold higher total bile acid levels than wild type without statin treatment (P < 0.01). Atorvastatin increased fecal total bile acid concentrations approximately 2-fold over respective controls for both wild-type and PCSK9-Y119X mice (Fig. 5, right panel; significance shown for male mice). Female PCSK9 mutant mice appeared to be more sensitive to atorvastatin than males. Further analysis by LC-MS of individual bile acids in the samples resolved 14 individual bile acid chemical species, including primary and secondary bile acids in conjugated and unconjugated forms. This method provided high resolution with relative quantification for the various species based on deuterated internal standard for deoxycholate only (the highest abundance bile acid in the samples). The sum of all bile acids resolved by LC-MS, as well as individual bile acids, was plotted by treatment group (Fig. 6). These results show patterns similar to those in the enzymatic total bile acids assay (Fig. 5). Within each group, female PCSK9-Y119X mice tended to be more sensitive than males to the inducing effect of atorvastatin, including the primary cholic acid and secondary deoxycholic acid in both unconjugated and conjugated forms (Fig. 6). Similar patterns between groups were seen for several other bile acids (not shown): ursodeoxycholic acid and ϖ-muricholic acid, chenodeoxycholic acid, α-muricholic acid, β-muricholic acid, glyco-deoxycholic acid, glyco-ursocholic acid, tauro-chenodeoxycholic acid, tauro-ursodeoxycholic acid, and tauro-ursocholic acid.
Fig. 6.
Individual fecal bile acids by LC-MS for male and female mice at 9 weeks. Data represent the individual bile acids assayed by LC-MS from 48 h fecal samples of mice. The scatterplot (first panel) shows individual animal data for male mice (M) (open symbols) and female mice (F) (closed symbols) for the sum of all 14 bile acid species resolved by the method. The other panels show each of the 7 highest abundance bile acids with separate bars for male (n = 6) and female (n = 4) mice. Bars represent mean and SD for relative concentrations normalized to deuterated deoxycholic acid internal standard.
Gene regulation in liver
Liver RNA samples were isolated at the end of the study (week 12) for assay of specific mRNAs for selected genes. Assays were conducted by RT-PCR using quantitative conditions with standard curves. The genes were selected based on biological roles in respective pathways, the statin mechanism, and data from a previous experiment using Affymetrix profiling to verify expression in the mouse model (Affymetrix GeneChip HT MG-430 PM, ∼39,000 transcripts). Atorvastatin strongly induced liver mRNA expression for the major regulatory genes in the sterol synthesis pathway in both wild-type and PCSK9-knockout mice (Table 1). Expression of the sterol regulatory binding protein (SREBP)2 was increased 2-fold in both groups, while SRBEP1 was unaffected. Several SREBP2 response genes which control the cholesterol synthesis pathway were strongly upregulated by atorvastatin to similar degrees in both wild-type and PCSK9-Y119X mice. The LDL receptor gene was also induced significantly in both groups. In PCSK9-Y119X mutant mice without statin treatment, no differences in expression of any of these genes was found compared with wild-type mice, with the exception of PCSK9 mRNA which was 9-fold lower than in wild-type mice, consistent with nonsense-mediated decay of the mutant mRNA.
TABLE 1.
Liver expression of genes regulating sterol pathway and bile acid synthesis, conjugation, and transport
| mRNA concentration by RT-PCR (normalized to 18S RNA) |
||||||
| Function | Protein | Gene | WT | WT + Statin | PCSK9-Y119X | PCSK9-Y119X + Statin |
| Fatty acid and TG pathway | SREBP1 | SREBF1 | 100 ± 10.0 | 107 ± 11.8 | 96.2 ± 9.3 | 97.4 ± 10.9 |
| Cholesterol pathway | SREBP2 | SREBF2 | 100 ± 37.1 | 202 ± 59.4* | 94.3 ± 26.0 | 189 ± 37.1* |
| Cholesterol synthesis | HMGS | HMGCS1 | 100 ± 33.8 | 360 ± 99.4* | 76.3 ± 28.1 | 343 ± 45.5* |
| Cholesterol synthesis | HMGR | HMGCR | 100 ± 52.8 | 546 ± 185* | 77.1 ± 31.4 | 110 ± 52.8* |
| LDL receptor | LDLR | LDLR | 100 ± 13.0 | 156 ± 25.1* | 95.8 ± 13.0 | 146 ± 16.1* |
| LDLR regulation | PCSK9 | PCSK9 | 100 ± 25.1 | 205 ± 50.4* | 10.7 ± 3.5 | 21.9 ± 5.1* |
| BA synthesis main pathway, cholesterol substrate | CYP7α hydroxylase | CYP7A1 | 100 ± 61.7 | 180 ± 78.7* | 63.6 ± 50.2 | 143 ± 64.7* |
| BA synthesis, steroids and oxysterol substrates | CYP7α hydroxylase | CYP7B1 | 100 ± 70.4 | 113 ± 87.8 | 129.5 ± 121 | 103 ± 79.2 |
| BA synthesis, alternative pathway | CYP27α hydroxylase | CYP27A1 | 100 ± 13.6 | 120 ± 17.1 | 113 ± 13.1 | 113 ± 16 |
| Balance between cholic and deoxycholic acid synthesis | Sterol 12-α hydroxylase | CYP8B1 | 100 ± 42.6 | 215 ± 33.0* | 78.5 ± 28.1 | 199 ± 32.1* |
| Amino acid conjugation | BAT | BAAT | 100 ± 9.6 | 113 ± 11 | 99.3 ± 11.4 | 112 ± 10.6 |
| BA activation to CoA-ester | BACS | SLC27A5 | 100 ± 15 | 105 ± 14.7 | 95.1 ± 14.2 | 104 ± 8.2 |
| Sodium taurocholate cotransport, extraction of BA from portal blood | NTCP | SLC10A1 | 100 ± 6.2 | 101 ± 19.7 | 106 ± 16.6 | 92.7 ± 6.8 |
| Bile salt export pump, ATP-dependent | BSEP | ABCB11 | 100 ± 12.6 | 84.5 ± 12.9 | 116 ± 18.8 | 93.9 ± 9.1 |
| Apical sodium bile acid transporter family, BA reuptake transporter | ASBT | SLC10A7 | 100 ± 12.9 | 107 ± 14.4 | 104 ± 14.0 | 101 ± 4.2 |
| Na independent organic anion transporter | OATP1B2 | SLCO1B1 | 100 ± 9.2 | 105 ± 12.2 | 101 ± 14.3 | 93.7 ± 14.1 |
Values are RT-PCR assayed mean mRNA concentrations in arbitrary units ± SD for 16 selected liver genes, averaged for n = 10 mice per group. Individual values were first normalized to 18S RNA in each sample before averaging. Group mean values were then normalized to mean wild-type control = 100%. *P < 0.01 versus control for respective genotype. WT, wild type; TG, triglyceride; BA, bile acid.
Atorvastatin treatment increased liver mRNA levels significantly by over 2-fold compared with statin-free controls for the rate-limiting enzymes of the main (neutral pH) bile acid pathway, CYP7A1 and CYP8B1, in both wild-type and PCSK9-Y119X mice (Table 1). No differences were seen in expression of enzymes of the minor (acidic pH) pathway (CYP27A and 7B1). The enzymes regulating bile acid-amino acid conjugation were unaffected. These findings suggest that liver cholesterol flux is diverted into bile acid production and secretion into bile in liver of both wild-type and PCSK9 knockout mice through upregulation of key bile acid synthesis enzymes to similar degree. Without statin treatment, PCSK9-Y119X mice showed little or no difference from wild-type mice in the expression of these genes.
Expression of the bile acid transporter genes for export or reuptake were similar for both wild type and mutants with or without atorvastatin, with only slight decreases (<20%) noted in the bile salt export pump (BSEP) mRNA with atorvastatin for both genotypes (Table 1). Expression of an apical sodium bile acid transporter (ASBT) family member (SLC10A7), the sodium taurocholate cotransporting polypeptide (NTCP), and the sodium independent organic anion transporter (OATP1B2) were unaffected in any of the groups. These data suggest that the capacity and rate of transhepatocellular movement of bile acids across hepatocytes appears to be sufficient without further upregulation of the transporter genes in the presence of combined PCSK9 and HMG-CoA reductase inhibition. The efficiency of both export from the hepatocyte and the extraction and reuptake of bile acids from blood into the liver appears to be sufficient to accommodate the increased flux of bile acids encountered following prolonged statin treatment of both wild-type and PCSK9 knockout mice.
Four of the 16 liver genes assayed showed sex-dependent differences in expression: SREBP2, CYP7A1, CYP7B1, and CYP8B1. In particular, SREBP2 and CYP7A1 baseline mRNA concentrations were higher and were induced by statins to higher levels in females than males for both genotypes (Table 2). These differences may at least partly explain the trends toward higher levels of fecal cholesterol and bile acids in female PCSK9-Y119X mice treated with statins as shown in Fig. 6.
TABLE 2.
Liver gene expression with different responses for male versus female mice
| mRNA Concentration by RT-PCR (Normalized to 18S RNA) |
||||
| Gene | WT (n = 10) | WT + Statin (n = 10) | PCSK9-Y119X (n = 10) | PCSK9-Y119X + Statin (n = 10) |
| SREBP2 | ||||
| Male | 100 ± 10.4 | 197 ± 23.7 | 95.1 ± 16.7 | 199 ± 16.7 |
| Female | 157 ± 28.2 | 326 ± 39.8* | 147 ± 20.0 | 283 ± 13.6* |
| CYP7A1 | ||||
| Male | 100 ± 46.4 | 257 ± 83.0 | 88.3 ± 80.7 | 179 ± 52.0 |
| Female | 275 ± 70.1 | 381 ± 173 | 138 ± 94.9 | 337 ± 108 |
| CYP7B1 | ||||
| Male | 100 ± 32.3 | 115 ± 44.4 | 131 ± 82.6 | 109 ± 26.1 |
| Female | 19.9 ± 2.36 | 19.5 ± 2.89 | 24.3 ± 4.77 | 12.2 ± 1.13 |
| CYP8B1 | ||||
| Male | 100 ± 16.4 | 169 ± 26.3 | 72.7 ± 17.2 | 141 ± 19.3 |
| Female | 41.9 ± 7.8 | 160 ± 26.5* | 41.5 ± 10.8 | 171 ± 22.8* |
Table lists data for four genes from those summarized in Table 1 which showed significant quantitative differences for female versus male mice. Shown are mean ± SD for n = 6 (male) and n = 4 (female) mice per group. *P < 0.01 versus control for respective genotype. WT, wild type.
DISCUSSION
The results described here show that PCSK9 inactivation through ENU mutagenesis in mice, creating an inactivating point mutation in PCSK9 and apparent destabilization of its mRNA, has a profound effect on hepatic cholesterol metabolism. This is consistent with previous studies utilizing alternative knockout technologies, PCSK9-inhibiting monoclonal antibodies, or antisense approaches (5, 6, 8). PCSK9 genetic suppression and atorvastatin individually resulted in lower plasma LDL-C and total cholesterol levels, and the combination decreased levels further compared with controls, to average levels as low as 3 mg/dl (LDL-C) after statin treatment for 8 weeks in the mutant mice. Though LDL-C is a minor component of mouse plasma, the regulation through PCSK9 and LDLR were pronounced in the mouse, and changes in non-LDL cholesterol were also observed. The latter may reflect clearance of apoE-containing high density lipoproteins by the LDLR. The mice continued to thrive despite the very low LDL, and there was no evidence of liver malfunction (ALT and AST were normal).
Statins potently suppress cholesterol synthesis, leading to feedback upregulation through the SREBP2 response mechanism, which includes increased expression of both LDLR and PCSK9. In our studies, chronic treatment of mice with a high dosage of atorvastatin resulted in 4- to 7-fold increases in liver expression of the key sterol synthesis genes HMG-CoA synthase and HMG-CoA reductase in both wild-type and PCSK9-Y119X mice. Interestingly, SREBP2 mRNA levels also increased in liver, potentiating expression of these genes. LDLR and PCSK9 expression were each induced ∼2-fold by atorvastatin for both genotypes, although the basal levels of PCSK9 mRNA were ∼10-fold lower in the Y119X mutant mice, consistent with nonsense-mediated decay of the mRNA.
The fundamental molecular physiology controlling cholesterol balance in mice and larger mammals including humans is similar, although absolute rates of cholesterol synthesis and LDL-C clearance were shown by Dietschy's group to be much higher in the mouse, typical of its high metabolic activity in many pathways (14). In that comprehensive study, it was found that ∼80% of whole-body LDL-C clearance from circulation was directed into the liver and ∼90% of this was mediated by LDLR endocytosis in mice. They concluded that extrahepatic tissues biosynthesize nearly all of their sterol requirements while most LDL-C returns directly to the liver. Changes in LDLR activity profoundly alter plasma LDL-C, but have little effect on cholesterol balance across the extrahepatic organs. In contrast, the unique ability of the liver to catabolize cholesterol to bile acids and excrete both into bile constitutes the main mechanism for whole-body cholesterol homeostasis. Regulation of hepatic bile acid synthesis ensures this balance and also provides critical emulsification activity in the intestine. When hepatocyte levels of bile acids are in excess they repress their own biosynthesis, and when deficient their synthesis is increased. In mice, increased hepatic cholesterol influx can drive activation of the bile acid biosynthetic pathway (15). Data from studies in humans led to an understanding that the pool of cholesterol for liver bile acid synthesis was primarily derived from LDL and VLDL and not de novo synthesis, while biliary cholesterol was derived mainly from HDL, suggesting cholesterol pool compartmentalization within the liver (16). Our findings that increased LDL uptake led to higher bile acid production are therefore not surprising but consistent with this view.
Atorvastatin increased excreted cholesterol in wild-type and in PCSK9-Y119X mice, while PCSK9 suppression alone had no effect versus wild-type mice. Atorvastatin had significant effects on fecal bile acid concentrations in both normal and PCSK9 knockout mice, while PCSK9 genetic suppression alone had only modest effects versus wild-type mice. LDL-derived cholesterol in liver appeared to be efficiently converted and excreted as bile acids as well as being excreted as free cholesterol. Female PCSK9-Y119X mice exhibited greater increases in excreted cholesterol and bile acids than males following atorvastatin. Baseline fecal total bile acid concentrations in wild-type mice in our study were similar to previously reported values for normal mice (17). Importantly, liver concentrations of free and esterified cholesterol and total bile acids were essentially identical in all four groups, indicating that hepatic homeostasis was maintained for PCSK9 suppression either without or with maximal HMG-CoA reductase inhibition. The observed metabolic changes were associated with significant upregulation of key bile acid synthesis pathway gene expression in liver. However, both strains of mice expressed similar baseline levels of these enzymes, and each increased to similar extent with statin treatment. Together these findings suggest that in the absence of PCSK9, the intracellular cholesterol pool or flux providing substrate to CYP7A is greater, or is more bioavailable or in closer kinetic proximity to the enzyme.
The overall pathway of bile acid synthesis includes up to 17 enzymes in liver (18). It has not been resolved whether the intracellular pool of cholesterol substrate for the initial ring hydroxylation step mediated by the CYP7A enzyme is preferentially derived from LDL endocytosis and hydrolysis, or endogenous synthesis, or both. Intracellular trafficking of de novo synthesized cholesterol from the endoplasmic reticulum to the plasma membrane appears to be distinct from movement of LDLR-endocytosed cholesterol which is routed through the lysosomes and ultimately to intracellular membranes (19). Metabolic regulation through changes in expression levels and kinetic activity of key enzymes enable the liver to accommodate the increased influx of cholesterol following the stimulated LDLR-dependent cholesterol uptake seen with the treatment. In our study, the findings of increased mRNA expression of the important liver bile acid synthesis gatekeeper genes, CYP7A1 and CYP8B1 (20), along with higher fecal bile acid levels, and lack of liver cholesterol or bile acid accumulation suggests that the conversion to bile acids in liver enabled liver cholesterol homeostasis in the combination treatment. The covariation of CYP7A1 and CYP8B1 expression also is consistent with the observed increases in both cholic acid and chenodeoxycholic acid. In other studies in mice the oxysterol nuclear receptor, liver X receptor (LXR)α, was found to be the key driver of bile acid synthesis gene transcriptional activation (21). However, in our study mRNA for the sensitive LXR response gene SREBP1 (22, 23) was unchanged by atorvastatin and the same in all groups, suggesting that the increased expression of CYP7A1 and CYP8B1 may not be LXR-mediated in this study. While oxysterols activate CYP7A1 and other genes through LXR, bile acids repress CYP7A1 through the farnesoid X receptor (FXR), forming a feedback control loop in liver. Other FXR response genes were not measured in our study, thus it is presently unclear whether this feedback system contributes to the CYP7A1 and CYP8B1 expression pattern observed. It is also possible that the bile acid response elements in the promoters of CYP7A1 and CYP8B1 may respond to other transcription factors including hepatocyte nuclear factor 4 (HNF-4) which were not assessed in this work (24).
While plasma LDL-C and total cholesterol were affected we observed no differences in circulating total bile acids or plasma ALT and AST in any group, suggesting that hepatocyte integrity and normal metabolic functions were maintained. Bile acids synthesized in the liver are secreted into the small intestine where they facilitate absorption of fat-soluble nutrients. Normally most (over 90%) of bile acids are efficiently reabsorbed from the intestine and returned to the liver via the portal vein, forming an enterohepatic circuit, and a minor fraction is excreted in the feces (25). Bile acids are transported out of hepatocytes at the canalicular membrane through the activity of the ATP-dependent BSEP and across the liver biliary epithelium mediated by members of the ASBT SLC10 gene family (25, 26). Changes in expression or activity can affect export or extraction efficiency and affect circulating total bile acid concentrations, and the latter has been considered a biomarker of altered hepatic function or potential metabolic or structural problems such as cholestasis. The lack of changes in liver expression of BSEP or an ASBT family member in our study suggests that normal liver export of bile acids was maintained throughout the study. Furthermore, expression of the NTCP, and the sodium independent organic anion transporter (OATP1B2) were the same in all groups, suggesting that reuptake of bile acids into liver was not impaired. These results along with the constancy of liver cholesterol and total bile acid concentrations support the interpretation that efficient enterohepatic circulation of bile acids was maintained throughout the study.
Because species differences exist in the regulation of bile acid metabolism, it is presently not clear how well these pharmacodynamic results in a mouse model predict human responses. Still, the data suggest that metabolic regulation through enzyme pathway responses to increases in substrate concentration, together with increased gene expression for the key regulatory enzymes in liver, accommodate the increased flux of cholesterol into liver following stimulated LDL-C clearance with combined statin/PCSK9 suppression in mice. The findings suggest that the liver handles its responsibilities very well even when faced with the profound LDL lowering action of the combined PCSK9-statin mechanism.
Acknowledgments
We thank George Psaltis for breeding, Jinwen Huang for genotyping, and Daniel Meyers, Hossain Monshizadegan, and Richard Yang for phenotyping the PCSK9-Y119X mouse model (Bristol-Myers Squibb Research and Development).
Footnotes
Abbreviations:
- ALT
- alanine aminotransferase
- ASBT
- apical sodium bile acid transporter
- AST
- aspartate aminotransferase
- BSEP
- bile salt export pump
- ENU
- N-ethyl N-nitrosourea
- FXR
- farnesoid X receptor
- LDL-C
- low density lipoprotein cholesterol
- LDLR
- LDL receptor
- LXR
- liver X receptor
- NTCP
- sodium taurocholate cotransporting polypeptide
- PCSK9
- proprotein convertase subtilisin-kexin-9
- SREBP
- sterol regulatory binding protein
Studies were funded by Bristol-Myers Squibb R & D.
REFERENCES
- 1.Abifadel M., Varret M., Rabes J. P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D. 2003. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 34: 154–156. [DOI] [PubMed] [Google Scholar]
- 2.Cohen J. C., Boerwinkle E., Mosley T. H., Jr, Hobbs H. H. 2006. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354: 1264–1272. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Z., Tuakli-Wosornu Y., Lagace T. A., Kinch L., Grishin N. V., Horton J. D., Cohen J. C., Hobbs H. H. 2006. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am. J. Hum. Genet. 79: 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan J. C., Piper D. E., Cao Q., Liu D., King C., Wang W., Tang J., Liu Q., Higbee J., Xia Z., et al. 2009. A PCSK9 neutralizing antibody reduces serum cholesterol in mice and nonhuman primates. Proc. Natl. Acad. Sci. USA. 106: 9820–9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rashid S., Curtis D. E., Garuti R., Anderson N. N., Bashmakov Y., Ho Y. K., Hammer R. E., Moon Y. A., Horton J. D. 2005. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking PCSK9. Proc. Natl. Acad. Sci. USA. 102: 5374–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denis M., Marcinkiewicz J., Zaid A., Gauthier D., Poirier S., Lazure C., Seidah N. G., Prat A. 2012. Gene inactivation of PCSK9 reduces atherosclerosis in mice. Circulation. 125: 894–901. [DOI] [PubMed] [Google Scholar]
- 7.Maxwell K. N., Fisher E. A., Breslow J. L. 2005. Overexpression of PCSK9 accelerates THE degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc. Natl. Acad. Sci. USA. 102: 2069–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seidah N. G., Prat A. 2012. The biology and therapeutic targeting of the proprotein convertases. Nat. Rev. Drug Discov. 11: 367–383. [DOI] [PubMed] [Google Scholar]
- 9.Dias C. S., Shaywitz A. J., Wasserman S. M., Smith B. P., Gao B., Stolman D. S., Crispino C. P., Smirnakis K. V., Emery M. G., Colbert A., et al. 2012. Effects of AMG 145 on low-density lipoprotein cholesterol levels. J. Am. Coll. Cardiol. 60: 1888–1898. [DOI] [PubMed] [Google Scholar]
- 10.Stein E. A., Mellis S., Yancopoulos G. D., Stahl N., Logan D., Smith W. B., Lisbon E., Gutierrez M., Webb C., Wu R., et al. 2012. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N. Engl. J. Med. 366: 1108–1118. [DOI] [PubMed] [Google Scholar]
- 11.Cordes S. P. N-ethyl-N-nitrosourea mutagenesis: boarding the mouse mutant express. 2005. Microbiol. Mol. Biol. Rev. 69: 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gälman C., Ostlund-Lindqvist A. M., Björquist A., Schreyer S., Svensson L., Angelin B., Rudling M. 2003. Pharmacological interference with intestinal bile acid transport reduces plasma cholesterol in LDL receptor/apoE deficiency. FASEB J. 17: 265–267. [DOI] [PubMed] [Google Scholar]
- 13.Alnouti Y., Csanaky I., Klaassen C. 2008. Quantitative profiling of bile acids and their conjugates in mouse liver, bile, plasma, and urine using LC-MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 873: 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osono Y., Woollett L. A., Herz J., Dietschy J. M. 1995. Role of the low density lipoprotein receptor in the flux of cholesterol through the plasma and across the tissues of the mouse. J. Clin. Invest. 95: 1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72: 137–174. [DOI] [PubMed] [Google Scholar]
- 16.Carey M. C. 1997. Homing-in on the origin of biliary steroids. Gut. 41: 721–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchida K., Takase H., Nomura Y., Takeda K., Takeuchi N., Ishikaw Y. 1984. Changes in biliary and fecal bile acids in mice after treatments with diosgenin and β-sitosterol. J. Lipid Res. 25: 236–245. [PubMed] [Google Scholar]
- 18.Hofmann A. F., Hagey L. R. 2008. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell. Mol. Life Sci. 65: 2461–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liscum L., Dahl N. 1992. Intracellular cholesterol transport. J. Lipid Res. 33: 1239–1254. [PubMed] [Google Scholar]
- 20.Jelinek D. F., Andersson S., Slaughter C. A., Russell D. W. 1990. Cloning and regulation of cholesterol 7α-hydroxylase, the rate-limiting enzyme in bile acid synthesis. J. Biol. Chem. 265: 8190–8197. [PMC free article] [PubMed] [Google Scholar]
- 21.Peet D. J., Turley S. D., Ma W., Janowski B. A., Lobaccaro J. M., Hammer R. E., Mangelsdorf D. J. 1998. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXRα. Cell. 93: 693–704. [DOI] [PubMed] [Google Scholar]
- 22.Yoshikawa T., Shimano H., Amemiya-Kudo M., Yahagi N., Hasty A. 2001. Identification of LXR-RXR as an activator of the sterol regulatory element binding protein 1c gene promoter. Mol. Cell. Biol. 21: 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D. J., et al. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14: 2831–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crestani M., Sadeghpour A., Stroup D., Galli G., Chiang J. Y. 1998. Transcriptional activation of the cholesterol 7α-hydroxylase gene (CYP7A) by nuclear hormone receptors. J. Lipid Res. 39: 2192–2200. [PubMed] [Google Scholar]
- 25.Dawson P. A., Lan T., Rao A. 2009. Thematic review series, bile acids: bile acid transporters. J. Lipid Res. 50: 2340–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Claro da Silva T., Polli J. E., Swaan P. W. 2013. The solute carrier family 10 (SLC10): Beyond bile acid transport. Mol. Aspects Med. 34: 252–269. [DOI] [PMC free article] [PubMed] [Google Scholar]