Abstract
Scavenger receptor class B type I (SR-BI) is a multi-ligand receptor that binds a variety of lipoproteins, including high density lipoprotein (HDL) and low density lipoprotein (LDL), but lipoprotein(a) [Lp(a)] has not been investigated as a possible ligand. Stable cell lines (HEK293 and HeLa) expressing human SR-BI were incubated with protein- or lipid-labeled Lp(a) to investigate SR-BI-dependent Lp(a) cell association. SR-BI expression enhanced the association of both 125I- and Alexa Fluor-labeled protein from Lp(a). By confocal microscopy, SR-BI was also found to promote the internalization of fluorescent lipids (BODIPY-cholesteryl ester (CE)- and DiI-labeled) from Lp(a), and by immunocytochemistry the cellular internalization of apolipoprotein(a) and apolipoprotein B. When dual-labeled (3H-cholesteryl ether,125I-protein) Lp(a) was added to cells expressing SR-BI, there was a greater relative increase in lipid uptake over protein, indicating that SR-BI mediates selective lipid uptake from Lp(a). Compared with C57BL/6 control mice, transgenic mice overexpressing human SR-BI in liver were found to have increased plasma clearance of 3H-CE-Lp(a), whereas mouse scavenger receptor class B type I knockout (Sr-b1-KO) mice had decreased plasma clearance (fractional catabolic rate: 0.63 ± 0.08/day, 1.64 ± 0.62/day, and 4.64 ± 0.40/day for Sr-b1-KO, C57BL/6, and human scavenger receptor class B type I transgenic mice, respectively). We conclude that Lp(a) is a novel ligand for SR-BI and that SR-BI mediates selective uptake of Lp(a)-associated lipids.
Keywords: lipoprotein receptors, atherosclerosis, apolipoprotein(a), oxidized lipids, selective uptake
Scavenger receptor class B type I (SR-BI) is a multi-ligand receptor. It binds a variety of oxidized as well as native lipoproteins, including high density lipoprotein (HDL) and low density lipoprotein (LDL) (1–4). SR-BI preferentially mediates the cellular uptake of neutral lipids over protein from lipoproteins by a process termed selective uptake. In the liver, the selective uptake of cholesteryl esters (CEs) from HDL potentiates the reverse cholesterol transport pathway, by increasing the hepatic excretion of cholesterol (5).
Lipoprotein(a) [Lp(a)] is a pro-atherogenic lipoprotein particle that contains one copy of apolipoprotein(a) [apo(a)] covalently linked to apoB-100 by a disulfide bond. It is found in some primates but is not present in most other mammalian species (6). Variation in the plasma level of Lp(a) is largely under genetic control, with polymorphisms in the kringle 4 repeat of apo(a) accounting for 40% of the variation (7). In general, apo(a) size is inversely associated with plasma Lp(a) level, but the relationship between apo(a) size and Lp(a) concentration varies among ethnic groups (8). In addition, a pentanucleotide repeat polymorphism in the 5′ control region of apo(a) accounts for 10–14% of the variation in Lp(a) concentrations in Caucasians (9).
Human tracer studies have shown that plasma concentrations of Lp(a) were not only positively correlated with the secretion rates of Lp(a) protein, but also negatively correlated with the fractional catabolic rates (FCRs) of Lp(a) proteins, both apo(a) and apoB-100, indicating that plasma Lp(a) levels are also regulated by its rate of catabolism (10). However, the receptors that bind and mediate the catabolism of Lp(a) are not well characterized. Although Lp(a) is structurally and compositionally similar to LDL, it is known to be a relatively poor ligand for the LDL receptor (11), as well as for the lipoprotein receptor-related protein (12). Lp(a) can bind to the VLDL receptor, but mice defective in this receptor showed only a modest delay in the catabolism of heterologous Lp(a) (13), suggesting the presence of other receptor(s). Because of the close structural similarity of Lp(a) with LDL, a known ligand for SR-BI (1–4), we investigated in this study whether SR-BI can also serve as a receptor for Lp(a).
MATERIALS AND METHODS
Generation of stable cell lines
Human SR-BI cDNA, generated by RT-PCR from total RNA of human monocytes, was inserted into a pcDNA3 vector (Invitrogen, Carlsbad, CA) and sequence confirmed. HEK293 and HeLa cells were purchased from American Type Culture Collection. Cells were cultured in DMEM growth medium (DMEM, 4 mM glutamine, 10% fetal bovine serum, 100 u/ml penicillin, and 100 μg/ml streptomycin). The SR-BI cDNA-containing plasmid was transfected into 80% confluent HEK293 and HeLa cells (14) using Effectene transfection reagent (Qiagen, Valencia, CA). Cell clones were selected in DMEM growth medium containing 2 mg/ml G418 for 2 weeks and maintained with 0.25 mg/ml G418. To verify SR-BI expression, cells before or after transfection were lysed and analyzed by Western blotting with rabbit anti-human polyclonal SR-BI or rabbit anti-human polyclonal VLDL receptor antibodies (Abcam, Cambridge, UK). Western blotting was performed as previously described (15).
Preparation of radiolabeled and fluorescently labeled Lp(a)
Lp(a) was purchased from Meridian Life Science, Inc. (Saco, MA). Each lot of Lp(a) was prepared from a single donor with documented high Lp(a) concentration to enhance yield and purity. Beginning with plasmapheresis, plasma was drawn directly into a Trasylol (aprotinin)-containing receiver, giving 100 KIU aprotinin/ml plasma. Immediately thereafter, sodium azide (0.01%) and EDTA (0.001%) were added and the Lp(a) was isolated by ultracentrifugation at 4°C followed by Lysine-Sepharose chromatography (16), with elution by aminocaproic acid to complete the purification. The purified Lp(a) was analyzed by SDS-PAGE without reducing agents for the appearance of a single band, indicating the absence of other proteins and lipoproteins. Lp(a) was sent the day of preparation, stored in 15 mM NaCl and 1 mM EDTA, pH 7.4, and used within 10 days to minimize oxidation and aggregation. The purity of Lp(a) preparations was confirmed by fast protein liquid chromatography in our laboratory. The protein molecular mass of the Lp(a) used in this study was approximately 1,000 kDa [apo(a) predominantly 550 kDa and apoB 500 kDa], and the protein concentration was 0.6 mg/ml.
The Chloramine-T method was used for 125I iodination of Lp(a) protein. A labeling kit, including carrier-free 125I, was purchased from MP Biomedicals LLC (Solon, OH). Labeled lipoproteins were purified by a desalting column (PD-10 column, GE Healthcare, Buckinghamshire, UK). The integrity and purity of all labeled lipoproteins was confirmed by fast protein liquid chromatography. The 125I-labeled protein-specific activities ranged between 400 and 456 cpm/ng protein. To fluorescently label Lp(a) proteins, an Alexa Fluor® 488 protein labeling kit (Invitrogen) was used according to the manufacturer's instructions.
[3H]cholesteryl ether (CEth) is a nonhydrolyzable analog of CE (17). [3H]CEth-labeled Lp(a), used in cell culture studies, was prepared as previously described (16). Briefly, 25 ml human donor serum was adjusted to a density of 1.215 g/ml with potassium bromide (Sigma-Aldrich, St. Louis, MO), and ultracentrifuged at 40,000 rpm (Beckman L8-55 ultracentrifuge) at 8°C for 48 h to obtain a lipoprotein-free serum fraction containing cholesteryl ester transfer protein (CETP) (1.215 g/ml bottom fraction). Cholesteryl hexadecyl ether [cholesteryl-1,2-3H(N)-] (200–250 μCi) (Perkin Elmer, Waltham, MA) was mixed with 22 nmol butylated hydroxytoluene (Sigma), and evaporated under nitrogen in a glass tube. After addition of 1 ml TSE buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, and 2 mM EDTA) and 80 μl egg l-phosphatidylcholine (100 mg/ml, Sigma), the 3H-lipid mixture was sonicated. Lp(a) (600 μg) was added to 3 ml of the d > 1.215 g/ml bottom fraction containing CETP. One milliliter of the 3H-lipid mixture was added, and then incubated at 37°C overnight under nitrogen. The labeled Lp(a) was then dialyzed in TSE buffer, adjusted to a density of 1.050 g/ml, and centrifuged (Beckman Optima TL ultra-centrifuge) at 85,000 rpm at 4°C for 12 h to separate any unincorporated lipid tracer. The bottom fraction was dialyzed and readjusted to a density of 1.125 g/ml and centrifuged for 24 h. The upper fraction, which contained the final 3H-Lp(a) preparation, was dialyzed with PBS buffer. The specific activities of the radiolabeled Lp(a) preparations were approximately 200–265 dpm/ng Lp(a) protein. For dual-labeled Lp(a), Lp(a) was first labeled with [3H]CEth followed by 125I labeling.
[3H]CE-labeled Lp(a), used for kinetic studies in mice (Fig. 7), was generated as previously described (18). Two hundred and fifty microcuries of 3H-cholesteryl oleate {cholesteryl-[1,2,6,7- 3H (N)] oleate} (60 Ci/mmol; American Radiolabeled Chemicals, Inc., St. Louis, MO) was dried under nitrogen, resuspended in 10 μl of absolute ethanol, and added dropwise over 3 min to the lipoprotein solution (0.6 mg/ml protein concentration) with shaking and vortexing as previously described (18). [3H]CE-Lp(a) was then reisolated by ultracentrifugation, dialyzed overnight against 1× PBS, 0.01% EDTA, and finally analyzed by agarose gel electrophoresis to ensure integrity of the particle.
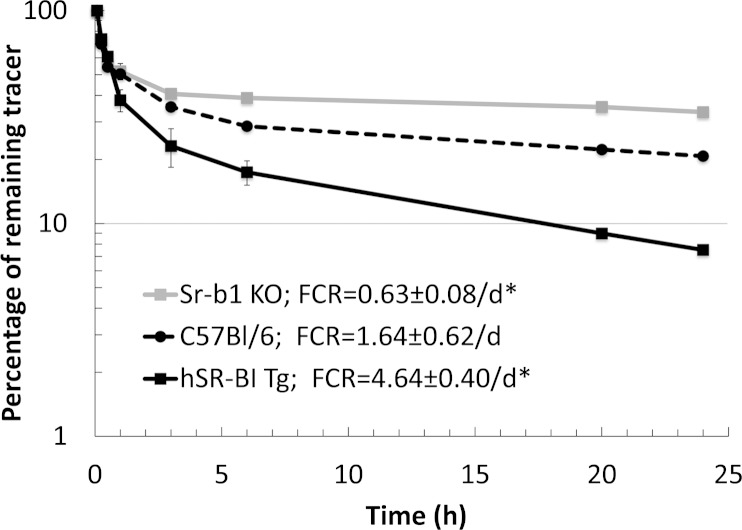
Fig. 7.
SR-BI mediates plasma clearance of Lp(a). [3H]CE-Lp(a) was injected into the plasma of Sr-b1-KO mice (n = 3), hSR-BI-Tg mice (n = 3), and C57BL/6 mice (n = 3) and remaining counts of the tracer in the plasma compartment were monitored at the indicated time points. Fifty microliters of blood were collected in each time point, plasma (25 ul) was separated and counted for 5 min. Plasma decay curves and FCR were determined as described in Materials and Methods. *P < 0.01 compared with C57BL/6 mice. Results are expressed as the mean ± SD of triplicates.
HDL (d = 1.063 to 1.21) and LDL (d = 1.019 to 1.063) were isolated by density gradient ultracentrifugation, as previously described (19) and used within 10 days of preparation.
Cell association studies
To quantitate cellular Lp(a) cell association using [125I]Lp(a), HEK293 cells grown to 95% confluence in 24-well plates were incubated with the indicated concentration of [125I]Lp(a) for 1.5 h either in the absence of unlabeled Lp(a) (for calculating total cell association) or in the presence of a 40-fold excess of unlabeled Lp(a) (for calculating nonspecific cell association) at 37°C. Cells were washed four times with PBS, and harvested with 0.01 N NaOH and 0.1% SDS solution. Specific cell association was calculated by subtracting nonspecific cell association from total cell association value and expressed as ng Lp(a)/mg cellular protein.
To observe cellular Lp(a) cell association by flow cytometry, HEK293 cells were incubated with 10 μg/ml (10 nM) Alexa Fluor® 488-labeled Lp(a) in DMEM containing 2 mg/ml BSA and 20 mM HEPES at 37°C for 1.5 h. Cells were washed with PBS, detached with a cell dissociation solution (Mediatech, Herndon, VA), fixed with 4% paraformaldehyde in PBS, and analyzed by a fluorescence-activated cell sorter (FACS, model A; Hitachi).
To observe cellular Lp(a) cell association by confocal microscopy, HeLa cells on coverslips were incubated with 5 μg/ml Alexa Fluor® 488-labeled Lp(a) in DMEM containing 2 mg/ml BSA and 20 mM HEPES at 37°C for 1 h, followed by a chase without labeled Lp(a) for 1 h, then fixed and examined by confocal microscopy.
For the competition assay, unlabeled HDL and LDL were prepared by density gradient ultracentrifugation using fresh human donor plasma, and used within 10 days of preparation. To convert the protein mass of lipoproteins to moles, protein concentrations (in mg/ml) of purified LDL, Lp(a), and HDL were first determined by the BCA Protein Assay Kit (Pierce), using BSA as the standard. Molar concentrations were determined using estimated average protein molecular mass of approximately 500 kDa for LDL (one 500 kDa apoB/particle), approximately 1,000 kDa for Lp(a) [one 500 kDa apoB + one 550 kDa apo(a)/particle], and 200 kDa for HDL [400 kDa average molecular weight, 50% average protein content (20)], respectively.
For fluorescent competition studies, HEK293 cells expressing SR-BI were incubated with 10 nM Alex Fluor 488-labeled Lp(a) in the presence or absence of 40-fold excess unlabeled competitors [Lp(a), HDL, or LDL] for 1.5 h, then followed by the flow cytometry assay.
For 3H-Lp(a) competition studies, 3H-CEth-Lp(a) plus competitors [Lp(a), HDL, or LDL] at the indicated concentrations were added to the cell culture medium [DMEM with 5% lipoprotein-deficient serum (LPDS) (Sigma)] and incubated with the cells for 1.5 h. Cells were washed five times with PBS, and cell lysates, as prepared above, were used to measure protein and lipid uptake.
Fluorescent lipid labeling and uptake studies
Fluorescent CE-labeled Lp(a) [BODIPY-CE-Lp(a)] was prepared as previously described (21). Briefly, 0.4 mg of BODIPY-cholesteryl FLC12 (Invitrogen) was mixed with 0.2 mg of phosphatidylcholine (100 mg/ml, Sigma) and 22 nmol butylated hydroxytoluene, and dried under nitrogen. After sonication in 1 ml of TBS, the mixture of fluorescent lipids was added to 200 μg of Lp(a) in 2 ml of 1.215 g/ml bottom fraction containing CETP with ampicillin (final volume 4 ml), and gently shaken for 12 h at 37°C. The labeled BODIPY-CE-Lp(a) fraction (density 1.050–1.125 g/ml) was collected by sequential ultracentrifugation of the sample at 85,000 rpm for 12 and 24 h respectively, and dialyzed against PBS buffer.
DiI-labeled lipoprotein was prepared as previously described (22). Briefly, 200 μg of Lp(a) was mixed with 0.3 mg DiI [DiIC18 (3), Invitrogen] (final concentration 100 μg/ml), 2 ml of LPDS (Sigma), and 100 μg/ml ampicillin, and gently shaken at 37°C overnight. The labeled Lp(a) was adjusted to a density of 1.125 g/ml with potassium bromide (Sigma), and centrifuged at 85,000 rpm (Beckman Optima TL ultracentrifuge) for 24 h to remove unincorporated dye. The supernatant was collected and dialyzed against PBS buffer.
To observe cellular uptake of fluorescent lipids, HeLa cells transfected with vector without (Mock) or with SR-BI cDNA were seeded onto glass coverslips coated with 0.1 mg/ml poly-l-lysine in 6-well plates. After culture in DMEM with 5% LPDS for 24 h, the cells were replaced with fresh medium with 10 μg/ml fluorescent-labeled Lp(a) at 37°C for 1 h or 3 h. The medium was discarded, and the coverslips were washed five times with PBS buffer and fixed with 4% paraformaldehyde. After mounting and sealing, the coverslips were examined under a Zeiss 510 confocal microscope.
Immunohistochemistry
Sheep anti-human polyclonal apo(a) antibody was purchased from Meridian Life Science Inc., and goat anti-human apoB polyclonal antibody from R and D Systems (Minneapolis, MN). The antibodies for immunostaining were labeled with either the Alexa Fluor 488 or 546 monoclonal antibody labeling kits (Molecular Probes, Eugene, OR). For direct immunostaining, HeLa cells were cultured in DMEM medium with 5% LPDS serum for 24 h; the media was then replaced with fresh DMEM (containing 2 mg/ml BSA and 20 mM HEPES) with Lp(a) (final concentration 15 μg/ml) and cells were incubated at 37°C for 2 h. After washing six times, the cells were fixed with 4% paraformaldehyde for 10 min, and permeabilized with methanol at −20°C for 10 min. Immunostaining was then performed with the fluorescent-labeled antibodies. For indirect immunostaining, the cells were incubated with 15 μg/ml Lp(a) at 4°C for 1 h, washed, and then incubated at 37°C for 2 h. The cells were fixed, permeabilized, blocked, and incubated with primary antibodies for 2 h, followed by detection with Northern Lights™ donkey anti-sheep IgG-NL637 and anti-goat IgG-NL493 (R & D Systems).
Protein degradation studies
Protein degradation was monitored by determining the release of the 125I-labeled protein degradation product (125I-monoiodotyrosine containing peptides) into the conditioned cell culture medium, as previously described (23). Briefly, 0.25 ml of 50% trichloroacetic acid was added to 1 ml of conditioned medium in a glass tube; the mixture was incubated at 4°C for 30 min and then centrifuged to precipitate undegraded 125I-lipoprotein. Supernatant (0.5 ml) was then mixed with 5 μl of 40% potassium iodide, followed by addition of 20 μl of 30% H2O2. Chloroform (1 ml) was added and vortexed to extract free 125I-iodine. The upper aqueous layer was removed and 0.2 ml was γ-counted to measure the release of 125I-labeled peptides. Protein degradation in cell lysates was similarly measured.
Generation of human SR-BI transgenic mice
The liver-specific expression vector pLIV.11 (24) was used to develop mice expressing human SR-BI (hSR-BI). The full-length (1.7 kb) human SR-BI cDNA (GenBank: BC112037.1) was flanked by Not I linkers and inserted into the unique Not I site of pLIV.11 and correctly oriented clones were identified after digestion with Sph I and Aat II. The plasmid, pLIV11-hSR-BI, was then digested with Sal I and Spe I, and a 11.6 kb DNA fragment containing the complete hSR-BI expression cassette was isolated from a 0.8% agarose gel and purified by CsCl density gradient ultracentrifugation (Beckman TL-100 tabletop ultracentrifuge, 95,000 rpm, 24 h, 20°C). After dialysis against 10 mM Tris-HCl pH 7.4 and 0.1 mM EDTA, the DNA fragment was microinjected into pronuclei of fertilized eggs from C57BL/6J females (Jackson Laboratory, Bar Harbor, ME). Genotyping and expression analysis of the transgenic mice was performed as previously described (25). A founder line of mice containing approximately 25 copies of the human SR-BI gene per heterozygous genome was used to establish a colony of human SR-BI transgenic (hSR-BI-Tg) mice. Similar to previous reports (4), the hSR-BI-Tg mice had decreased plasma levels of HDL cholesterol compared with control mice (HDL cholesterol: hSR-BI-Tg <3.0 mg/dl vs. C57BL/6 50.2 ± 1.5 mg/dl, n = 21, P < 0.001).
In vivo tracer kinetics of 3H-CE-Lp(a)
[3H]CE-Lp(a) (total of 1 × 106 cpm) was injected into the retro-orbital sinus of C57BL/6 mice (n = 3), mouse scavenger receptor class B type I knockout (Sr-b1-KO) mice (n = 3), and hSR-BI-Tg mice (n = 3). Approximately 50 μl of blood was collected at the indicated time points and plasma (25 μl) was then separated and counted for 5 min in a Tri-Carb 2500 TR liquid scintillation counter (Packard Instrument Co., Downers Grove, IL). Plasma decay curves for [3H]CE-Lp(a) were generated by dividing the plasma radioactivity at each time point by the plasma radioactivity at the initial 1 min time point. This initial time point did not differ among the study groups (P > 0.4). The FCR was determined from the area under the plasma radioactivity curves, using a multi-exponential curve fitting technique on the SAAM II program.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA). Unless otherwise indicated all results are expressed as the mean ± 1 SD of at least triplicates, and the results shown represent two or more independent experiments. A two-tailed Student's t-test was used for statistical analysis, and P < 0.05 was considered significant.
RESULTS
Stable cell lines expressing SR-BI
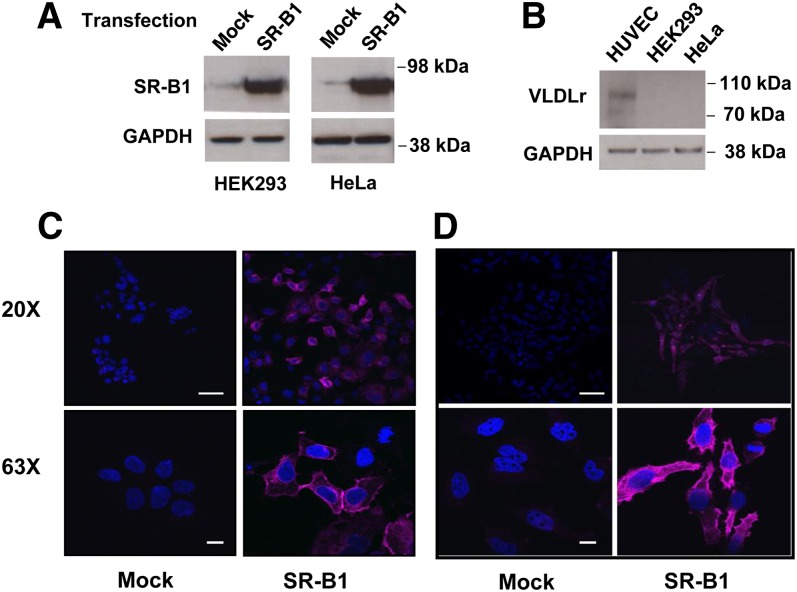
By Western blotting, HEK293 and HeLa cells do not express significant amounts of endogenous SR-BI (Fig. 1A) or VLDL receptor (Fig. 1B), as previously reported (26). After transfection with human SR-BI cDNA, both cell lines expressed significantly increased amounts of SR-BI protein (Fig. 1A). Similarly, SR-BI was not detected by confocal microscopy in mock-transfected HEK293 or HeLa cells (Fig. 1C, D) after staining with a rabbit anti-human SR-BI polyclonal antibody. In contrast, stably transfected HEK293 and HeLa cells showed abundant staining for SR-BI, which was primarily present on the cell surface, but also in intracellular vesicles, particularly in the case of HeLa cells.
Fig. 1.
Expression of SR-BI in transfected HEK293 cells and HeLa cells. A: HEK293 or HeLa cells before (mock) or after transfection (SR-BI) with SR-BI were analyzed by immunoblots for SR-BI or GAPDH. B: Lysates from human umbilical vein endothelial cell (HUVEC), HEK293, and HeLa cells were analyzed by immunoblots for VLDL receptor or GAPDH. Twenty micrograms of protein were loaded per lane in all immunoblots. C, D: HEK293 cells (C) or HeLa cells (D) before (mock) or after transfection (SR-BI) were stained with a primary human SR-BI antibody and analyzed by confocal microscopy (upper panel 20× image, lower panel 63× image; scale bars are 50 um for 20× and 10 um for 63×). Cells were counterstained with DAPI.
SR-BI mediates the association of Lp(a) with cells
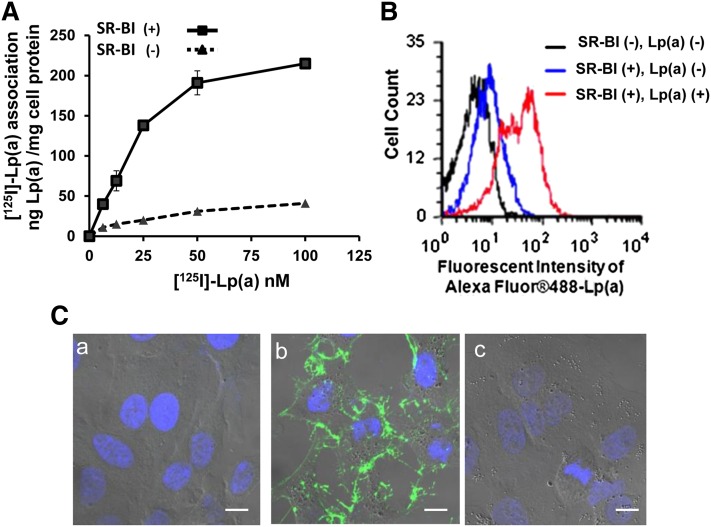
Compared with control HEK293 cells, a HEK293 cell clone stably expressing human SR-BI showed a 3- to 4-fold increase in its association with Lp(a), radiolabeled in its protein moiety by 125I (Fig. 2A). Similarly, by flow cytometry, stably transfected HEK293 cells showed an approximate 5-fold increased association with Alexa-fluorescent-labeled Lp(a) protein compared with control cells (Fig. 2B).
Fig. 2.
Enhanced Lp(a) association with SR-BI-transfected cells. A: HEK293 cells stably transfected with (solid line) or without (dashed line) SR-BI were incubated with increasing concentrations of [125I]Lp(a) at 37°C for 1.5 h. Specific cell association of [125I]Lp(a) (shown) is expressed as ng Lp(a)/mg cellular protein. Specific cell association was determined by subtracting nonspecific cell association {residual [125I]Lp(a) cell association in the presence of a 40-fold excess of unlabeled Lp(a)} from total cell association. B: HEK293 cells stably transfected with SR-BI were incubated with [SR-BI (+), Lp(a) (+)] or without [SR-BI (+), Lp(a) (−)] 10 ug/ml Alexa Fluor 488-labeled Lp(a) for 1.5 h and analyzed by flow cytometry. SR-BI (−) cells without Lp(a) [shown as SR-BI (−), Lp(a) (−)] were included as a control to define background staining. C: HeLa cells transfected with empty vector (a), SR-BI (b), or LDL receptor (c) were incubated with 5 μg/ml Alexa Fluor 488 Lp(a) at 37°C for 1 h followed by Lp(a)-free chase for 1 h, fixed, and examined by microscopy. Confocal photographs with differential interference contrast (DIC) are shown (63×). Scale bars are 10 um for 63× magnification. Cells were counterstained with DAPI.
Stably transfected HeLa cells expressing SR-BI, which were better than HEK293 cells for visualization by confocal microscopy, also associated with increased amounts of Lp(a) that was fluorescently labeled in its protein moiety by Alexa Fluor 488 (Fig. 2C). Cell-bound Lp(a) (green) could be detected in SR-BI transfected HeLa cells (Fig. 2C, panel b) but not in mock-transfected HeLa cells (Fig. 2C, panel a) or in HeLa cells transfected with the human LDL receptor (Fig. 2C, panel c).
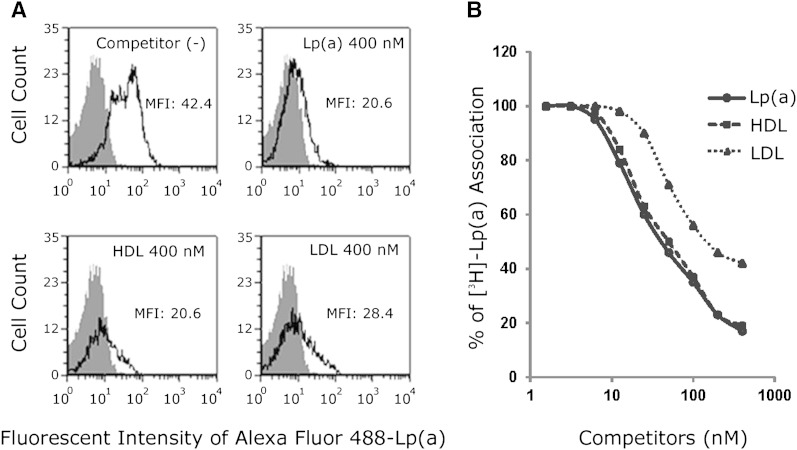
A 40-fold molar excess of unlabeled Lp(a), HDL, or LDL almost completely inhibited the association of fluorescent-labeled Lp(a) protein with HEK293 cells transfected with SR-BI (Fig. 3A). Similar results were obtained when unlabeled Lp(a), HDL, or LDL were used to compete for association of Lp(a) labeled with 3H-CEth, a nonhydrolyzable analog of CE (Fig. 3B), although LDL was found to be slightly less effective than the other lipoproteins tested.
Fig. 3.
Unlabeled Lp(a), HDL, and LDL compete with fluorescent- or 3H-CEth-labeled Lp(a) cell association. A: HEK293 cells expressing SR-BI were incubated with 10 nM Alexa Fluor 488-labeled Lp(a) in the presence or absence of 40-fold excess unlabeled competitors for 1.5 h, and analyzed by flow cytometry. Gray-shaded area shows cells incubated in the absence of fluorescent-labeled Lp(a). MFI, mean fluorescent intensity. B: HEK293 cells transfected with SR-BI were incubated with 10 μg/ml (10 nM) [3H]CEth-Lp(a) at 37°C in the presence of the indicated concentration (nM) of competitors for 1.5 h, and cell lysates were counted for radioactivity.
SR-BI mediates the uptake of lipid and protein components of Lp(a)
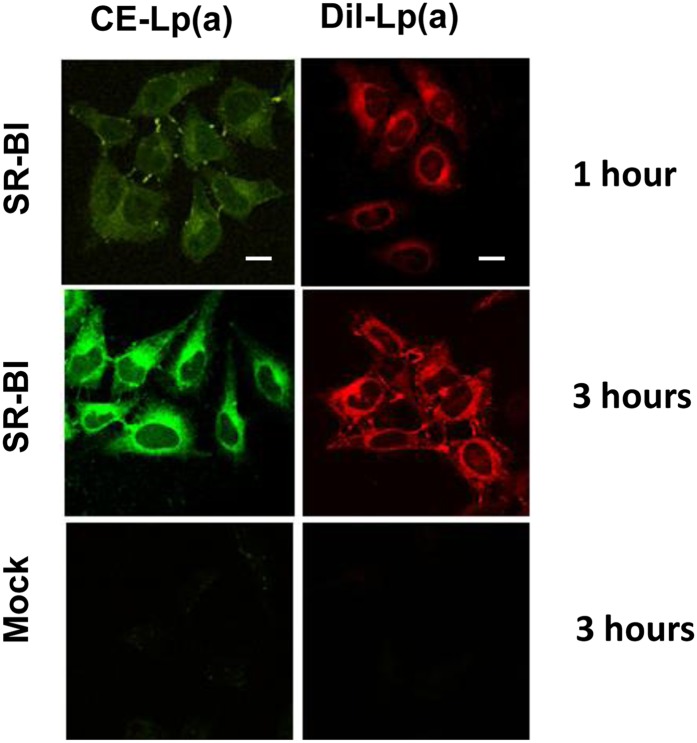
The uptake of the various lipid and protein components of Lp(a) by SR-BI-expressing cells was further investigated by confocal microscopy. After incubating SR-BI-transfected HeLa cells with Lp(a) labeled with either BODIPY-CE (green) (a marker for a neutral core lipid) or DiI [DiIC18 (3)] (red) (a marker for a surface amphipathic lipid), a time-dependent uptake of both fluorescent lipids was observed (Fig. 4, top panels). After 1–3 h of incubation, fluorescent lipids were primarily observed in intracellular vesicles around the Golgi body and nucleus (Fig. 4, top and middle panels). In contrast, no significant lipid uptake was observed in control cells after 3 h (Fig. 4, lower panels).
Fig. 4.
SR-BI promotes uptake of fluorescent lipids from Lp(a). HeLa cells stably transfected with SR-BI (top and middle panels) or without SR-BI (bottom panels) were incubated with BODIPY-CE (green) or DiI [DiIC18 (3)] (red) labeled Lp(a) for 1 h or 3 h. After fixation, the cells were examined under confocal microscopy (63×). Scale bars are 10 um for 63× magnification. Cells were counterstained with DAPI.
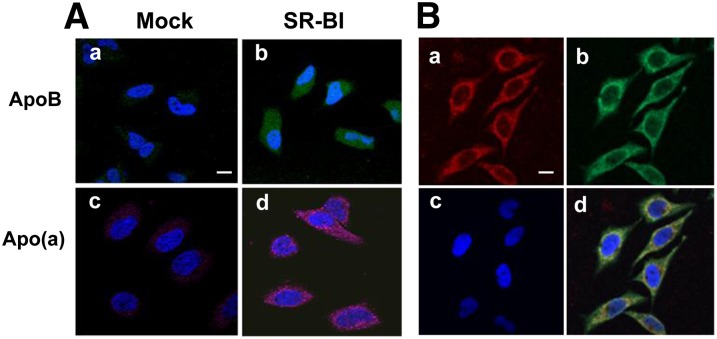
After incubation with Lp(a), HeLa cells were indirectly stained with either anti-apoB (green) or anti-apo(a) (pink) antibodies (Fig. 5A). Readily observable apoB (green) and apo(a) (pink) staining was seen only in HeLa cells transfected with SR-BI and not in control cells (Fig. 5A). When cells were stained directly with both fluorescent-labeled antibodies (apoB-green:apo(a)-red) significant intracellular colocalization (yellow) of the two proteins was seen, particularly in the perinuclear region (Fig. 5B).
Fig. 5.
Detection of cellular internalization of apo(a) and apoB . A: HeLa cells stably transfected with SR-BI (b, d) or without SR-BI (a, c) were incubated with 15 μg/ml Lp(a) at 4°C for 1 h. After washing, the cells were incubated at 37°C for 2 h, then fixed and exposed to goat anti-apoB antibody (a, b) or sheep anti-apo(a) antibody (c, d), and detected with fluorescent-labeled donkey anti-goat (green) or donkey anti-sheep (pink) antibodies and analyzed by confocal microscopy. B: HeLa cells stably transfected with SR-BI were cultured in the presence of 15 μg/ml of Lp(a) for 2 h and washed, then stained with fluorescent-labeled anti-apoB (green) and anti-apo(a) (red) antibodies and analyzed by confocal microscopy (63×). Scale bars are 10 um for 63× magnification. Merged image (yellow) is shown in (d). Cells were counterstained with DAPI. Apo(a) staining (a), apoB staining (b), DAPI staining (c), and merge (d).
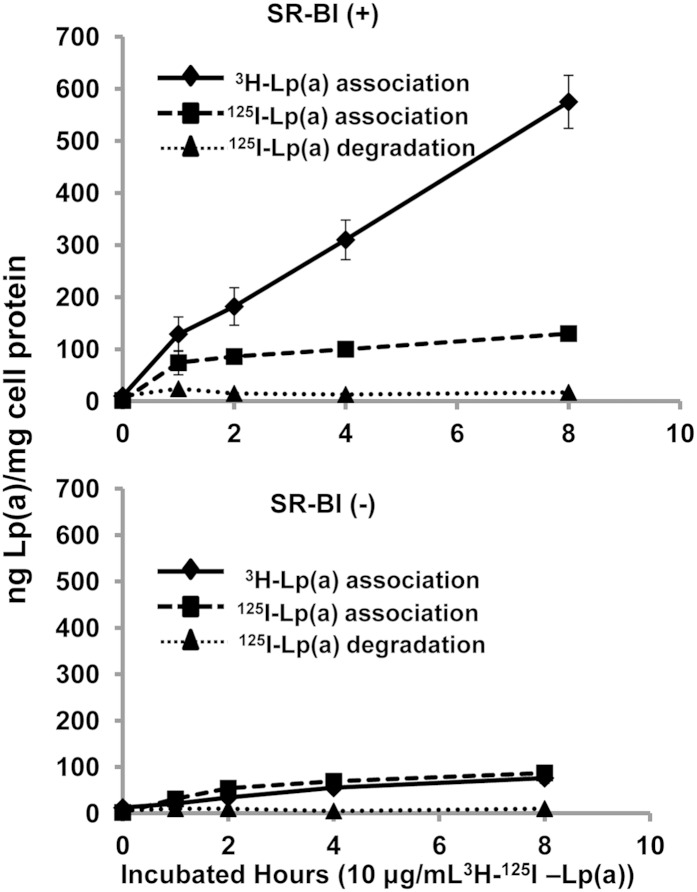
SR-BI mediates selective lipid uptake from Lp(a)
HEK293 cells were incubated with 10 μg/ml of dual-labeled (3H-CEth, 125I-protein) Lp(a) (Fig. 6). Compared with mock-transfected cells (lower panel), HEK293 cells expressing SR-BI (upper panel) showed a marked increase in the association of 3H-CEth-labeled Lp(a) (Fig. 6). Although SR-BI-transfected cells also showed an increase in the association of 125I-labeled Lp(a) protein, the increase in protein binding relative to control cells was not as large (Fig. 6). The level of 125I-protein counts associated with the SR-BI (+) cells also reached a plateau at 2 h, whereas the cellular uptake of 3H-CEth from Lp(a) continued to increase over the 8 h incubation period. Negligible amounts of degraded Lp(a) protein were observed in the conditioned cell media from either cell line.
Fig. 6.
SR-BI mediates selective uptake of CEth from Lp(a). HEK293 cells stably transfected with SR-BI (top panel) or without SR-BI (bottom panel) were incubated with 10 μg/ml (10 nM) 3H-CEth, 125I-protein dual-labeled Lp(a) and harvested at the indicated time points. Cell-associated radioactivity (3H, solid line; 125I, dashed line) was expressed as ng Lp(a)/mg cell protein. Conditioned media were also collected and the 125I-Lp(a) degraded protein (dotted line) was also determined and expressed as ng Lp(a)/mg cell protein. Results are expressed as the mean ± SD of triplicates.
SR-BI mediates the plasma clearance of Lp(a)
In order to investigate the ability of SR-BI to interact with Lp(a) in vivo, 3H-CE-Lp(a) was injected into three lines of mice with various levels of SR-BI, namely control C57BL/6 mice, Sr-b1-KO mice, and hSR-BI-Tg mice, which overexpress the human SR-BI transgene in the liver of C57BL/6 mice. An approximate 3-fold increase in the plasma clearance of 3H-CE-Lp(a) was observed in the hSR-BI-Tg mice compared with the control mice (Fig. 7). In contrast, a marked delay in the plasma clearance of 3H-CE-Lp(a) was evident in Sr-b1-KO mice compared with the control C57BL/6 mice.
DISCUSSION
A major finding from this study is that Lp(a) is a ligand for the SR-BI receptor. This was shown to occur in two different cell lines, namely HEK293 cells and HeLa cells. Both cell lines when transfected with SR-BI showed increased association of Lp(a) (Figs. 2, 3). This was true for both the lipid and protein components of Lp(a). Based on the competition studies (Fig. 3), all of the lipoproteins competed with Lp(a) for SR-BI binding. This was not unexpected, because SR-BI is a scavenger receptor and is already known to bind to LDL and HDL. Interestingly, LDL was slightly less effective than the unlabeled Lp(a) as a competitor, which suggests that there may be a possible interaction of the apo(a) component of Lp(a) with SR-BI, which is lacking in LDL.
Another major finding from this study is that SR-BI can promote the selective lipid uptake of CEs from Lp(a), as it does from other lipoproteins (27, 28). Although SR-BI was found to mediate the cellular uptake of all the major protein and lipid components of Lp(a) (Figs. 4, 5), it appears to preferentially promote the uptake of CE, a neutral core lipid (Fig. 4). When SR-BI-expressing cells were incubated with dual-labeled Lp(a), a greater relative increase in the uptake of labeled CEth was observed over the protein label (Fig. 6). Furthermore, although some apoB and apo(a) protein labels could be observed in SR-BI-expressing cells compared with control cells (Fig. 5), negligible amounts of Lp(a) protein degradation were detected (Fig. 6).
The cell culture experiments showing that SR-BI can serve as a receptor for Lp(a) were further confirmed in plasma turnover studies in mice (Fig. 7). hSR-BI-Tg mice, with increased expression of hepatic SR-BI, showed increased clearance of plasma 3H-CE-Lp(a) compared with C57BL/6 mice, whereas Sr-b1-KO mice had decreased clearance. Mice do not normally express Lp(a), but the modulation of hepatic levels of SR-BI in mice resulted in changes in the catabolism of exogenously added Lp(a) in a manner predicted based on the cell culture binding studies. It is important to note, however, that although there was a delay in Lp(a) catabolism in Sr-b1-KO mice, approximately 65% of the 3H-CE-Lp(a) was still removed from the plasma of Sr-b1-KO mice 24 h after injection (Fig. 7). This must indicate that there are other receptors and/or pathways for Lp(a) catabolism from the circulation.
Compared with LDL, Lp(a) is typically present in the plasma at a much lower concentration (6); nevertheless, Lp(a) has been shown on a per particle basis to be more pro-atherogenic (6). Various modifications of LDL in the vessel wall, such as oxidation and aggregation, make it a good ligand for several different types of scavenger receptors on macrophages (29), and perhaps Lp(a) can also undergo some of these same modifications. Interestingly, Lp(a) and not LDL has also been shown to be the principal carrier of negatively charged oxidized lipids in plasma (30). Enrichment of anionic charged lipids has previously been shown to enhance the uptake of lipoproteins by SR-BI (31); thus, negatively charged oxidized lipids could potentially explain the affinity of Lp(a) for SR-BI. This is an important area of future investigation not only for possibly explaining the uptake of Lp(a) by SR-BI but also for explaining why Lp(a) may be more pro-atherogenic than LDL (30). The delivery of oxidized lipids to cells is believed to be pro-inflammatory (32, 33). Also of interest, CE hydroperoxides from oxidized lipoproteins have been shown to be better substrates than CEs for selective lipid uptake by SR-BI (31). Given the myriad effects of oxidized cholesterol on gene regulation and inflammation, the delivery of oxidized lipids from Lp(a) to cells by SR-BI could be a contributing factor to the atherogenicity of Lp(a). The atherogenic risk from Lp(a) is in part related to its plasma level, which is known to be inversely related to the size of its apo(a) isoform (34). Whether apo(a) contributes to the binding of Lp(a) to SR-BI and whether different size isoforms of apo(a) can also affect the binding of Lp(a) to SR-BI is not known but would also be an important area of future investigation.
In summary, SR-BI was found to bind and promote the cellular uptake of Lp(a). In particular, SR-BI was found to favor the selective lipid uptake of CEs from Lp(a). These results indicate a possible new physiologic role for SR-BI as a receptor for Lp(a) and suggests several novel mechanisms for the atherogenic properties of Lp(a).
Acknowledgments
The authors thank the National Heart, Lung, and Blood Institute Light Microscopy Core for the use of their confocal microscopes.
Footnotes
Abbreviations:
- CE
- cholesteryl ester
- CEth
- cholesteryl ether
- CETP
- cholesteryl ester transfer protein
- FCR
- fractional catabolic rate
- hSR-BI-Tg
- human scavenger receptor class B type I transgenic
- Lp(a)
- lipoprotein(a)
- LPDS
- lipoprotein-deficient serum
- SR-BI
- human scavenger receptor class B type I
- Sr-b1-KO
- mouse scavenger receptor class B type I knockout
This work was supported by grants HL-72518 (L.C.B.), HL-59684 (L.C.B.), and HL-65608 (L.C.B.) from the National Heart, Lung, and Blood Institute and intramural research funding of National Heart, Lung, and Blood Institute (A.T.R.).
REFERENCES
- 1.Rhainds D., Brissette L. 2004. The role of scavenger receptor class B type I (SR-BI) in lipid trafficking. Defining the rules for lipid traders. Int. J. Biochem. Cell Biol. 36: 39–77. [DOI] [PubMed] [Google Scholar]
- 2.Acton S., Rigotti A., Landschulz K. T., Xu S., Hobbs H. H., Krieger M. 1996. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science. 271: 518–520. [DOI] [PubMed] [Google Scholar]
- 3.Murao K., Terpstra V., Green S. R., Kondratenko N., Steinberg D., Quehenberger O. 1997. Characterization of CLA-1, a human homologue of rodent scavenger receptor BI, as a receptor for high density lipoprotein and apoptotic thymocytes. J. Biol. Chem. 272: 17551–17557. [DOI] [PubMed] [Google Scholar]
- 4.Wang N., Arai T., Ji Y., Rinninger F., Tall A. R. 1998. Liver-specific overexpression of scavenger receptor BI decreases levels of very low density lipoprotein ApoB, low density lipoprotein ApoB, and high density lipoprotein in transgenic mice. J. Biol. Chem. 273: 32920–32926. [DOI] [PubMed] [Google Scholar]
- 5.Rader D. J., Alexander E. T., Weibel G. L., Billheimer J., Rothblat G. H. 2009. The role of reverse cholesterol transport in animals and humans and relationship to atherosclerosis. J. Lipid Res. 50 (Suppl.): S189–S194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Utermann G. 1989. The mysteries of lipoprotein(a). Science. 246: 904–910. [DOI] [PubMed] [Google Scholar]
- 7.Gavish D., Azrolan N., Breslow J. L. 1989. Plasma lp(a) concentration is inversely correlated with the ratio of Kringle IV/Kringle V encoding domains in the apo(a) gene. J. Clin. Invest. 84: 2021–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanktree M. B., Anand S. S., Yusuf S., Hegele R. A. 2010. Comprehensive analysis of genomic variation in the LPA locus and its relationship to plasma lipoprotein(a) in South Asians, Chinese, and European Caucasians. Circ. Cardiovasc. Genet. 3: 39–46. [DOI] [PubMed] [Google Scholar]
- 9.Trommsdorff M., Kochl S., Lingenhel A., Kronenberg F., Delport R., Vermaak H., Lemming L., Klausen I. C., Faergeman O., Utermann G., et al. 1995. A pentanucleotide repeat polymorphism in the 5′ control region of the apolipoprotein(a) gene is associated with lipoprotein(a) plasma concentrations in Caucasians. J. Clin. Invest. 96: 150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenner J. L., Seman L. J., Millar J. S., Lamon-Fava S., Welty F. K., Dolnikowski G. G., Marcovina S. M., Lichtenstein A. H., Barrett P. H., deLuca C., et al. 2005. The metabolism of apolipoproteins (a) and B-100 within plasma lipoprotein (a) in human beings. Metabolism. 54: 361–369. [DOI] [PubMed] [Google Scholar]
- 11.Rader D. J., Mann W. A., Cain W., Kraft H. G., Usher D., Zech L. A., Hoeg J. M., Davignon J., Lupien P., Grossman M., et al. 1995. The low density lipoprotein receptor is not required for normal catabolism of Lp(a) in humans. J. Clin. Invest. 95: 1403–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reblin T., Niemeier A., Meyer N., Willnow T. E., Kronenberg F., Dieplinger H., Greten H., Beisiegel U. 1997. Cellular uptake of lipoprotein[a] by mouse embryonic fibroblasts via the LDL receptor and the LDL receptor-related protein. J. Lipid Res. 38: 2103–2110. [PubMed] [Google Scholar]
- 13.Argraves K. M., Kozarsky K. F., Fallon J. T., Harpel P. C., Strickland D. K. 1997. The atherogenic lipoprotein Lp(a) is internalized and degraded in a process mediated by the VLDL receptor. J. Clin. Invest. 100: 2170–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vishnyakova T. G., Bocharov A. V., Baranova I. N., Chen Z., Remaley A. T., Csako G., Eggerman T. L., Patterson A. P. 2003. Binding and internalization of lipopolysaccharide by Cla-1, a human orthologue of rodent scavenger receptor B1. J. Biol. Chem. 278: 22771–22780. [DOI] [PubMed] [Google Scholar]
- 15.Yang X. P., Mattagajasingh S., Su S., Chen G., Cai Z., Fox-Talbot K., Irani K., Becker L. C. 2007. Fractalkine upregulates intercellular adhesion molecule-1 in endothelial cells through CX3CR1 and the Jak Stat5 pathway. Circ. Res. 101: 1001–1008. [DOI] [PubMed] [Google Scholar]
- 16.Fless G. M., Snyder M. L. 1996. Quantitation of lipoprotein (a) after lysine-sepharose chromatography and density gradient centrifugation. Methods Enzymol. 263: 238–251. [DOI] [PubMed] [Google Scholar]
- 17.Stein Y., Dabach Y., Hollander G., Halperin G., Stein O. 1983. Metabolism of HDL-cholesteryl ester in the rat, studied with a nonhydrolyzable analog, cholesteryl linoleyl ether. Biochim. Biophys. Acta. 752: 98–105. [DOI] [PubMed] [Google Scholar]
- 18.Shamburek R. D., Pentchev P. G., Zech L. A., Blanchette-Mackie J., Carstea E. D., VandenBroek J. M., Cooper P. S., Neufeld E. B., Phair R. D., Brewer H. B., Jr, et al. 1997. Intracellular trafficking of the free cholesterol derived from LDL cholesteryl ester is defective in vivo in Niemann-Pick C disease: insights on normal metabolism of HDL and LDL gained from the NP-C mutation. J. Lipid Res. 38: 2422–2435. [PubMed] [Google Scholar]
- 19.Redgrave T. G., Roberts D. C., West C. E. 1975. Separation of plasma lipoproteins by density-gradient ultracentrifugation. Anal. Biochem. 65: 42–49. [DOI] [PubMed] [Google Scholar]
- 20.Gotto A. M., Pownall H. J. 2003. Manual of Lipid Disorders: Reducing the Risk for Coronary Disease. Lippincott, Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 21.Reaven E., Tsai L., Azhar S. 1996. Intracellular events in the “selective” transport of lipoprotein-derived cholesteryl esters. J. Biol. Chem. 271: 16208–16217. [DOI] [PubMed] [Google Scholar]
- 22.Pitas R. E., Innerarity T. L., Weinstein J. N., Mahley R. W. 1981. Acetoacetylated lipoproteins used to distinguish fibroblasts from macrophages in vitro by fluorescence microscopy. Arteriosclerosis. 1: 177–185. [DOI] [PubMed] [Google Scholar]
- 23.Goldstein J. L., Basu S. K., Brown M. S. 1983. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 98: 241–260. [DOI] [PubMed] [Google Scholar]
- 24.Fan J., Wang J., Bensadoun A., Lauer S. J., Dang Q., Mahley R. W., Taylor J. M. 1994. Overexpression of hepatic lipase in transgenic rabbits leads to a marked reduction of plasma high density lipoproteins and intermediate density lipoproteins. Proc. Natl. Acad. Sci. USA. 91: 8724–8728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaisman B. L., Demosky S. J., Stonik J. A., Ghias M., Knapper C. L., Sampson M. L., Dai C., Levine S. J., Remaley A. T. 2012. Endothelial expression of human ABCA1 in mice increases plasma HDL cholesterol and reduces diet-induced atherosclerosis. J. Lipid Res. 53: 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stockinger W., Hengstschlager-Ottnad E., Novak S., Matus A., Huttinger M., Bauer J., Lassmann H., Schneider W. J., Nimpf J. 1998. The low density lipoprotein receptor gene family. Differential expression of two alpha2-macroglobulin receptors in the brain. J. Biol. Chem. 273: 32213–32221. [DOI] [PubMed] [Google Scholar]
- 27.Stangl H., Cao G., Wyne K. L., Hobbs H. H. 1998. Scavenger receptor, class B, type I-dependent stimulation of cholesterol esterification by high density lipoproteins, low density lipoproteins, and nonlipoprotein cholesterol. J. Biol. Chem. 273: 31002–31008. [DOI] [PubMed] [Google Scholar]
- 28.Stangl H., Hyatt M., Hobbs H. H. 1999. Transport of lipids from high and low density lipoproteins via scavenger receptor-BI. J. Biol. Chem. 274: 32692–32698. [DOI] [PubMed] [Google Scholar]
- 29.Tabas I., Williams K. J., Boren J. 2007. Subendothelial lipoprotein retention as the initiating process in atherosclerosis: update and therapeutic implications. Circulation. 116: 1832–1844. [DOI] [PubMed] [Google Scholar]
- 30.Tsimikas S., Witztum J. L. 2008. The role of oxidized phospholipids in mediating lipoprotein(a) atherogenicity. Curr. Opin. Lipidol. 19: 369–377. [DOI] [PubMed] [Google Scholar]
- 31.Fluiter K., Sattler W., De Beer M. C., Connell P. M., van der Westhuyzen D. R., van Berkel T. J. 1999. Scavenger receptor BI mediates the selective uptake of oxidized cholesterol esters by rat liver. J. Biol. Chem. 274: 8893–8899. [DOI] [PubMed] [Google Scholar]
- 32.Jamieson D. G., Usher D. C., Rader D. J., Lavi E. 1995. Apolipoprotein(a) deposition in atherosclerotic plaques of cerebral vessels. A potential role for endothelial cells in lesion formation. Am. J. Pathol. 147: 1567–1574. [PMC free article] [PubMed] [Google Scholar]
- 33.Leonarduzzi G., Gamba P., Gargiulo S., Biasi F., Poli G. 2012. Inflammation-related gene expression by lipid oxidation-derived products in the progression of atherosclerosis. Free Radic. Biol. Med. 52: 19–34. [DOI] [PubMed] [Google Scholar]
- 34.Kamstrup P. R., Tybjaerg-Hansen A., Nordestgaard B. G. 2013. Extreme lipoprotein(a) levels and improved cardiovascular risk prediction. J. Am. Coll. Cardiol. 61: 1146–1156. [DOI] [PubMed] [Google Scholar]