Abstract
Class I alcohol dehydrogenases (ADH1s) are the rate-limiting enzymes for ethanol and vitamin A (retinol) metabolism in the liver. Because previous studies have shown that human ADH1 enzymes may participate in bile acid metabolism, we investigated whether the bile acid-activated nuclear receptor farnesoid X receptor (FXR) regulates ADH1 genes. In human hepatocytes, both the endogenous FXR ligand chenodeoxycholic acid and synthetic FXR-specific agonist GW4064 increased ADH1 mRNA, protein, and activity. Moreover, overexpression of a constitutively active form of FXR induced ADH1A and ADH1B expression, whereas silencing of FXR abolished the effects of FXR agonists on ADH1 expression and activity. Transient transfection studies and electrophoretic mobility shift assays revealed functional FXR response elements in the ADH1A and ADH1B proximal promoters, thus indicating that both genes are direct targets of FXR. These findings provide the first evidence for direct connection of bile acid signaling and alcohol metabolism.
Keywords: farnesoid X receptor, gene regulation, ethanol, liver metabolism
Alcohol dehydrogenases (ADHs) (EC 1.1.1.1) are enzymes that are able to reversibly oxidize a wide variety of endogenous and xenobiotic alcohols to the corresponding aldehydes (1). Mammals have evolved different ADHs that can be grouped into five classes based on similarities in amino acid sequences and biochemical properties (2). Class I ADH (ADH1) constitutes the classical hepatic form that accounts for most of the ethanol-oxidizing capacity in the liver (3). Only one ADH1 enzyme and gene (Adh1) is found in mice and rats. By contrast, humans have three ADH1s (ADH1A, ADH1B, and ADH1C; traditionally designated α, β, and γ isoenzymes, respectively) that are encoded by unique genes that are clustered head-to-tail within 80 kb on chromosome 4 and are very similar to each other in the exon-intron structure and nucleotide sequences (1). All three human ADH1 enzymes are highly expressed in the adult liver and at lower levels in a variety of extrahepatic tissues (4). A different pattern of temporal expression for each of the three isoenzymes has been observed during liver development (4). Considerable efforts have been devoted to identify the factors involved in controlling human ADH1 gene expression, most of them focused on the analysis of tissue-specific and temporal regulation. The three ADH1 genes share 80–84% sequence identity for about 270 bp upstream of the transcription start site, and significant similarity among them extends to −795 bp (5). However, in vitro binding analysis and transfection studies in liver and nonliver cancer cell lines revealed regulatory sequences mediating differential transcriptional mechanisms for each of the ADH1 genes to account for the distinct expression patterns (6). Thus, transcription factors CCAAT/enhancer-binding protein (C/EBP)α, C/EBPβ, and HNF-1 were found to selectively contribute to the liver-specific expression and the induction of each of the ADH1 genes during liver development (7, 8).
Bile acids may function as signaling molecules in regulating their own synthesis and transport and controlling lipid and glucose homeostasis (9). Bile acid signaling in the liver is mostly mediated via the farnesoid X receptor (FXR) α (FXRα NR1H4, hereafter referred to as FXR), a member of the nuclear receptor superfamily of ligand-activated transcription factors (10). FXR heterodimerizes with the retinoid X receptor (RXR) and binds to consensus sequences, most commonly an inverted hexameric nucleotide repeat separated by one nucleotide (IR1) and, in the presence of specific agonists, activates transcription of target genes involved in bile acid, cholesterol, lipoprotein, and glucose metabolism (10). FXR has been shown to have relevance in the attenuation of clinically important conditions such as gallstone disease (11, 12), cholestasis (13), and fatty liver disease (14). Most of these hepatoprotective functions of FXR can be attributed to the induction of genes involved in bile acid detoxification and xenobiotic metabolism, including phase I oxidation enzyme CYP3A4 (15) and a number of phase II conjugation enzymes (16) and phase III efflux transporters (10).
In addition to the rate-limiting step in ethanol metabolism, human ADH1 isoenzymes catalyze steps in several metabolic pathways including vitamin A (retinol) oxidation, which is the rate-limiting step in the conversion of retinol to retinoic acid (17) and, interestingly, bile acid metabolism. Thus, ADH1B has been shown to oxidize the bile alcohol 5β-cholestane-3α,7α,12α,26-tetrol to the 3α,7α,12α-trihydroxy-5β-cholestanoic acid, an intermediate in bile acid synthesis (18), whereas ADH1C was identified as the sole bile acid 3β-hydroxysteroid dehydrogenase present in human liver cytosol that promotes epimerization of iso bile acids to 3α-hydroxy bile acids, which are subsequently secreted by hepatocytes into bile (19, 20). In this study, we investigated the effect of both the endogenous bile acid chenodeoxycholic acid (CDCA) and the synthetic FXR agonist GW4064 on ADH1 gene expression. The data presented herein identify the nuclear receptor FXR as a regulator of human ADH1 genes, thus linking bile acid signaling and alcohol metabolism.
MATERIALS AND METHODS
Animal experiments
Experimental protocols with mice were performed with the approval of the animal ethics committee of the University of Barcelona (Spain). Male 10-week-old C57BL/6 mice were injected intraperitoneally with either vehicle (corn oil 5% DMSO) or GW4064 (GlaxoSmithKline, Research Triangle Park, NC) dissolved in vehicle (10 mg/ml) at a dose of 50 mg/kg. After 8 h, mice were sacrificed, and livers were excised, snap-frozen in liquid nitrogen, and stored at −80°C until analysis.
Cell culture and treatment conditions
Human hepatoma HepG2 and Huh7 cells and rat hepatoma FAO cells were cultured in DMEM supplemented with 10% FBS. Primary mouse hepatocytes were isolated by the collagenase method as previously described (21) and cultured for 6 h before treatments. Human primary hepatocytes were obtained commercially (Ready HepsTM Fresh Hepatocytes, Lonza, Basel, Switzerland) and maintained in Hepatocyte Complete Medium (HCMTM bulletkit, Lonza). Cells were treated with ligands in the same medium supplemented with 10% charcoal stripped FBS (Biological Industries).
RNA isolation and real-time quantitative PCR analysis
Total RNA was isolated from cells or livers by using Tri-Reagent (Ambion) and further treated with DNase I (Ambion). cDNA was synthesized from total RNA (1 μg) by murine leukemia virus reverse transcriptase (Invitrogen) and p(dN)6 random primers (Roche Diagnostics) according to the manufacturer's instructions. cDNA was subjected to real-time quantitative PCR using Platinum® Quantitative PCR SuperMix-UDG with ROX (Invitrogen) and the specific TaqMan® Gene Expression Assay probes (Applied Biosystems): ADH1A, Hs00605167_g1; ADH1B, Hs00605175_m1; ADH1C, Hs00817827_m1; CYP2E, Hs00559368_m1; ALDH1A, Hs00946916_m1; ALDH2 Hs00355914_m1; PLTP, Hs01067287_m1; NR0B2, Hs00222677_m1; NR1H4, Hs00231968_m1; mouse Adh1, Mm00507711_m1; mouse Nr0b2, Mm00442278_m1; rat Adh1, Rn00570670_m1; and rat Nr0b2, Rn00589173_m1. Relative mRNA abundance was obtained by normalizing to 18S levels.
Small interfering RNA transfection
The human FXR siGENOME SMARTpool (M-003414-01) or siGENOME NonTargeting small interfering RNA (siRNA) #1 (D-001210-01) (Dharmacon, Lafayette, CO) were used to transfect Huh7 cells with the DharmaFECT 4 transfection reagent at a final concentration of 25 nM each for 48 h, followed by 24 h of treatments as described above.
Adenoviral infection
Huh7 cells were infected at a multiplicity of infection of 25 or 50 for 48 h with adenoviruses expressing VP16 (AdVP16) or VP16FXR chimeras (AdVP16FXR) previously described (22).
Plasmids
The ADH1A proximal promoter (−1877 to +78 relative to the transcription start site) was amplified by PCR from genomic DNA using primers 5′-agtcacgcgtggagctaggtatagttgatg-3′ and 5′-agtcctcgagtccttgtggatttcttcc-3′, MluI/XhoI digested, and subcloned into pGL3-basic vector (Promega) to generate pGL3-ADH1A. Similarly, the ADH1B promoter (−2850 to +59) was amplified using primers 5′-agtcggtaccattattctgcctagcacacc-3′ and 5′-agtcgctagcagactgtgagtctttgtgg-3′, while the ADH1C promoter (−2519 to +35) was amplified using primers 5′-agtcggtacctcgtactatccctgattgg-3′ and 5′-agtcgctagcttcttctctgcttgagtgc-3′, and the PCR products were subcloned into KpnI/NheI sites of the pGL3bv to generate pGL3-ADH1B and pGL3-ADH1C, respectively. Mutagenesis were performed using the QuickChangeTM site-directed mutagenesis kit (Stratagene) and oligonucleotides containing mutations corresponding respectively to nt −97A→G/−92C→G/−91C→G of ADH1A promoter, nt −120A→G/−115C→G/−114C→G of ADH1B promoter, and nt −122G→A/−121G→T/−115A→T/−114T→C of ADH1C promoter. The reporter plasmids pADH1AIR1-TK, pADH1BIR1-TK, and pADH1CIR1-TK were generated by insertion of four copies of a double-stranded oligonucleotide-containing sequence spanning nt −108 to −82 of ADHA1 (5′-gatccattgctggttcattgccctcttcttta-3′), nt −131 to −106 of ADH1B (5′-gatccattgctggttcagtacccttttatcta-3′), or nt −133 to −107 of ADH1C (5′-gatccagtgctggttcggtgcccatttcttta-3′) into the BglII site of pGL3-TK (23). Plasmids expressing human RXRα (NR2B1) and FXR into pSG5 have been described (23).
Cell transfection and reporter assays
Huh7 cells were transiently transfected as previously described (23). After 6 h, cells were treated for 24 h with the vehicle (DMSO), 100 μM CDCA, or 1 μM GW4064 as described above. Luciferase activities were assayed as previously described (23). All transfections were performed in triplicate, and similar results were obtained in at least three independent experiments.
Western blot analysis
Whole protein cell extracts were obtained from Huh7 cells in Nonidet P-40 lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 1% Nonidet P-40) supplemented with a mixture of protease inhibitors (Sigma-Aldrich) and 0.1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich). Fifty micrograms of protein were resolved in 10% polyacrylamide gels, transferred onto Immobilon-P membranes (Millipore, Bedford, MA) and probed with antibodies: anti-pan ADH1 (1:200, Santa Cruz Biotechnology, Santa Cruz, CA, catalog #sc-22750); anti-FXR (1:1000, Invitrogen, catalog #A9033A); anti-actin (1:1000, Sigma-Aldrich, catalog #A2066).
In silico analysis of FXR response elements
The analysis of genomic sequences for the identification of putative FXR response elements (FXREs) was performed using the NUBIScan computer algorithm.
In vitro transcription/translation and electrophoretic mobility shift assay
Double-stranded oligonucleotides corresponding to the sequences spanning nt −108 to −82 of ADHA1, nt −131 to −106 of ADH1B, or nt −133 to −107 of ADH1C were radiolabeled and used in an electrophoretic mobility shift assay (EMSA) as previously described (23). A probe containing the FXRE of the human ileal-bile acid-binding protein (I-BABP) gene promoter was used as a control. For competition experiments, increasing fold molar excess of unlabeled probes was included during a 15 min preincubation on ice. The probe mutADH1AIR1 corresponds to the sequence spanning nt −108 to −82 of ADHA1 harboring mutations in nt −97A→G/−92C→G/−91C→G.
Alcohol dehydrogenase activity assay
ADH enzymatic activity was determined using Alcohol Dehydrogenase Assay kit (catalog #K787, BioVision, Milpitas, CA) following the manufacturer's instructions, and normalized to protein content in each sample.
Statistical analysis
Data are expressed as mean ± SEM as determined by analysis of multiple independent samples, as indicated in figure legends. Significant differences were assessed using a two-tailed Student's t-test. Values of P < 0.05 were considered to be significant.
RESULTS
ADH1 gene expression is induced in human primary hepatocytes and hepatoma cell lines in response to FXR ligands
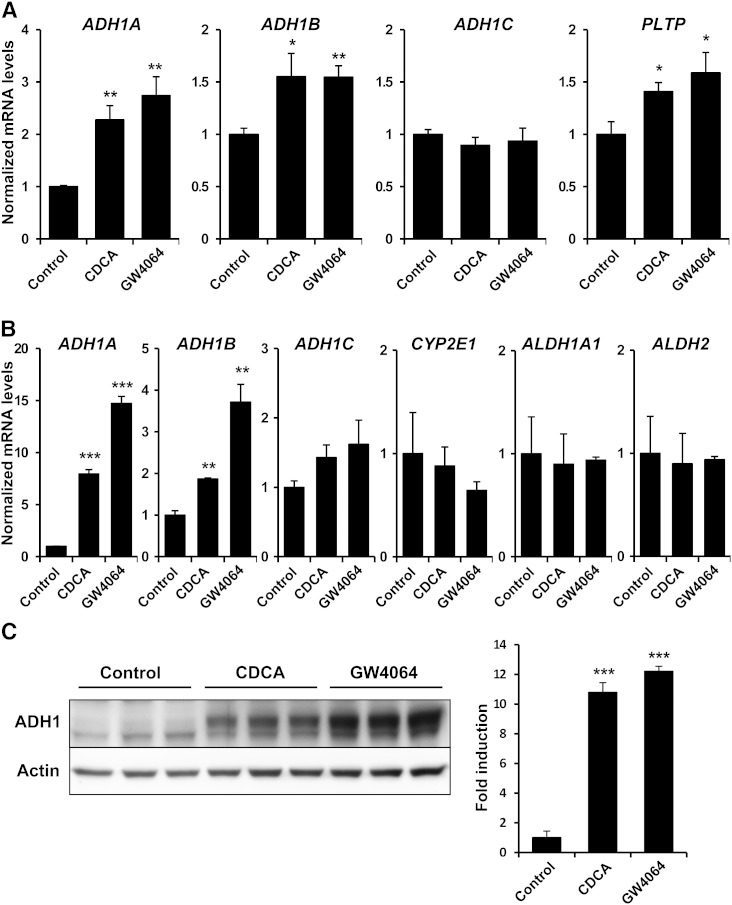
Inasmuch as FXR regulates the expression of genes involved in detoxification and drug metabolism (15, 16), and because ADH1 enzymes are reported to take part in bile acid metabolism (18–20), we evaluated whether this nuclear receptor modulates ADH1 gene expression. Human primary hepatocytes were exposed for 24 h to CDCA, a natural bile acid agonist of FXR, or GW4064, a synthetic and specific FXR agonist, and analyzed for the expression of the three ADH1 genes. As illustrated in Fig. 1A, both FXR ligands significantly increased ADH1A and ADH1B mRNA levels. As expected, the well-characterized FXR target gene PLTP (24) was also induced when primary human hepatocytes were treated with FXR agonists. In contrast, ADH1C mRNA was not induced after activation of FXR (Fig. 1A). To confirm these results in human hepatoma cell lines, HepG2 cells were also incubated in the presence of vehicle (DMSO) or FXR agonists. Again, no significant induction of ADH1C was observed in response to FXR ligands, whereas ADH1A and ADH1B mRNA levels were strongly increased (Fig. 1B). In addition, FXR activation failed to induce cytochrome P450 2E1 (CYP2E1), cytosolic aldehyde dehydrogenase (ALDH1A1), or mitochondrial aldehyde dehydrogenase (ALDH2) mRNA levels. Next, Western blot analyses performed on cell lysates from Huh7 treated with FXR agonists revealed that the quantity of ADH1 protein was robustly increased compared with cells receiving vehicle (Fig. 1C).
Fig. 1.
Bile acids and synthetic FXR ligand GW4064 increase human ADH1 mRNA and protein levels. Human primary hepatocytes (A) and HepG2 cells (B) were incubated with vehicle (Control), 50 μM CDCA or 5 μM GW4064 for 24 h. Total RNA was extracted and reverse transcribed for analysis by real-time quantitative PCR. Specific mRNA levels normalized to 18S content are expressed relative to controls, which were set as 1 (mean ± SEM). C: Western blot analysis of total protein lysates from Huh7 cells treated as in (A). Protein signals were quantified and pan ADH1 levels were normalized to actin content. *P < 0.05; **P < 0.01; ***P < 0.001 versus untreated controls. The results are representative of three independent experiments from triplicate dishes.
Overexpression of constitutively active FXR increases ADH1A and ADH1B mRNA levels
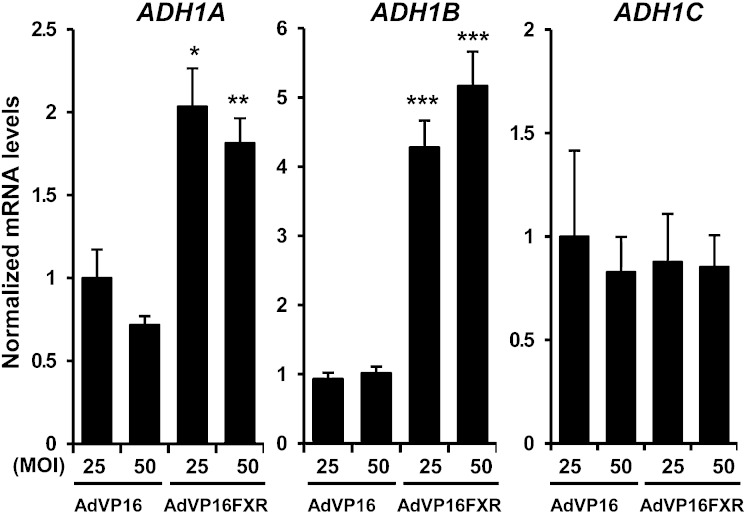
To further confirm the role of FXR in the regulation of the ADH1 gene cluster, we performed gain-of-function studies by ectopically expressing a constitutively active form of FXR (VP16FXR) (22) in Huh7 cells. The expression of ADH1A and ADH1B, but not ADH1C, was induced by FXR overexpression (Fig. 2). Collectively, these data suggest that FXR induces specifically the expression of ADH1A and ADH1B isoenzymes.
Fig. 2.
Constitutive active FXR induces ADH1A and ADH1B expression. Huh7 cells were infected with increasing multiplicity of infection (MOI) of adenovirus expressing either VP16 (AdVP16) or a constitutively active chimera of VP16 and FXR (AdVP16FXR) for 48 h. Specific mRNA levels of ADH1A, ADH1B, and ADH1C were determined by real-time quantitative PCR, normalized to 18S content, and expressed relative to controls, which were set as 1 (mean ± SEM). *P < 0.05; **P < 0.01; ***P < 0.001 versus 25 MOI AdVP16. The results are representative of two independent experiments from triplicate dishes.
The inductions of ADH1A and ADH1B by CDCA and GW4064 require FXR expression
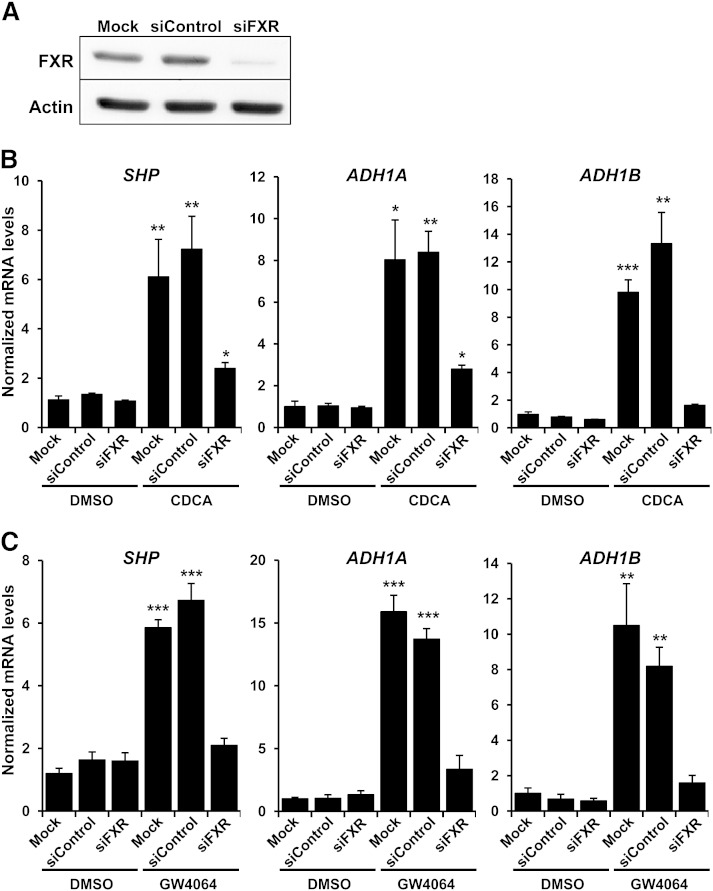
Because bile acids may exert their signaling actions through FXR-independent pathways (9, 25), we silenced FXR by siRNA to ascertain whether the inductions observed upon treatment with CDCA were dependent on FXR. Huh7 cells were transfected with nontargeting siRNA or siRNA complexes directed against FXR (siFXR) before treatments. siFXR-mediated knockdown of endogenous FXR levels (Fig. 3A) almost completely eliminated the induction of ADH1A and ADH1B by bile acid CDCA (Fig. 3B). As a control, CDCA-dependent induction of small heterodimer partner (SHP), a well-known FXR target gene (10), was similarly attenuated in siFXR-transfected cells. Likewise, as depicted in Fig. 3C, we also confirmed by FXR knockdown the requirement of FXR in the responses of these genes to GW4064 to rule out any effect of this synthetic ligand that could relate to weak agonistic effects on other receptors (26).
Fig. 3.
The inductions of ADH1A and ADH1B expression by CDCA and GW4064 are FXR dependent. Huh7 cells were transfected with either control siRNA (siControl) or siFXR. At 48 h posttransfection, cells were treated with vehicle (Control) (A–C), 50 μM CDCA (B), or 5 μM GW4064 (C) for 24 h. Specificity of the knockdown was checked by determination of FXR protein levels by Western blot (A). Specific mRNA levels of SHP, ADH1A, and ADH1B were normalized to 18S content and expressed relative to values of mock-transfected cells (Mock) treated with vehicle, which were set as 1 (mean ± SEM). *P < 0.05; **P < 0.01; ***P < 0.001 versus the corresponding controls. The results are representative of two independent experiments from triplicate dishes.
FXR regulation of ADH1 does not occur in rodents
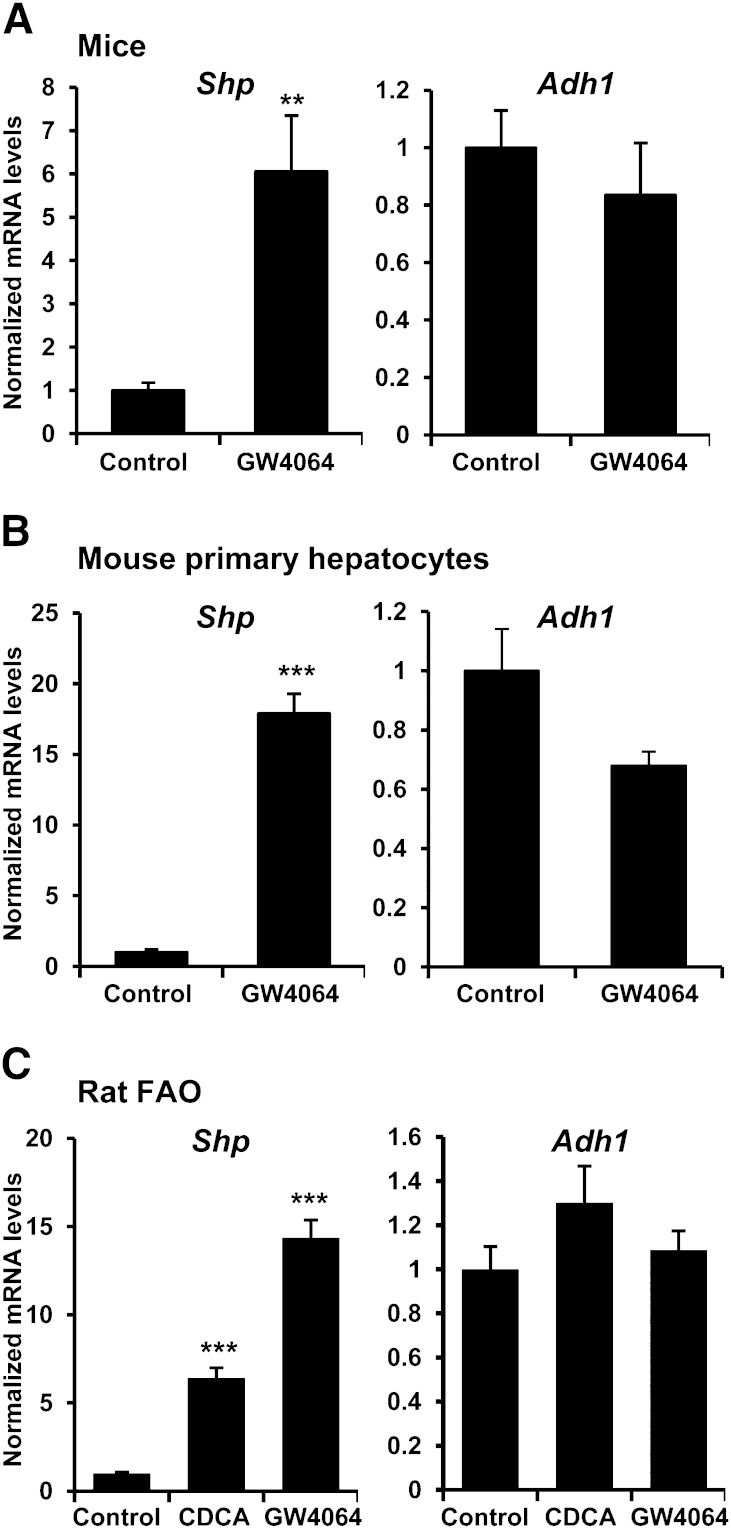
To examine whether FXR regulation of ADH1 also occurs in rodents, C57BL/6 mice were treated with either vehicle or FXR agonist GW4064 and analyzed for hepatic transcript levels. Unexpectedly, no significant change of Adh1 mRNA levels was observed following activation of FXR, in spite of the marked induction of the known FXR target Shp (Fig. 4A). To exclude the possibility that the dissimilar findings in human hepatocytes and in mouse liver were attributable to methodological differences, mouse primary hepatocytes were incubated in the presence of vehicle or GW4064. Again, Shp was highly induced after FXR activation, whereas no significant change was detected in Adh1 transcript levels (Fig. 4B). Likewise, analyses performed on rat hepatoma FAO cells treated with CDCA, GW4064, or vehicle showed induction of Shp in response to FXR ligands, but no change in rat Adh1 transcript levels (Fig. 4C). Taken together, these data indicate that induction of ADH1 genes by activation of FXR is species specific because it was observed in human but not in rodent-derived hepatocytes.
Fig. 4.
Induction of ADH1 by FXR does not occur in rodents. A: C57BL/6 mice (n = 5/group) were treated with either vehicle (Control) or 50 mg/kg GW4064 for 8 h. Adh1 and Shp expression was measured by quantitative real-time PCR in liver. B: Primary mouse hepatocytes were incubated with vehicle (Control) or 5 μM GW4064 for 24 h. C: Rat hepatoma FAO cells were treated with vehicle (Control), 50 μM CDCA, or 5 μM GW4064 for 24 h. RNA was extracted and mRNA levels for Adh1 and Shp were determined. Specific Shp and Adh1 mRNA levels normalized to 18S content are expressed relative to controls, which were set as 1 (mean ± SEM). **P < 0.01; ***P < 0.001 versus untreated controls.
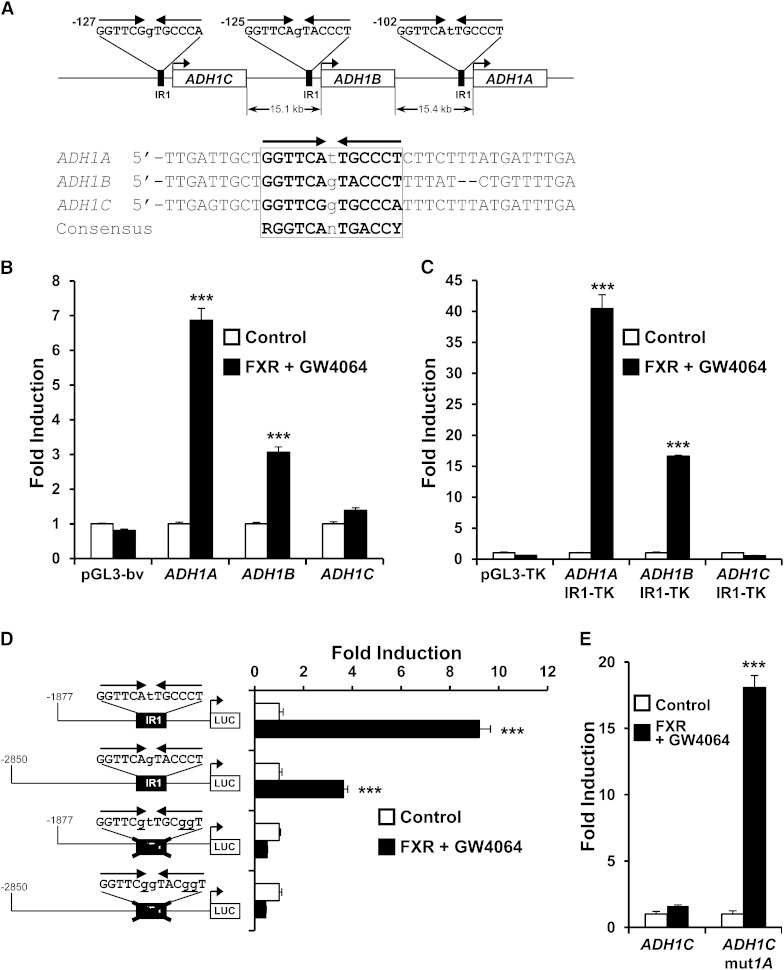
Identification and functional characterization of putative FXREs in the promoters of ADH1 genes
In order to determine whether FXR directly controls ADH1 expression and also with the aim to understand the molecular mechanisms involved in the differences observed in the FXR-dependent regulation of the distinct human ADH1 genes, the promoters of ADH1A, ADH1B, and ADH1C were in silico analyzed for putative FXREs using the NUBIScan computer algorithm. Most known FXREs consist of an IR of the RGGTCA hexad with minor variants (27, 28). Accordingly, an IR1 motif was identified as a putative FXRE in the proximal promoter of each of the three genes. As illustrated in Fig. 5A, IR1 elements in ADH1A, ADH1B, and ADH1C promoters differ from 2, 3, and 4 nucleotides, respectively, from the consensus FXRE. The presence of a T at position 3 and a C at position 10 of the IR1 element are permissive (e.g., FXREs of FGF19 and ALAS1, respectively) (10, 29). In contrast, A into G conversion at the sixth position, as it occurs in the ADH1C element, is detrimental to receptor binding (27, 28). To determine if the ADH1 proximal promoters were able to confer a response to FXR, we performed transient transfection assays in Huh7 cells with luciferase reporter constructs under the control of ∼2–2.9 kb fragments corresponding to ADH1 gene promoters in the presence or absence of a plasmid encoding FXR and the agonist GW4064. The data in Fig. 5B show that reporter activity of ADH1A and ADH1B promoter constructs was increased by activated FXR, whereas no significant effects were observed on the ADH1C promoter construct or the promoterless pGL3 basic vector. These results are consistent with the higher similarity of ADH1A and ADH1B IR1 elements to the consensus FXRE. To further confirm this observation, the IR1 element for each promoter was cloned upstream of the thymidine kinase promoter-driven luciferase reporter gene. As expected, transient transfection assays showed that the reporter constructs containing the IR sequences of ADH1A or ADH1B, but not of ADH1C, were strongly transactivated by FXR (Fig. 5C).
Fig. 5.
Characterization of a functional IR1 in the proximal promoter of ADH1A and ADH1B. A: Schematic representation of human ADH1 gene cluster and localization of the IR1 elements identified by NUBIScan in each proximal promoter. Alignment of the three IR1 and the FXRE consensus is shown below. The RGGTCA half-sites are indicated by arrows. B: The promoters of ADH1A and ADH1B, but not of ADH1C, respond to activated FXR. Huh7 cells were transfected with a plasmid containing luciferase reporter constructs driven by ∼2–2.9 kb fragments corresponding to ADH1 gene promoters (pGL3-ADH1A, pGL3-ADH1B, and pGL3-ADH1C, respectively), or the empty pGL3-basic vector (bv) as negative control, along with a plasmid expressing FXR, or the empty expression vector pSG5 as control, and then treated for 24 h with vehicle (Control) or 1 μM GW4064 and luciferase activities were measured. C: IR1 elements in ADH1A and ADH1B, but not ADH1C, confer FXR responsiveness to a heterologous promoter. Experiments were performed as in (B) with reporter constructs containing four copies of the IR1 site identified in the proximal promoter of ADH1 genes cloned in front of a heterologous thymidine kinase (TK) promoter-driven luciferase gene. pGL3-TK reporter vector was used as negative control. D: Disruption of ADH1A and ADH1B IR1 elements by site-directed mutagenesis abrogates the response to FXR. Experiments were performed as in (B) with the indicated reporter constructs containing wild-type or IR1 mutated sequence. E: Conversion of ADH1C IR1 to ADH1A IR1 element by site-directed mutagenesis confers FXR responsiveness to ADH1C promoter. Experiments were performed as in (B) with the constructs containing wild-type ADH1C promoter or a modified version containing the IR1 converted to ADH1A IR1 element. Results are expressed as -fold induction over control. ***P < 0.001 versus untreated controls. The results are representative of three independent experiments from triplicate dishes.
To assess the importance of the identified IR1 elements in FXR-dependent activation, we introduced single nucleotide mutations to disrupt the IR1 in the context of ADH1 promoters to generate pGL3-ADH1Amut and pGL3-ADH1Bmut constructs, respectively. As depicted in Fig. 5D, these point mutations completely abrogated the response to FXR. We next investigated the effects of performing site-directed mutations to convert the ADH1C IR1 element into the ADH1A IR1 element in the context of the of ∼2.6 kb ADH1C promoter. In contrast to the wild type, the activity of the construct bearing the mutated IR1 (pGL3-ADH1Cmut1A) was markedly increased by FXR (Fig. 5E). Collectively, these findings indicate that ADH1A and ADH1B are direct target genes of FXR and that the IR1 elements located at their respective proximal promoters are essential for the response to FXR.
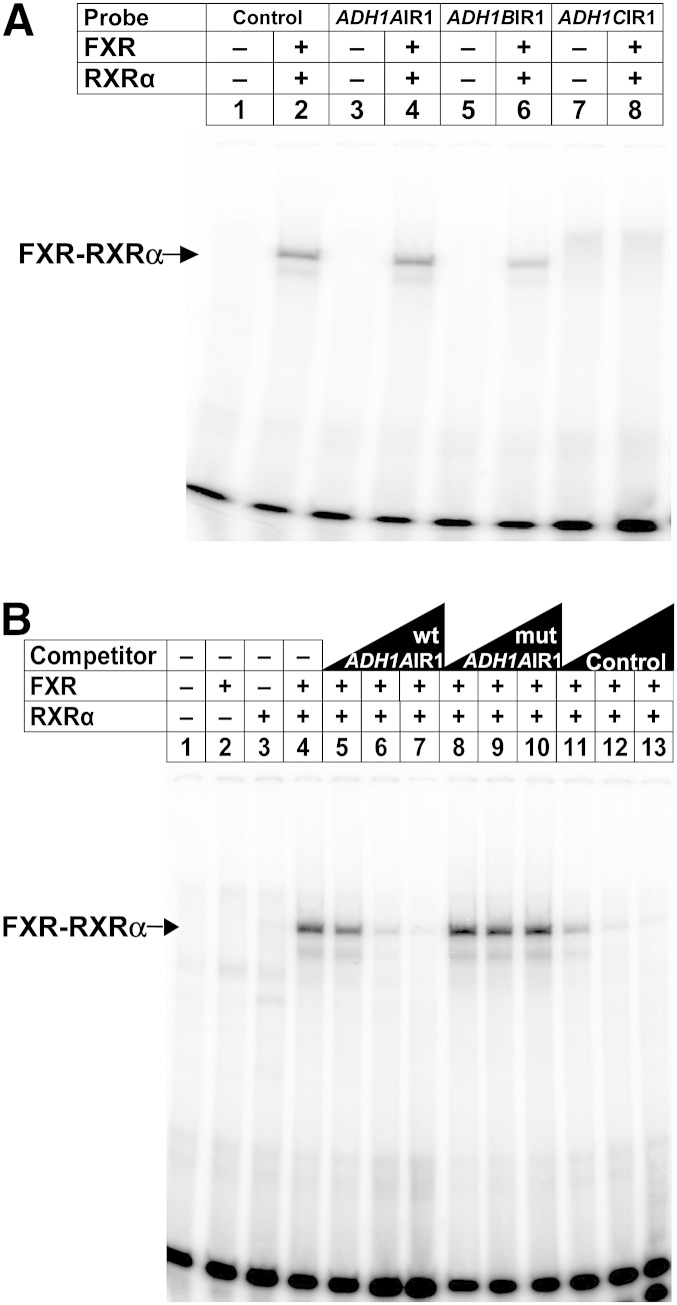
Binding analysis of FXR to the IR1s in the ADH1A and ADH1B promoters
To determine whether FXR-RXR heterodimers directly bind the IR1s identified in the proximal promoters of ADH1 genes, EMSAs were performed. Incubation of in vitro translated FXR and RXR together with radiolabeled double-stranded oligonucleotides containing the IR1 located at the proximal promoter of ADH1A or ADH1B resulted in a specific retarded complex (Fig. 6A, lanes 4 and 6, respectively). In contrast, the probe containing the IR1 in the ADH1C promoter failed to form a specific DNA protein complex (Fig. 6A, lane 8). The well-characterized IR1 from I-BABP served as a positive control (Fig. 6A, lane 2) (30). As expected, more detailed gel-shift analyses demonstrated that FXR indeed binds as a heterodimer with RXR to these IR1s because the appearance of a robust retarded complex with ADH1AIR1 (Fig. 6B, lane 4) or ADH1BIR1 (data not shown) required the presence of both FXR and RXR proteins. The specificity of these complexes was demonstrated by competition with increasing concentrations of either the unlabeled oligonucleotide dimers corresponding to the wild-type IR1 or I-BABP FXRE probes (Fig. 6B, lanes 5–7 and lanes 11–13), whereas no disappearance of the retarded complexes was appreciated when a mutated IR1 was used as a competitor (Fig. 6B, lanes 8–10).
Fig. 6.
The FXR/RXR heterodimer binds to the IR1 element in the proximal promoter of ADH1A and ADH1B. A: EMSAs were performed using in vitro transcribed/translated FXR, RXRα, or unprogrammed reticulocyte lysate (−), when indicated, and labeled double-stranded oligonucleotides containing the ADH1A, ADH1B, or ADH1C IR1 element. Control, FXRE of I-BABP. Note that the faint retarded bands of probe ADH1CIR1 (lanes 7 and 8) are nonspecific. The specific FXR-RXRα-FXRE complex is indicated by an arrow. B: EMSAs were performed as in (A) with a radiolabeled probe for ADH1A IR1 element. Competition experiments were performed by adding a 10-, 50-, and 250-fold molar excess of the indicated unlabeled probes. Mutations present in the modified version of ADH1A IR1 (mutADH1AIR1) are shown in Fig. 5D. Control, FXRE of I-BABP.
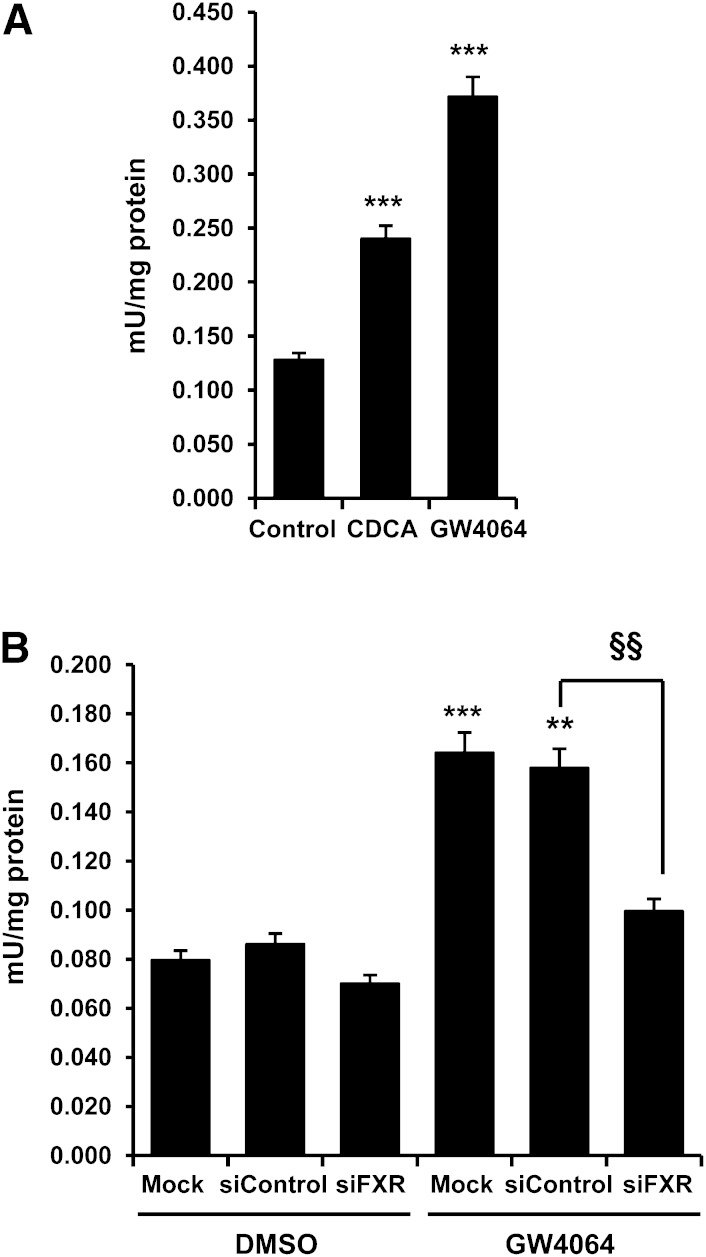
Activation of FXR increases ADH1 enzymatic activity
Having shown that activation of FXR increases ADH1 mRNA and protein levels, we next investigated whether the activity of the enzyme was also increased. Exposure of Huh7 cells to CDCA or GW4064 resulted in an increase of alcohol dehydrogenase activity (Fig. 7A). To confirm that such increase was dependent on FXR, we transfected Huh7 cells with FXR-specific or nontargeting siRNA prior to treatments with GW4064. As depicted in Fig. 7B, when FXR expression was silenced, the stimulatory effect of GW4064 on ADH activity was completely abolished. Thus, we conclude that FXR regulates not only ADH1 expression but also the overall cellular ADH activity.
Fig. 7.
Activation of FXR increases ADH1 enzymatic activity. A: FXR agonists increase ADH1 activity. Huh7 cells were treated with vehicle (Control), 50 μM CDCA, or 5 μM GW4064 for 48 h. Cells lysates were prepared and assayed for alcohol dehydrogenase activity as described in Materials and Methods. ***P < 0.001 versus untreated control. B: Knockdown of FXR abrogates the stimulatory effects of GW4064 treatment on ADH1 activity. Huh7 cells were transfected with either control siRNA (siControl) or siFXR. At 48 h posttransfection, cells were treated with vehicle or 5 μM GW4064 for 24 h, and ADH1 activity was determined. Values are normalized to protein content and expressed as mean ± SEM. **P < 0.01; ***P < 0.001 versus untreated control; §§P < 0.01 versus siControl. Mock indicates untransfected cells. The results are representative of at least two independent experiments from triplicate dishes.
DISCUSSION
The present study identifies the human ADH1A and ADH1B genes as direct targets of the bile acid-activated nuclear receptor FXR. The evidence for this finding is the following: a) the expression of ADH1A and ADH1B is induced in HepG2 cells, Huh7 cells, and primary human hepatocytes in response to natural and synthetic FXR ligands; b) overexpression of constitutively active FXR augments ADH1A and ADH1B mRNA levels; c) IR1 elements identified at the proximal promoters of ADH1A and ADH1B are able to confer FXR response and bind FXR-RXR heterodimers; d) alcohol dehydrogenase activity is increased upon FXR activation; and e) silencing of FXR abolished the effects of FXR agonists on ADH1 expression and activity.
At the present time, the reason why FXR regulates ADH1 genes is not immediately obvious. Previous studies have shown that 3β-dehydrogenation of iso bile acids is catalyzed by ADH1C isoenzyme (19, 20), yet we have failed to observe significant changes in ADH1C expression in response to FXR agonists. Moreover, although ADH1B has been shown to catalyze a step of bile acid synthesis (18), it is counterintuitive to presume that this is the explanation for the FXR-dependent regulation of ADH1B given the inhibitory effects that FXR exerts on the biosynthesis of bile acids (10). In fact, more recent reports have concluded that ADH1 activity may be dispensable for bile acid biosynthesis, because the bile acid pool is unchanged when ADH1 is absent (31), and because mitochondrial CYP27 performs all steps in the formation of 3α,7α,12α-trihydroxy-5β-cholestanoic acid from the corresponding 3α,7α,12α-triol (32, 33). Nevertheless, given the bile acid detoxifying role of FXR (16), we cannot exclude that the regulation of ADH1 genes by FXR responds to protective pathways whereby ADH1A and ADH1B isoenzymes metabolize yet unknown bile alcohols and/or bile acids to less toxic products. In agreement with this hypothesis, several bile alcohols and bile acids that are intermediates in the bile acid synthetic pathway have been identified as highly efficacious ligands for FXR (34). Also in this regard, it is interesting to consider the sequential turn-on of ADH1 genes during liver development. Whereas ADH1A isoenzyme is found during early stages of fetal liver development and ADH1B appears at later fetal stages, ADH1C is only detected several months postnatally (4). Therefore, it is tempting to speculate that the isoenzyme-specific regulation by FXR that we observe may correspond to a mechanism of protection during fetal liver development. In this context, it is worth noting that fetal bile acid synthesis differs markedly from that of the adult, and “atypical” bile acids are found in human fetal bile. As an example, C-4 hydroxylated bile acids, which account for 5–15% of the total biliary bile acids of the fetus, are exclusive to early human development (35). Hence, it will be of considerable interest for future liver development studies to determine whether human ADH1 isoenzymes metabolize fetal specific bile acids.
Strikingly, the FXR-dependent induction of ADH1 genes is species specific, because rodent hepatic Adh1 mRNA levels were not increased after activation of FXR (Fig. 4). Such a species-dependent regulation has also been observed for other human FXR target genes, including syndecan-1 (36), fibrinogen (37), αA-crystallin (38), peroxisome proliferator-activated receptor α (39), hepatic lipase (40), ALAS1 (29), fetuin-b (41), and PCSK9 (42). The reason for such expansion in the repertoire of FXR targets in humans, compared with rodents, remains obscure. Probably, it is a reflection of species differences in bile acid composition as well as the novel roles that bile acids have acquired as signaling molecules in humans. Interestingly, a number of studies have focused on the evolutionary history of ADH1 in order to understand specific processes regarding primate adaptation to dietary alcohols (43). The three human ADH1 paralogs mainly originated from sequential duplications of an ancestral ADH1 gene after the divergence between rodents and primates. Several lines of evidence indicate that the first split was between ADH1C and the gene that gave rise ADH1B and ADH1A (43). The current findings that FXR induces ADH1A and ADH1B but not ADH1C correlates with this evolutionary mechanism that places ADH1C as the outgroup. Consequently, it is tempting to postulate that the FXR response appeared after the first split and before the split between ADH1A and ADH1B.
Inasmuch as ADH1 enzymes catalyze the rate-limiting steps of retinol and ethanol metabolism in humans (3, 17), the current data suggest that the activation of FXR is likely to enhance the metabolism of retinol and ethanol. On the other hand, we did not observe a significant effect of FXR activators in the mRNA levels of CYP2E1, an enzyme that also contributes to ethanol metabolism in chronic alcohol ingestion. In addition, incubation of human cells with FXR agonists failed to increase ALDH1A1 or ALDH2 mRNA levels. Therefore, additional studies will be required to define whether treatments with FXR ligands might effectively modify ethanol metabolism in vivo.
In conclusion, our data reveal a direct stimulatory role for FXR on ADH1, which points to a possible involvement of ADH1 in bile alcohol and/or bile acid catabolism. Furthermore, the FXR-dependent activation of ADH1 also suggests a potential effect on ethanol metabolism that should be taken into account in future clinical trials with FXR modulators.
Acknowledgments
The authors thank GlaxoSmithKline for providing expression plasmids and GW4064, and Dr. Antonio Moschetta (Consorzio Mario Negri Sud, Chieti, Italy) for the generous gift of adenoviruses. The authors also thank Dr. Xavier Parés (Autonomous University of Barcelona, Bellaterra, Barcelona, Spain) and Dr. Ángel Baldán (Saint Louis University, Saint Louis, MO) for helpful discussions and Albert Pérez-Martí (University of Barcelona, Barcelona, Spain) for help with cell culture experiments.
Footnotes
Abbreviations:
- ADH
- alcohol dehydrogenase
- ADH1
- class I alcohol dehydrogenase
- CDCA
- chenodeoxycholic acid
- EMSA
- electrophoretic mobility shift assay
- FXR
- farnesoid X receptor
- FXRE
- farnesoid X receptor response element
- I-BABP
- ileal-bile acid-binding protein
- IR1
- inverted hexameric nucleotide repeat separated by one nucleotide
- RXR
- retinoid X receptor
- SHP
- small heterodimer partner
- siFXR
- small interfering RNA complexes directed against farnesoid X receptor
- siRNA
- small interfering RNA
This work was supported by grants from the Spanish government (SAF2008-04606) and the Catalan government (2009SGR163). E.P-C. was supported by an Ajut pel Personal Investigador en Formació (APIF) Scholarship from the University of Barcelona. J.C.R. was supported by the Ramón y Cajal Program from the Spanish government. The authors declare that they have no competing financial interests.
REFERENCES
- 1.Edenberg H. J. 2000. Regulation of the mammalian alcohol dehydrogenase genes. Prog. Nucleic Acid Res. Mol. Biol. 64: 295–341. [DOI] [PubMed] [Google Scholar]
- 2.Duester G., Farres J., Felder M. R., Holmes R. S., Hoog J. O., Pares X., Plapp B. V., Yin S. J., Jornvall H. 1999. Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem. Pharmacol. 58: 389–395. [DOI] [PubMed] [Google Scholar]
- 3.Lee S. L., Chau G. Y., Yao C. T., Wu C. W., Yin S. J. 2006. Functional assessment of human alcohol dehydrogenase family in ethanol metabolism: significance of first-pass metabolism. Alcohol. Clin. Exp. Res. 30: 1132–1142. [DOI] [PubMed] [Google Scholar]
- 4.Smith M., Hopkinson D. A., Harris H. 1971. Developmental changes and polymorphism in human alcohol dehydrogenase. Ann. Hum. Genet. 34: 251–271. [DOI] [PubMed] [Google Scholar]
- 5.Stewart M. J., McBride M. S., Winter L. A., Duester G. 1990. Promoters for the human alcohol dehydrogenase genes ADH1, ADH2, and ADH3: interaction of CCAAT/enhancer-binding protein with elements flanking the ADH2 TATA box. Gene. 90: 271–279. [DOI] [PubMed] [Google Scholar]
- 6.Brown C. J., Zhang L., Edenberg H. J. 1996. Gene expression in a young multigene family: tissue-specific differences in the expression of the human alcohol dehydrogenase genes ADH1, ADH2, and ADH3. DNA Cell Biol. 15: 187–196. [DOI] [PubMed] [Google Scholar]
- 7.van Ooij C., Snyder R. C., Paeper B. W., Duester G. 1992. Temporal expression of the human alcohol dehydrogenase gene family during liver development correlates with differential promoter activation by hepatocyte nuclear factor 1, CCAAT/enhancer-binding protein alpha, liver activator protein, and D-element-binding protein. Mol. Cell. Biol. 12: 3023–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su J. S., Tsai T. F., Chang H. M., Chao K. M., Su T. S., Tsai S. F. 2006. Distant HNF1 site as a master control for the human class I alcohol dehydrogenase gene expression. J. Biol. Chem. 281: 19809–19821. [DOI] [PubMed] [Google Scholar]
- 9.Lefebvre P., Cariou B., Lien F., Kuipers F., Staels B. 2009. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89: 147–191. [DOI] [PubMed] [Google Scholar]
- 10.Kalaany N. Y., Mangelsdorf D. J. 2006. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu. Rev. Physiol. 68: 159–191. [DOI] [PubMed] [Google Scholar]
- 11.Moschetta A., Bookout A. L., Mangelsdorf D. J. 2004. Prevention of cholesterol gallstone disease by FXR agonists in a mouse model. Nat. Med. 10: 1352–1358. [DOI] [PubMed] [Google Scholar]
- 12.Kovacs P., Kress R., Rocha J., Kurtz U., Miquel J. F., Nervi F., Mendez-Sanchez N., Uribe M., Bock H. H., Schirin-Sokhan R., et al. 2008. Variation of the gene encoding the nuclear bile salt receptor FXR and gallstone susceptibility in mice and humans. J. Hepatol. 48: 116–124. [DOI] [PubMed] [Google Scholar]
- 13.Jonker J. W., Liddle C., Downes M. 2012. FXR and PXR: potential therapeutic targets in cholestasis. J. Steroid Biochem. Mol. Biol. 130: 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adorini L., Pruzanski M., Shapiro D. 2012. Farnesoid X receptor targeting to treat nonalcoholic steatohepatitis. Drug Discov. Today. 17: 988–997. [DOI] [PubMed] [Google Scholar]
- 15.Gnerre C., Blattler S., Kaufmann M. R., Looser R., Meyer U. A. 2004. Regulation of CYP3A4 by the bile acid receptor FXR: evidence for functional binding sites in the CYP3A4 gene. Pharmacogenetics. 14: 635–645. [DOI] [PubMed] [Google Scholar]
- 16.Lee F. Y., de Aguiar Vallim T. Q., Chong H. K., Zhang Y., Liu Y., Jones S. A., Osborne T. F., Edwards P. A. 2010. Activation of the farnesoid X receptor provides protection against acetaminophen-induced hepatic toxicity. Mol. Endocrinol. 24: 1626–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mezey E., Holt P. R. 1971. The inhibitory effect of ethanol on retinol oxidation by human liver and cattle retina. Exp. Mol. Pathol. 15: 148–156. [DOI] [PubMed] [Google Scholar]
- 18.Okuda A., Okuda K. 1983. Physiological function and kinetic mechanism of human liver alcohol dehydrogenase as 5 beta-cholestane-3 alpha,7 alpha,12 alpha,26-tetrol dehydrogenase. J. Biol. Chem. 258: 2899–2905. [PubMed] [Google Scholar]
- 19.McEvily A. J., Holmquist B., Auld D. S., Vallee B. L. 1988. 3 beta-Hydroxy-5 beta-steroid dehydrogenase activity of human liver alcohol dehydrogenase is specific to gamma-subunits. Biochemistry. 27: 4284–4288. [DOI] [PubMed] [Google Scholar]
- 20.Marschall H. U., Oppermann U. C., Svensson S., Nordling E., Persson B., Hoog J. O., Jornvall H. 2000. Human liver class I alcohol dehydrogenase gammagamma isozyme: the sole cytosolic 3beta-hydroxysteroid dehydrogenase of iso bile acids. Hepatology. 31: 990–996. [DOI] [PubMed] [Google Scholar]
- 21.Orellana-Gavaldà J. M., Herrero L., Malandrino M. I., Paneda A., Sol Rodriguez-Pena M., Petry H., Asins G., Van Deventer S., Hegardt F. G., Serra D. 2011. Molecular therapy for obesity and diabetes based on a long-term increase in hepatic fatty-acid oxidation. Hepatology. 53: 821–832. [DOI] [PubMed] [Google Scholar]
- 22.Modica S., Murzilli S., Salvatore L., Schmidt D. R., Moschetta A. 2008. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 68: 9589–9594. [DOI] [PubMed] [Google Scholar]
- 23.Prieur X., Coste H., Rodriguez J. C. 2003. The human apolipoprotein AV gene is regulated by peroxisome proliferator-activated receptor-alpha and contains a novel farnesoid X-activated receptor response element. J. Biol. Chem. 278: 25468–25480. [DOI] [PubMed] [Google Scholar]
- 24.Urizar N. L., Dowhan D. H., Moore D. D. 2000. The farnesoid X-activated receptor mediates bile acid activation of phospholipid transfer protein gene expression. J. Biol. Chem. 275: 39313–39317. [DOI] [PubMed] [Google Scholar]
- 25.Wang L., Lee Y. K., Bundman D., Han Y., Thevananther S., Kim C. S., Chua S. S., Wei P., Heyman R. A., Karin M., et al. 2002. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell. 2: 721–731. [DOI] [PubMed] [Google Scholar]
- 26.Dwivedi S. K., Singh N., Kumari R., Mishra J. S., Tripathi S., Banerjee P., Shah P., Kukshal V., Tyagi A. M., Gaikwad A. N., et al. 2011. Bile acid receptor agonist GW4064 regulates PPARgamma coactivator-1alpha expression through estrogen receptor-related receptor alpha. Mol. Endocrinol. 25: 922–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laffitte B. A., Kast H. R., Nguyen C. M., Zavacki A. M., Moore D. D., Edwards P. A. 2000. Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J. Biol. Chem. 275: 10638–10647. [DOI] [PubMed] [Google Scholar]
- 28.Thomas A. M., Hart S. N., Kong B., Fang J., Zhong X. B., Guo G. L. 2010. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology. 51: 1410–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peyer A. K., Jung D., Beer M., Gnerre C., Keogh A., Stroka D., Zavolan M., Meyer U. A. 2007. Regulation of human liver delta-aminolevulinic acid synthase by bile acids. Hepatology. 46: 1960–1970. [DOI] [PubMed] [Google Scholar]
- 30.Grober J., Zaghini I., Fujii H., Jones S. A., Kliewer S. A., Willson T. M., Ono T., Besnard P. 1999. Identification of a bile acid-responsive element in the human ileal bile acid-binding protein gene. Involvement of the farnesoid X receptor/9-cis-retinoic acid receptor heterodimer. J. Biol. Chem. 274: 29749–29754. [DOI] [PubMed] [Google Scholar]
- 31.Sjövall J., Andersson S. H., Lieber C. S. 1985. Bile acids in deermice lacking liver alcohol dehydrogenase. Biochim. Biophys. Acta. 836: 8–13. [DOI] [PubMed] [Google Scholar]
- 32.Holmberg-Betsholtz I., Lund E., Bjorkhem I., Wikvall K. 1993. Sterol 27-hydroxylase in bile acid biosynthesis. Mechanism of oxidation of 5 beta-cholestane-3 alpha,7 alpha,12 alpha,27-tetrol into 3 alpha,7 alpha,12 alpha-trihydroxy-5 beta-cholestanoic acid. J. Biol. Chem. 268: 11079–11085. [PubMed] [Google Scholar]
- 33.Pikuleva I. A., Babiker A., Waterman M. R., Bjorkhem I. 1998. Activities of recombinant human cytochrome P450c27 (CYP27) which produce intermediates of alternative bile acid biosynthetic pathways. J. Biol. Chem. 273: 18153–18160. [DOI] [PubMed] [Google Scholar]
- 34.Nishimaki-Mogami T., Une M., Fujino T., Sato Y., Tamehiro N., Kawahara Y., Shudo K., Inoue K. 2004. Identification of intermediates in the bile acid synthetic pathway as ligands for the farnesoid X receptor. J. Lipid Res. 45: 1538–1545. [DOI] [PubMed] [Google Scholar]
- 35.Setchell K. D., Dumaswala R., Colombo C., Ronchi M. 1988. Hepatic bile acid metabolism during early development revealed from the analysis of human fetal gallbladder bile. J. Biol. Chem. 263: 16637–16644. [PubMed] [Google Scholar]
- 36.Anisfeld A. M., Kast-Woelbern H. R., Meyer M. E., Jones S. A., Zhang Y., Williams K. J., Willson T., Edwards P. A. 2003. Syndecan-1 expression is regulated in an isoform-specific manner by the farnesoid-X receptor. J. Biol. Chem. 278: 20420–20428. [DOI] [PubMed] [Google Scholar]
- 37.Anisfeld A. M., Kast-Woelbern H. R., Lee H., Zhang Y., Lee F. Y., Edwards P. A. 2005. Activation of the nuclear receptor FXR induces fibrinogen expression: a new role for bile acid signaling. J. Lipid Res. 46: 458–468. [DOI] [PubMed] [Google Scholar]
- 38.Lee F. Y., Kast-Woelbern H. R., Chang J., Luo G., Jones S. A., Fishbein M. C., Edwards P. A. 2005. Alpha-crystallin is a target gene of the farnesoid X-activated receptor in human livers. J. Biol. Chem. 280: 31792–31800. [DOI] [PubMed] [Google Scholar]
- 39.Pineda Torra I., Claudel T., Duval C., Kosykh V., Fruchart J. C., Staels B. 2003. Bile acids induce the expression of the human peroxisome proliferator-activated receptor alpha gene via activation of the farnesoid X receptor. Mol. Endocrinol. 17: 259–272. [DOI] [PubMed] [Google Scholar]
- 40.Sirvent A., Verhoeven A. J., Jansen H., Kosykh V., Darteil R. J., Hum D. W., Fruchart J. C., Staels B. 2004. Farnesoid X receptor represses hepatic lipase gene expression. J. Lipid Res. 45: 2110–2115. [DOI] [PubMed] [Google Scholar]
- 41.Murakami T., Walczak R., Caron S., Duhem C., Vidal V., Darteil R., Staels B. 2007. The farnesoid X receptor induces fetuin-B gene expression in human hepatocytes. Biochem. J. 407: 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langhi C., Le May C., Kourimate S., Caron S., Staels B., Krempf M., Costet P., Cariou B. 2008. Activation of the farnesoid X receptor represses PCSK9 expression in human hepatocytes. FEBS Lett. 582: 949–955. [DOI] [PubMed] [Google Scholar]
- 43.Oota H., Dunn C. W., Speed W. C., Pakstis A. J., Palmatier M. A., Kidd J. R., Kidd K. K. 2007. Conservative evolution in duplicated genes of the primate class I ADH cluster. Gene. 392: 64–76. [DOI] [PubMed] [Google Scholar]