SUMMARY
Rice (Oryza sativa) produces momilactone diterpenoids as both phytoalexins and allelochemicals. Strikingly, the rice genome contains a biosynthetic gene cluster for momilactone production, located on rice chromosome 4, which contains two cytochromes P450 mono-oxygenases, CYP99A2 and CYP99A3, with undefined roles; although it has been previously shown that RNAi double knock-down of this pair of closely related CYP reduced momilactone accumulation. Here we attempted biochemical characterization of CYP99A2 and CYP99A3, which ultimately was achieved by complete gene recoding, enabling functional recombinant expression in bacteria. With these synthetic gene constructs it was possible to demonstrate that, while CYP99A2 does not exhibit significant activity with diterpene substrates, CYP99A3 catalyzes consecutive oxidations of the C19 methyl group of the momilactone precursor syn-pimara-7,15-diene to form, sequentially, syn-pimaradien-19-ol, syn-pimaradien-19-al and syn-pimaradien-19-oic acid. These are presumably intermediates in momilactone biosynthesis, as a C19 carboxylic acid moiety is required for formation of the core 19,6-γ-lactone ring structure. We further were able to detect syn-pimaradien-19-oic acid in rice plants, which indicates physiological relevance for the observed activity of CYP99A3. In addition, we found that CYP99A3 also oxidized syn-stemod-13(17)-ene at C19 to produce, sequentially, syn-stemoden-19-ol, syn-stemoden-19-al and syn-stemoden-19-oic acid, albeit with lower catalytic efficiency than with syn-pimaradiene. Although the CYP99A3 syn-stemodene derived products were not detected in planta, these results nevertheless provide a hint at the currently unknown metabolic fate of this diterpene in rice. Regardless of any wider role, our results strongly indicate that CYP99A3 acts as a multifunctional diterpene oxidase in momilactone biosynthesis.
Keywords: cytochrome P450, biosynthetic gene cluster, diterpenoid metabolism, natural products, phytoalexin, momilactone
INTRODUCTION
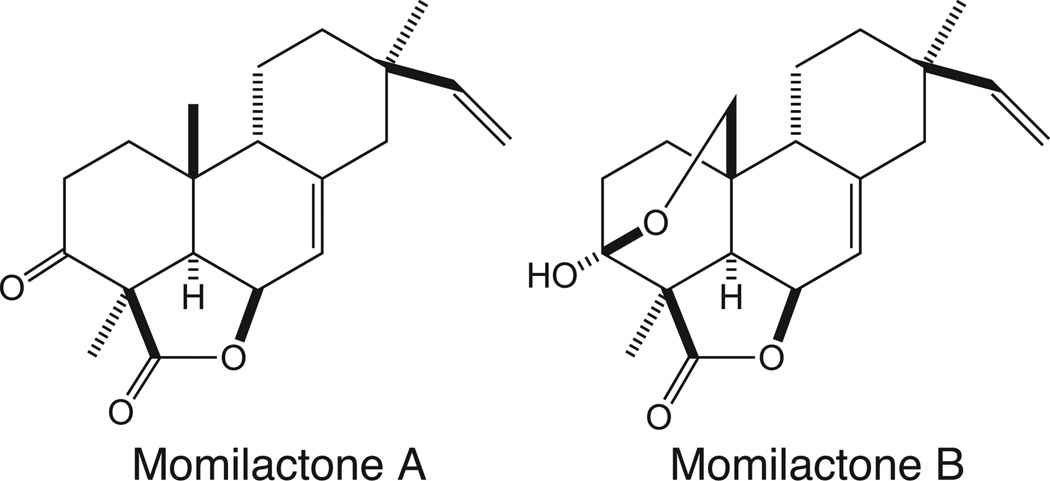
Rice is the single most important source of human food, providing ~20% of our caloric intake worldwide (FAO 2004). Unfortunately, 10–30% of the global rice harvest is lost every year to various microbial infections, with much of the loss resulting from the blast disease caused by the fungal pathogen Magnaporthe grisea (Talbot 2003). Rice produces a number of antimicrobial natural products in response to infection by M. grisea (Peters 2006), with the first such phytoalexins to be identified being momilactones A and B (Cartwright et al. 1981). These diterpenoid natural products had previously been reported to act as plant growth inhibitors (Kato et al. 1973), and more recent work indicates that the momilactones are constitutively secreted from the roots (Kato-Noguchi and Ino 2003). Thus, the momilactones also may act as allelochemicals as well as phytoalexins, and such dual activity would presumably provide increased selective pressure for their production.
With the sequencing of the rice genome (IRGSP 2005), it has been possible to take a functional genomics approach towards elucidating the biosynthetic machinery underlying the diterpenoid metabolism of this plant species (Peters 2006). Momilactones A and B (Figure 1), along with many other rice phytoalexins, are representatives of the labdane-related diterpenoid super-family of natural products, whose characteristic biosynthetic feature is an initial pair of sequential cyclization reactions. Specifically, bicyclization of the universal diterpenoid precursor (E,E,E)-geranylgeranyl diphosphate (GGPP) by copalyl/labdadienyl diphosphate synthases (CPS), which is followed by further cyclization and/or rearrangement to a polycyclic olefin catalyzed by enzymes often termed kaurene synthase like (KSL) for their relationship to the presumably ancestral kaurene synthase from gibberellin phytohormone metabolism. Production of the bioactive diterpenoid phytoalexins then further requires the action of cytochrome P450 (CYP) mono-oxygenases, as well as subsequently acting short chain alcohol dehydrogenases/reductases (SDR).
Figure 1.
Momilactones A and B.
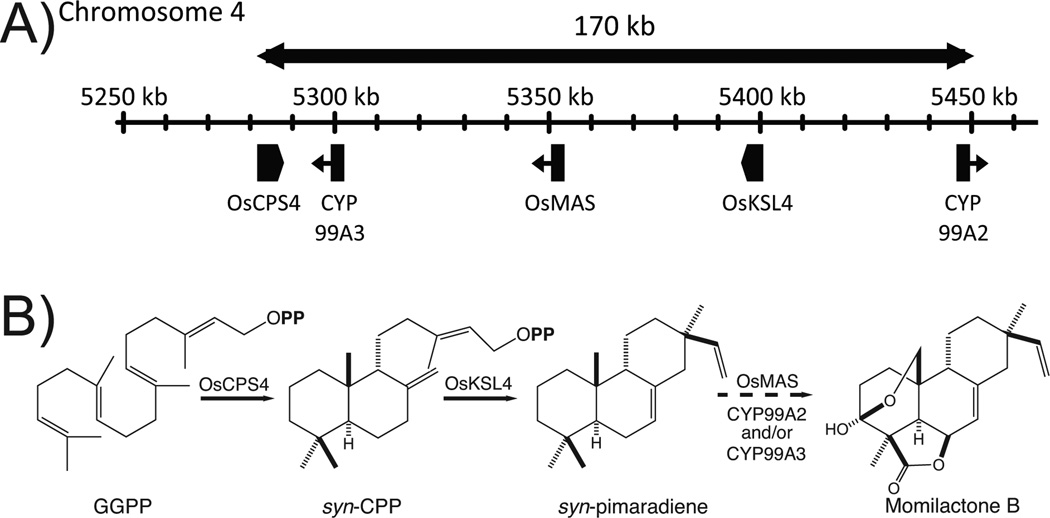
Surprisingly, it has been reported that there are two clusters of diterpenoid phytoalexin biosynthetic genes in the rice genome, which is an unusual occurrence in plants (Osbourn and Field 2009). It was first reported that the co-regulated (i.e., inducible) syn-CPP producing OsCPS4 and subsequently acting syn-pimara-7,15-diene synthase OsKSL4 involved in momilactone biosynthesis are found close together on chromosome 4 (Wilderman et al. 2004). Later work then demonstrated the neighboring CYP99A2, CYP99A3, and SDR (OsMAS), also are co-regulated and involved in momilactone biosynthesis (Figure 2); although the relevant biochemical function was only defined for the SDR, with an undefined role(s) for the CYP indicated by greatly reduced momilactone production in lines wherein RNAi had been used to knock-down transcription of both CYP99A2 and CYP99A3 (Shimura et al. 2007). Similarly, there is a CPS, KSL, and CYP containing gene cluster on chromosome 2, which contains the ent-CPP producing OsCPS2 and subsequently acting ent-cassa-12,15-diene synthase OsKSL7 (Prisic et al. 2004), as well as ent-cassadiene 11α-hydroxylase CYP76M7 (Swaminathan et al. 2009).
Figure 2.
Momilactone biosynthetic gene cluster and pathway. (A) Biosynthetic gene cluster on rice chromosome 4 associated with momilactone biosynthesis. (B) Momilactone B biosynthesis, shown with known transformations and enzymes.
Here we report functional identification of CYP99A3 as a diterpene oxidase that reacts with syn-pimaradiene to transform C19 from a methyl to carboxylic acid, forming syn-pimaradien-19-oic acid, which presumably is an early intermediate in momilactone biosynthesis. In addition, CYP99A3 carries out the same series of reactions to form a C19 carboxylic acid with syn-stemod-13(17)-ene, providing evidence regarding the currently unknown metabolic fate of this diterpene in rice.
RESULTS
Lack of recombinant activity with native gene constructs
We initiated our attempt to functionally characterize the role of CYP99A2 and CYP99A3 in momilactone biosynthesis by obtaining the corresponding full-length clones from the rice cDNA database (Kikuchi et al. 2003). These were then recombinantly expressed in insect (Spodoptera frugiperda) cell culture, just as described for CYP76M7 (Swaminathan, et al. 2009). However, despite being able to detect expression of the corresponding heterologous proteins (Figure S1), neither lysates nor microsomal preparations from these recombinant cells exhibited any activity with the syn-pimaradiene produced by the OsCPS4 and OsKSL4 that are co-clustered with CYP99A2 and CYP99A3, even with supplementation of a rice CYP-NADPH reductase (OsCPR1) to ensure provision of the necessary reducing equivalents. These recombinant preparations also did not react with several of the other diterpenes produced by rice (i.e. ent-sandaracopimaradiene and ent-cassadiene).
As an alternative approach, we turned to our modular metabolic engineering system (Cyr et al. 2007), which we have recently shown is amenable to functional co-expression of CYP with not only their CPR redox partner, but also with various combinations of diterpene synthases for the production of putative substrates (Swaminathan, et al. 2009). Unfortunately, when this was done with the native CYP99A2 and CYP99A3 gene constructs, which were each co-expressed with OsCPR1, we were unable to detect any enzymatic activity against any of the known rice diterpenes (i.e. syn-pimaradiene, syn-stemodene, syn-stemarene, ent-sandaracopimaradiene, ent-cassadiene, ent-isokaurene, ent-pimaradiene, and ent-kaurene). Nor was activity observed with N-terminally modified native CYP99A2 and CYP99A3 constructs designed on the basis of our previous functional bacterial expression of plant CYP (Morrone et al. in press, Swaminathan, et al. 2009), specifically replacement of the 5' sequence encoding the N-terminal membrane anchor region by a codon optimized sequence encoding a 10 amino acid long lysine and serine rich leader peptide.
Synthetic gene constructs enabled functional folding
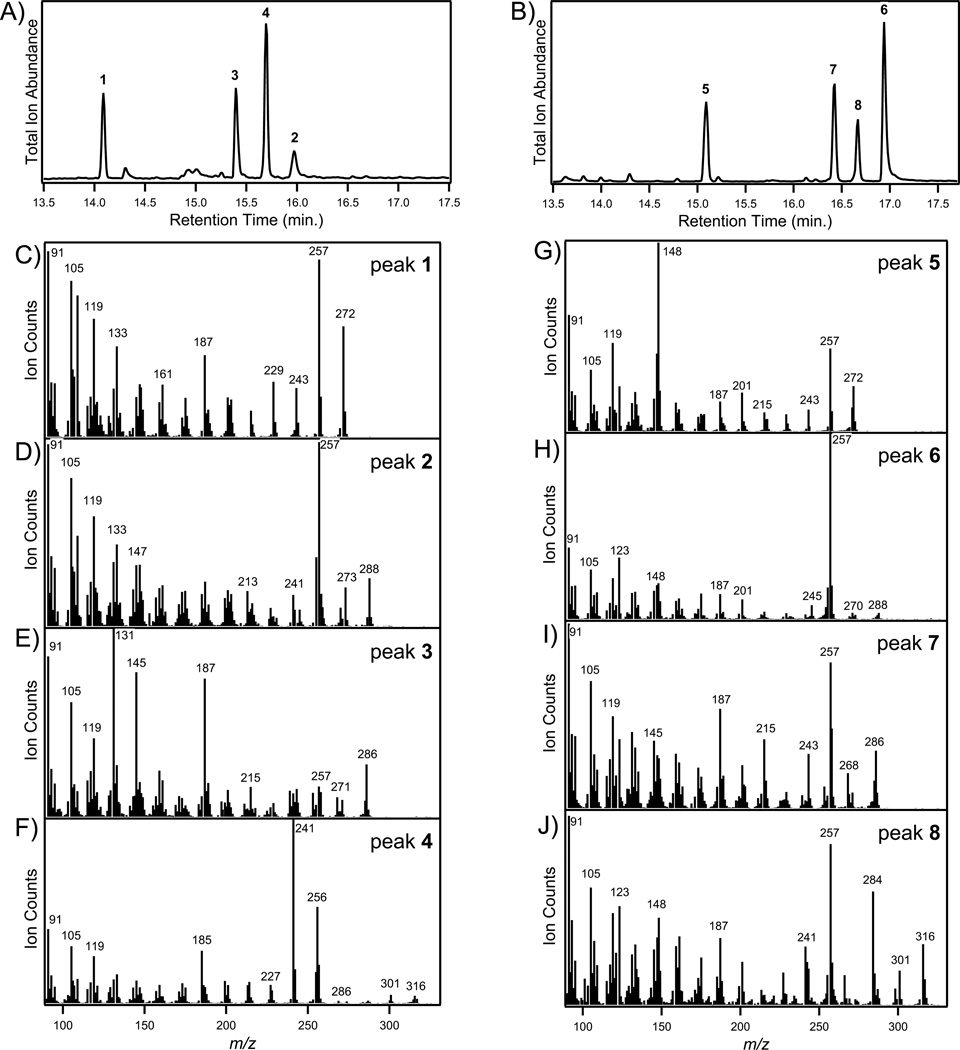
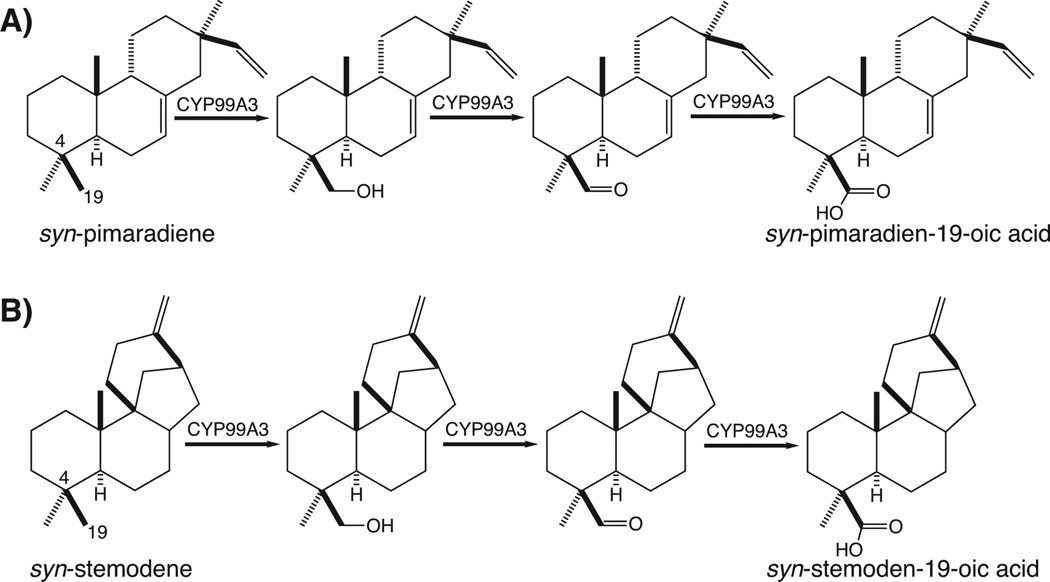
It has been reported that codon optimization improves activity in recombinant bacterial expression of plant CYP (Chang et al. 2007), which we also have observed in our own work with the CYP701A3 ent-kaurene oxidase from Arabidopsis thaliana (Morrone, et al. in press). Accordingly, we synthesized completely recoded genes, codon-optimized for expression in E. coli, for both CYP99A2 and CYP99A3 (see Supporting Information for the corresponding nucleotide sequences; Figures S2 and S3). These both were then N-terminally modified, as described above. Upon recombinant expression in E. coli along with OsCPR1, the resulting rCYP99A3 clearly oxidized syn-pimaradiene to a mixture of three products (Figure 3A), with masses corresponding to formation of a diterpenoid alcohol (MW = 288 Da), aldehyde (MW = 286 Da), and acid (detected as the methyl ester with MW = 316 Da), although only trace formation of two putative diterpenoid alcohols (one of which is the same as the alcohol made by rCYP99A3) was observed upon feeding syn-pimaradiene to the analogously modified rCYP99A2. In addition, while rCYP99A2 does not react with any other diterpene, rCYP99A3 reacts with syn-stemodene to produce a mixture of three products (Figure 3B), again with masses corresponding to formation of a diterpenoid alcohol (MW = 288 Da), aldehyde (MW = 286 Da), and acid (detected as the methyl ester with MW = 316 Da).
Figure 3.
CYP99A3 catalytic activity. GC-MS chromatograms showing production of oxidized products from, (A) syn-pimara-7,15-diene, and (B) syn-stemod-13(17)-ene. Mass spectra from (C) peak 1 (syn-pimaradiene), (D) peak 2 (syn-pimaradien-19-ol), (E) peak 3 (syn-pimaradien-19-al), (F) peak 4 (syn-pimaradien-19-oic acid, as its methyl ester derivative), (G) peak 5 (syn-stemodene), (H) peak 6 (syn-stemoden-19-ol), (I) peak 7 (syn-stemoden-19-al), and (J) peak 8 (syn-stemoden-19-oic acid, as its methyl ester derivative).
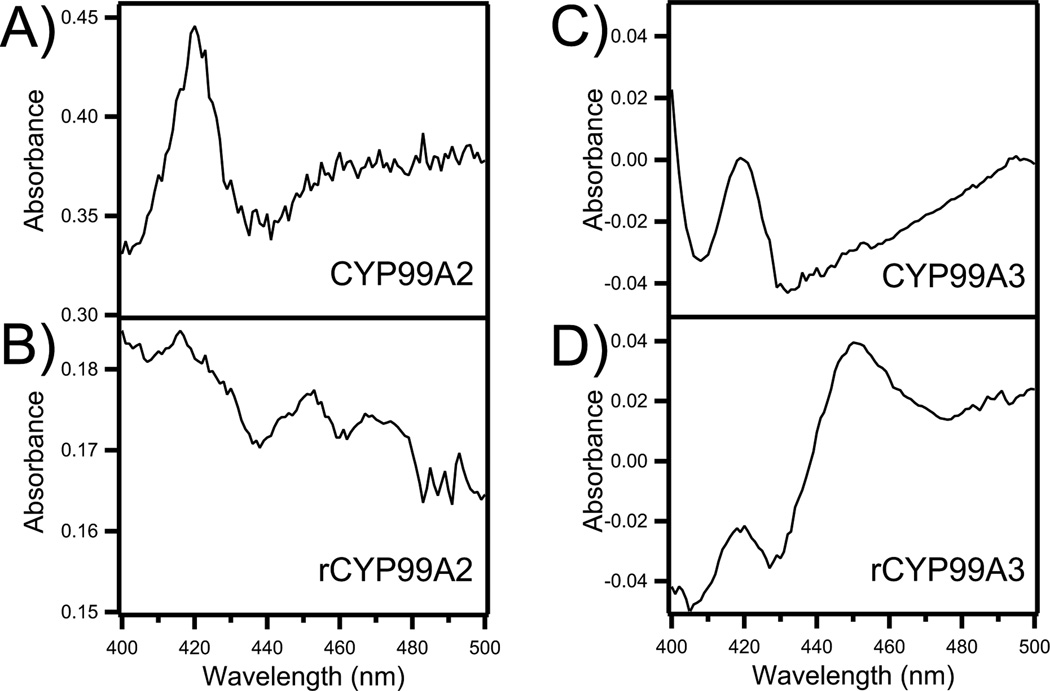
To investigate how codon optimization enabled functional expression, we compared CO-binding difference spectra from microsomal preparations of the native and synthetic versions of both CYP99A2 and CYP99A3, in each case with the N-terminal modification described above (Figure 4). Notably, spectra from both of the native gene constructs led to observation of a prominent peak at 420 nm, without the eponymous peak at 450 nm, indicating misfolding of the recombinant heme-proteins (Munro et al. 2007). By contrast, spectra from both synthetic gene constructs contained a peak at 450 nm, indicating that some proportion of the resulting enzyme is correctly folded. Consistent with the observed relative activity, rCYP99A3 exhibits a more prominent peak at 450 nm than does rCYP99A2, although we have previously observed robust activity even in the absence of such spectra (i.e. with CYP76M7; Swaminathan, et al. 2009). In addition, we also investigated the effect of N-terminal modification on product yield in the context of the engineered bacteria, and found that full-length synthetic CYP99A3 oxidized the same substrates (i.e., syn-pimaradiene and syn-stemodene), and to approximately the same extent, as rCYP99A3. This indicates that N-terminal modification does not significantly alter the enzymatic function, but also suggests that it does not lead to a manifold increase in specific activity, which is consistent with our observations with CYP701A3 where such modification increased activity only modestly (i.e. ~30%; Morrone, et al. in press). Nevertheless, the remaining experiments were all carried out with the N-terminally modified rCYP99A3.
Figure 4.
Characteristic CO-difference binding absorption spectra from reduced E. coli microsomes following recombinant expression of N-terminally modified (A) native CYP99A2, (B) synthetic rCYP99A2, (C) native CYP99A3, and (D) synthetic rCYP99A3.
Product identification
Identification of the rCYP99A3 products was enabled by straightforward scale-up of the metabolic engineered strains that produced the oxygenated diterpenoid products of interest, much as previously described (Swaminathan, et al. 2009). In particular, increasing flux into isoprenoid/terpenoid metabolism leads to overall yields of >10 mg/L of culture (Morrone et al. 2010) and, since greater than 80% of the produced diterpene olefin (i.e., syn-pimaradiene or syn-stemodene) was oxygenated to some degree (Figure 3), it was possible to produce and purify each of the relevant products in sufficient quantities for structural analysis by nuclear magnetic resonance (NMR) spectroscopy. This demonstrated that the products of CYP99A3 with both syn-pimaradiene and syn-stemodene were sequentially modified (i.e., to an alcohol, aldehyde, and carboxylic acid), at one of their C4 geminal methyl groups (Figure S4 and Tables S1–S6). Further analysis of the syn-pimaradienol demonstrated that this was in fact the C4β methyl, corresponding to C19, with similar results obtained for syn-stemodenol as well (Figure 5).
Figure 5.
CYP99A3 catalyzed reactions. (A) Oxidation of syn-pimara-7,15-diene to, sequentially, syn-pimara-7,15-dien-19-ol, syn-pimara-7,15-dien-19-al, and syn-pimara-7,15-dien-19-oic acid. (B) Oxidation of syn-stemod-13(17)-ene to, sequentially, syn-stemod-13(17)-en-19-ol, syn-stemod-13(17)-en-19-al, and syn-stemod-13(17)-en-19-oic acid.
Enzymatic characterization of rCYP99A3
It was possible to measure enzymatic activity in vitro with rCYP99A3. Specifically, using lysates or microsomal preparations from recombinant E. coli expressing rCYP99A3 and OsCPR1. While the full range of rice diterpenes were tested, activity in these in vitro assays was similarly confined to the syn-pimaradiene and syn-stemodene observed as substrates in the metabolically engineered bacterial experiments described above. Notably, the corresponding alcohol and aldehyde products were not observed from in vitro assays, but this is similar to our previous analysis of the analogous methyl to carboxylic acid transformation catalyzed by the kaurene oxidase CYP701A3 (Morrone, et al. in press), and presumably indicates retention of the intermediates during the three oxidation reactions necessary to convert the diterpene olefin substrates to carboxylic acid containing diterpenoid products. Steady state kinetic analysis carried out with both substrates demonstrated that rCYP99A3 exhibited ~4-fold higher catalytic efficiency with syn-pimaradiene than syn-stemodene (Table 1 and Figure S5).
Table 1.
rCYP99A3 steady-state kinetic constants
| Substrate | kcat (s−1) | KM (µM) | kcat/KM |
|---|---|---|---|
| syn-pimaradiene | 46 ± 2 | 2.0 ± 0.5 | 23 |
| syn-stemodene | 49 ± 5 | 9 ± 4 | 5 |
Physiological relevance of observed CYP99A3 activity
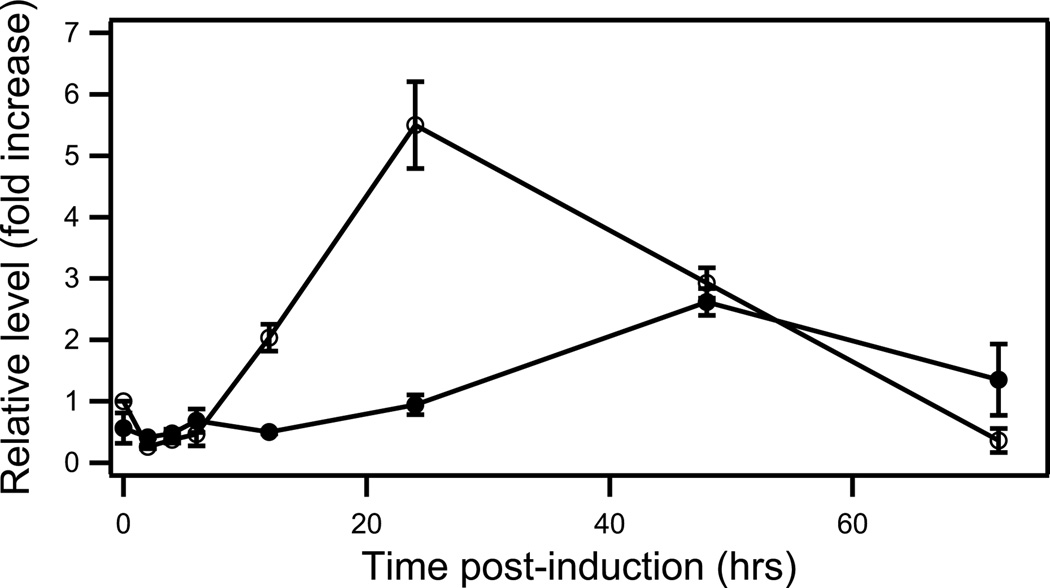
It has been shown that free jasmonic acid induces momilactone biosynthesis (Nojiri et al. 1996), and we have demonstrated that methyl jasmonate induces transcription of the relevant OsCPS4 and OsKSL4 (Wilderman, et al. 2004, Xu et al. 2004). Similarly, we found that the CYP99A3 end product syn-pimaradien-19-oic acid accumulated to a more significant extent in methyl jasmonate induced relative to control rice plants, although syn-stemoden-19-oic acid was not found in either (Figure S6). To investigate the relationship between CYP99A3 and syn-pimaradien-19-oic acid, a methyl jasmonate induction time course was carried, with analysis of both CYP99A3 mRNA level and syn-stemoden-19-oic acid accumulation (Figure 6), which indicated the expected correlation with transient CYP99A3 mRNA accumulation preceding that of the syn-stemoden-19-oic acid, which then seems likely to be an intermediate in a more complex biosynthetic pathway.
Figure 6.
Accumulation of CYP99A3 mRNA (open circles) and syn-pimaradien-19-oic acid (closed circles) in response to methyl jasmonate induction. Shown data points are the average values from duplicate samples, with error bars representing the corresponding standard deviation.
DISCUSSION
It has been previously demonstrated that CYP99A2 and/or CYP99A3 have an unknown role(s) in production of the momilactone phytoalexins/allelochemicals of rice, which helped define an associated biosynthetic gene cluster (Shimura, et al. 2007). Intrigued by this observation, we undertook biochemical characterization of these CYP, guided by the hypothesis that at least one should act on the momilactone precursor syn-pimaradiene that is produced by the co-clustered OsCPS4 and OsKSL4. Notably, while it has been previously reported that codon optimization can improve already observed activity of recombinantly expressed plant CYP (Chang, et al. 2007, Morrone, et al. in press), our results demonstrate that complete gene recoding can lead to activity even when none is observed with the native or even partially modified gene. This is particularly evident with CYP99A3, for which good activity was observed upon such recoding to optimize codon usage for the desired E. coli host. Further, although the similarly recoded CYP99A2 exhibited only trace activity with diterpene olefins, the observation that it is capable of hydroxylating syn-pimaradiene at a different position than CYP99A3 leaves open the possibility that CYP99A2 acts later in momilactone biosynthesis.
The activity exhibited by CYP99A3 with syn-stemodene provides evidence regarding the currently unknown metabolic fate of this particular labdane-related diterpene in rice. In particular, while clearly made by rice (Morrone et al. 2006), syn-stemodene derived natural products have not been reported from this plant species. Although the CYP99A3 syn-stemoden-19-oic acid product was not observed in rice plant extracts, it is possible that the ensuing biosynthetic step is highly efficient and does not allow accumulation of this particular intermediate. Thus, our results suggest that rice syn-stemodene derived natural products may include some in which C19 is a carboxylate or part of a derived structure (e.g., lactone ring).
Regardless of any putative role in the metabolism of syn-stemodene, the ability of CYP99A3 to oxidize C19 of the syn-pimaradiene produced by the co-clustered OsCPS4 and OsKSL4 has clear relevance to momilactone biosynthesis. It has already been demonstrated that CYP99A2 and/or CYP99A3 are required for the production of these dual action phytoalexins/allechemicals (Shimura, et al. 2007). Further, transformation of C19 from a methyl to carboxylic acid as catalyzed by CYP99A3 is required for momilactone biosynthesis, specifically prior to formation of the eponymous 19,6-γ-lactone ring (Figure 7). The observed correlation in transient transcription of CYP99A3 and accumulation of syn-pimaradien-19-oic acid (Figure 6) further supports physiological relevance for the enzymatic activity observed here. Accordingly, our results strongly indicate that CYP99A3 acts as a multifunction oxidase early in momilactone biosynthesis.
Figure 7.
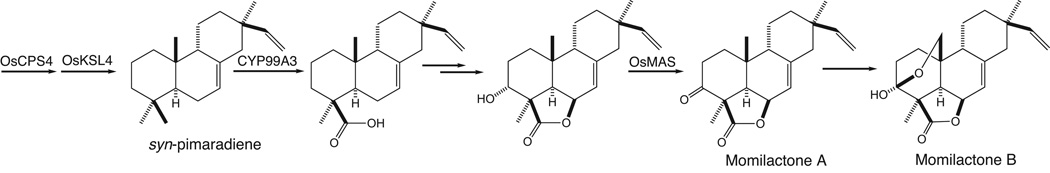
Proposed role for CYP99A3 in momilactone biosynthesis. Also shown is the subsequent formation of 3α-hydroxy-syn-pimaradien-19,6β-olide, ensuing oxidation to momilactone A catalyzed by OsMAS, and presumed downstream formation of momilactone B.
Production of the momilactones from the syn-pimaradien-19-oic acid produced by CYP99A3 presumably proceeds via additional hydroxylation at the C6β position to form the 19,6-γ-lactone ring, either before or after hydroxylation at the C3α position (Figure 7). The corresponding 3α-hydroxy-syn-pimaradien-19,6β-olide has been observed (Atawong et al. 2002), and is converted to momilactone A by OsMAS (Shimura, et al. 2007). Momilactone B is then presumably formed from momilactone A by C20 hydroxylation and hemi-ketal ring closure (Peters 2006). Thus, while CYP99A2 exhibits only trace activity with syn-pimaradiene, it is possible that this CYP catalyzes one of these later hydroxylation steps in momilactone biosynthesis. However, CYP99A2 and CYP99A3 are fairly closely related, sharing 84% identity at the amino acid level, and we cannot rule out the possibility that these two CYP are redundant or that CYP99A2 is non-functional.
In conclusion, regardless of any role for CYP99A2, we provide here clear evidence that CYP99A3 acts as a multifunctional oxidase in rice diterpenoid metabolism. This is consistent with its physical/genomic association with the relevant diterpene synthases, as well as previously reported genetic evidence (Shimura, et al. 2007). In particular, CYP99A3 presumably acts to oxidize the C19 methyl group of syn-pimaradiene to the carboxylic acid that is required for lactone ring formation in momilactone biosynthesis. Furthermore, CYP99A3 may also be involved in metabolism of the metabolically cryptic syn-stemodene as well. Thus, our results not only further elucidate momilactone biosynthesis, but also provide some additional insight into the complex diterpenoid metabolic network encoded by rice.
EXPERIMENTAL PROCEDURES
General
Unless otherwise noted, chemicals were purchased from Fisher Scientific (Loughborough, Leicestershire, UK), and molecular biology reagents from Invitrogen (Carlsbad, CA, USA). Sequence analyses were done with the Vector NTI software package (Invitrogen). Determination of the gene map and CYP nomenclature used here has been previously described (Swaminathan, et al. 2009). GC-MS was performed with a Varian (Palo Alto, CA) 3900 GC with Saturn 2100 ion trap mass spectrometer in electron ionization (70 eV) mode. Samples (1 µL) were injected in splitless mode at 50°C and, after holding for 3 min. at 50°C, the oven temperature was raised at a rate of 14°C/min. to 300°C, where it was held for an additional 3 min. MS data from 90 to 600 m/z were collected starting 12 min. after injection until the end of the run. Samples were routinely resuspended in diazomethane-saturated hexane to methylate carboxylic acid groups for GC-MS analysis, with the diazomethane generated by addition of KOH to diazald and collected in a hexane trap (Ngan and Toofan 1991). HPLC was carried out with an Agilent 1100 series instrument equipped with fraction collector and diode array detector.
Recombinant constructs
The native genes for CYP99A2 and CYP99A3 were obtained from the KOME rice cDNA database (GenBank accessions AK071546 and AK071864, respectively). These were transferred into the Gateway vector pENTR/SD/D-TOPO, verified by complete sequencing, and then recombined into the insect cell expression vector pDEST8. CYP99A2 and CYP99A3 were also completely recoded to optimize codon usage for E. coli expression via gene synthesis (Genscript). N-terminal modification of both native and synthetic genes was carried out via a two-stage PCR process, first removing 34 or 35 codons (for CYP99A2 and CYP99A3, respectively) from the 5' end of the open reading frame, and then adding ten new codons (encoding the amino acid sequence “MAKKTSSKGK” using the codon optimized sequence “ATG GCG AAA AAA ACC AGC AGC AAA GGT AAA”), with the resulting construct cloned back into pENTR/SD/D-TOPO and verified by complete sequencing. For recombinant bacterial expression the resulting modified and full length genes, both native and synthetic, were then recombined into the DEST cassette of a pCDF-Duet (Novagen) vector carrying a DEST cassette in its first multiple cloning site and OsCPR1 in its second multiple cloning site, which has been previously described (Swaminathan, et al. 2009).
Recombinant expression
Recombinant baculoviruses were constructed as previously described (Swaminathan, et al. 2009), starting from pDEST8 constructs, and used to express CYP99A2 and CYP99A3 in Sf21 insect cells. Microsomes or lysates were isolated from these recombinant cell cultures, and used for in vitro assays, again as previously described (Swaminathan, et al. 2009). After incubation at 28°C for 6 hours, the reaction mixture was extracted thrice with an equal volume of ethyl acetate. The organic extract was dried under a gentle stream of N2 gas, and dissolved in hexane for GC-MS analysis.
CYP99A2 and CYP99A3 were recombinantly expressed in E. coli using our previously described modular diterpene metabolic engineering system (Cyr, et al. 2007). Specifically, we co-expressed these CYP from the OsCPR1 co-expression constructs described above, with a GGPP synthase and CPS carried on co-compatible pGGxC vectors, and OsKSL expressed from the additionally co-compatible pDEST14 or pDEST15 (i.e., for expression as a fusion to GST), using the relevant constructs we have previously described (Xu et al. 2007), and the selective growth conditions for such multiply transformed bacteria we have previously described (Swaminathan, et al. 2009). Enzymatic products were extracted from 50 mL cultures with an equal volume of hexane, then ethyl acetate, and the pooled, concentrated, and methylated extract analyzed by GC-MS as previously described. In every case, the expected diterpene olefin product (i.e., given the co-expressed diterpene synthases) was easily observed, indicating that all potential substrates were present at sufficient levels for further transformation (i.e. by CYP99A2 or CYP99A3).
Diterpenoid production
The unknown enzymatic products were obtained in sufficient amounts for NMR analysis by increasing flux into isoprenoid metabolism and scaling up culture growth. Specifically, the pMBI vector encoding the bottom half of the mevalonate dependent isoprenoid precursor pathway, which is co-compatible with those mentioned above, was incorporated to increase precursor availability, given feeding of 20 mM mevalonolactone to the recombinant cultures, as previously described (Morrone, et al. 2010). The resulting diterpenoids were extracted from 3 L of culture (media and cells) with an equal volume of a 1:1 mixture of ethyl acetate and hexanes. These organic extracts were pooled and dried by rotary evaporation. The residue was dissolved in 5 mL 45% methanol/45% acetonitrile/10% dH2O, and the diterpenoids purified by HPLC over an Agilent ZORBAX Eclipse XDB-C8 column (4.6×150 mm, 5 µm) at a 0.5 mL/min flow rate. After loading, the column was washed with 20% acetonitrile/dH2O (0–2 min), and eluted with 20%–100% acetonitrile (2–7 min), followed by a 100% acetonitrile wash (7–27 min). The fractions containing the unknown diterpenoids were combined and dried under a gentle stream of N2 gas and then dissolved in 0.5 mL deuterated methanol (CD3OD; Sigma-Aldrich), with this evaporation-resuspension process repeated two more times to completely remove the protonated acetonitrile solvent, resulting in a final estimated ~10 mg of each unknown diterpenoid.
Chemical structure identification
NMR spectra for the syn-pimara-7,15-diene and syn-stemodene CYP450 products were recorded at 25 °C on a Bruker Avance 500 spectrometer equipped with a cryogenic probe for 1H and 13C. Structural analysis was performed using 1D 1H, 1D, DQF-COSY, HSQC, HMBC and NOESY spectra acquired at 500 MHz and 13C spectra (125.5 MHz) using standard experiments from the Bruker TopSpin v1.3 software. For syn-pimara-7,15-dien-19-al a Bruker Avance 700 spectrometer (1H 700.13 MHz; 13C 174 MHz) with a 5-mm HCN cryoprobe, in a Shigemi methanol-d4 NMR tube, was used for analysis. Chemical shifts were referenced to the known chloroform-d (13C 77.23, 1H 7.24 ppm) or methanol-d4 (13C 49.15, 1H 3.31 ppm) signals offset from TMS. Correlations from the HMBC spectra were used to propose the majority of the structures, while connections between protonated carbons were obtained from DQF-COSY to complete the partial structures and assign proton chemical shifts, etc (Fig. S4). The NOESY spectra from the syn-pimara-7,15-dien-19-ol and syn-stemoden-19-ol alcohols provided nuclear Overhauser effect (NOE) cross-peak signals to assign the stereochemistry of carbon-4 regarding which geminal methyl group (C18 or C19) was oxidized (Figure S4). Assignments for the annotated 13C and 1H 1-D for all six compounds spectra are presented in Tables S1–S6. Proton (500 MHz) chemical shifts and assignments for the physiologically relevant syn-pimara-7,15-dien-19-oic acid are as follows: 1.237 (1H, m, H1a), 1.487 (1H, m, H1b), 1.461 (1H, m, H2a), 1.520 (1H, m, H2b), 1.027 (1H, t, 9.5 Hz, H3a), 2.139 (1H, m, H3b), 1.461 (1H, d, 9.8, H5), 2.186 (1H, m, H6a), 2.421 (1H, t, 10.7, H6b), 5.303 (1H, d, 5.8, H7), 1.378 (1H, d, 9.8, H9), 1.230 (1H, m, H11a), 1.732 (1H, m, H11b), 1.437 (1H, m, H12a), 1.496 (1H, m, H12b), 1.800 (1H, d, 10.5), 1.984 (1H, d, 11.7, H14b), 5.801 (1H, q, 11.5, H15), 4.841 (1H, d, 9.2, H16E), 4.904 (1H, d, 17.2, H16Z), 0.880 (3H, s, H17), 1.238 (3H, s, H18), 0.857 (3H, s, H20). Carbon chemical shifts (125.5 MHz) and assignments for syn-pimara-7,15-dien-19-oic acid were: δ (ppm) 36.96 (C1), 19.73 (C2), 38.88 (C3), 44.25 (C4), 45.80 (C5), 24.81 (C6), 119.96 (C7), 136.10 (C8), 53.25 (C9), 35.56 (C10), 25.57 (C11), 38.08 (C12), 38.82 (C13), 48.16 (C14), 150.6 (C15), 109.41 (C16), 21.99 (C17), 29.38 (C18), 183.80 (C19), and 21.10 (C20).
Kinetic analysis
Kinetic analysis was carried out using rCYP99A3 expressed in E. coli C41 cells using the OsCPR1 co-expression construct described above. Expression cultures were grown in TB medium and induced with 1 mM IPTG upon reaching an A600 of 0.8–1.0, and complemented with addition of 1 mM thiamine, 5 mg/L riboflavin, and 75 mg/L delta-amino levulinic acid at this time as well. After 72 hours at 16°C, the cells were harvested and microsomes prepared for in vitro kinetic assays following measurement of CO-binding difference spectra as previously described (Morrone, et al. in press). The amount of active enzymes present was calculated from resulting peak at 450 nm using the standard extinction co-efficient of 91 mM-1 cm-1. The in vitro reactions were carried out using rCYP99A3 at 165 nM, with syn-pimardiene and syn-stemodene as substrates in concentrations ranging from 1 – 240 µM, but otherwise as previously described (Swaminathan, et al. 2009). After 30 minutes these 1 mL reactions were stopped by the addition 5 µL of 1 M HCl, 10 ng of abietic acid added as an internal standard, the reaction mixture extracted by ethyl acetate, methylated, and quantified by GC with flame ionization detection, with product verification by GC-MS.
Rice plant induction analyses
Rice plants (Orzya sativa L. ssp. Nipponbare) were cultivated in growth chambers under 12 hr light (28°C) and 12 hr dark (24°C) cycles to the 6th leaf stage. For induction, plants were treated with 1 mL 0.2% (v/v) methyl jasmonate per plant, while control/uninduced plants were only treated with the 0.1% Tween 20 carrier solution, and duplicate plants were harvested at 0, 2, 4, 6, 12, 24, 48 and 72 hrs after induction, with all subsequent analyses performed separately for each harvested plant to provide independent biological replicates. Reverse transcription (RT) was performed with SuperScript III RT (Invitrogen) according to the manufacturer's instructions using total RNA (1 µg) prepared from rice plant samples above. PCR was carried out using PrimeSTAR HS DNA Polymerase (Takara) with the following program: 2 min at 94 °C followed by 35 cycles of 30 s at 94 °C, 30 s at 55 °C, and 1 min 30 s at 72 °C, followed by cooling to 4 °C, using CYP99A3 gene-specific primers (forward: GAGCCTCCTCGTCTCGGA; reverse: AATTGCCTTGACGTGTGTTGA). CYP99A3 mRNA levels were determined by quantification of RT-PCR yield, with normalization against actin, using a Bio-Rad ChemiDoc XRS System. Metabolite analysis was carried out for each time point (and the control) using ~2 g rice plant (leaf) tissue, which was frozen and ground to powder in liquid nitrogen. This plant material was extracted with 50 mL ethyl acetate by stirring overnight at room temperature, with 50 ng abietic acid as an internal standard, and then methylated as described above. The mixture was clarified by centrifugation, and the ethyl acetate extract dried under nitrogen gas, with the residue redissolved in 0.2 mL 50% methanol/water, which was fractionated by HPLC using the same methods described for product purification above. Fractions were collected between 8–23 min of retention time over 1 min intervals. These fractions were dried under a gentle stream of N2 gas, resuspended in 100 µL hexanes, and quantified by single ion monitoring analysis via GC-MS. Specifically, these samples were run with the MS in chemical ionization mode using methanol as the reaction solvent, the molecular ion (m/z = 317) selected for fragmentation (0.5 eV), with quantification of the fragment at m/z = 257.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants from the USDA-CSREES-NRI (2008-35318-05027) and NIH (GM086281) to R.J.P.
Footnotes
Supporting Information:
Figure S1: Western blot analysis of CYP99A2 and CYP99A3 expressed in Sf21 insect cells.
Figure S2: Optimized gene sequence for CYP99A2.
Figure S3: Optimized gene sequence for CYP99A3.
Figure S4: Summary of NMR structural analyses.
Table S1: 1H and 13C NMR assignments for syn-pimara-7,15-dien-19-ol.
Table S2: 1H and 13C NMR assignments for syn-pimara-7,15-dien-19-al.
Table S3: 1H and 13C NMR assignments for syn-pimara-7,15-dien-19-oic acid.
Table S4: 1H and 13C NMR assignments for syn-stemod-13(17)-en-19-ol.
Table S5: 1H and 13C NMR assignments for syn- stemod-13(17)-en-19-al.
Table S6: 1H and 13C NMR assignments for syn- stemod-13(17)-en-19-oic acid.
Figure S5: Kinetic plots for rCYP99A3 with (A) syn-pimaradiene and (B) syn-stemodene.
Figure S6: Detection of syn-pimaradien-19-oic acid, but not syn-stemoden-19-oic acid, from rice plant metabolites.
REFERENCES
- Atawong A, Hasegawa M, Kodama O. Biosynthesis of rice phytoalexin: enzymatic conversion of 3β-hydroxy-9β-pimara-7,15-dien-19,6β-olide to momilactone A. Biosci. Biotechnol. Biochem. 2002;66:566–570. doi: 10.1271/bbb.66.566. [DOI] [PubMed] [Google Scholar]
- Cartwright DW, Langcake P, Pryce RJ, Leworthy DP, Ride JP. Isolation and characterization of two phytoalexins from rice as momilactones A and B. Phytochemistry. 1981;20:535–537. [Google Scholar]
- Chang MCY, Eachus RA, Trieu W, Ro D-K, Keasling JD. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat. Chem. Biol. 2007;3:274–277. doi: 10.1038/nchembio875. [DOI] [PubMed] [Google Scholar]
- Cyr A, Wilderman PR, Determan M, Peters RJ. A Modular Approach for Facile Biosynthesis of Labdane-Related Diterpenes. J. Am. Chem. Soc. 2007;129:6684–6685. doi: 10.1021/ja071158n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO. International year of rice 2004 - rice and human nutrition: Food and Agriculture Organization of the United Nations. 2004 [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Kato T, Kabuto C, Sasaki N, Tsunagawa M, Aizawa H, Fujita K, Kato Y, Kitahara Y, Takahashi N. Momilactones, growth inhibitors from rice. Oryza sativa L. Tetrahedron Lett. 1973;14:3861–3864. [Google Scholar]
- Kato-Noguchi H, Ino T. Rice seedlings release momilactone B into the environment. Phytochemistry. 2003;63:551–554. doi: 10.1016/s0031-9422(03)00194-8. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, Yazaki J, Ishikawa M, Yamada H, Ooka H, Hotta I, Kojima K, Namiki T, Ohneda E, Yahagi W, Suzuki K, Li CJ, Ohtsuki K, Shishiki T, Otomo Y, Murakami K, Iida Y, Sugano S, Fujimura T, Suzuki Y, Tsunoda Y, Kurosaki T, Kodama T, Masuda H, Kobayashi M, Xie Q, Lu M, Narikawa R, Sugiyama A, Mizuno K, Yokomizo S, Niikura J, Ikeda R, Ishibiki J, Kawamata M, Yoshimura A, Miura J, Kusumegi T, Oka M, Ryu R, Ueda M, Matsubara K, Kawai J, Carninci P, Adachi J, Aizawa K, Arakawa T, Fukuda S, Hara A, Hashidume W, Hayatsu N, Imotani K, Ishii Y, Itoh M, Kagawa I, Kondo S, Konno H, Miyazaki A, Osato N, Ota Y, Saito R, Sasaki D, Sato K, Shibata K, Shinagawa A, Shiraki T, Yoshino M, Hayashizaki Y. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- Morrone D, Chen X, Coates RM, Peters RJ. Characterization of the kaurene oxidase CYP701A3, a multifunctional cytochrome P450 from gibberellin biosynthesis. Biochem. J. doi: 10.1042/BJ20100597. (in press) [DOI] [PubMed] [Google Scholar]
- Morrone D, Jin Y, Xu M, Choi S-Y, Coates RM, Peters RJ. An unexpected diterpene cyclase from rice: Functional identification of a stemodene synthase. Arch. Biochem. Biophys. 2006;448:133–140. doi: 10.1016/j.abb.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Morrone D, Lowry L, Determan MK, Hershey DM, Xu M, Peters RJ. Increasing diterpene yield with a modular metabolic engineering system in E. coli: comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl. Microbiol. Biotechnol. 2010;85:1893–1906. doi: 10.1007/s00253-009-2219-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro AW, Girvan HM, McLean KJ. Variations on a (t)heme--novel mechanisms, redox partners and catalytic functions in the cytochrome P450 superfamily. Nat. Prod. Rep. 2007;24:585–609. doi: 10.1039/b604190f. [DOI] [PubMed] [Google Scholar]
- Ngan F, Toofan M. Modification of preparation of diazomethane for methyl esterification of environmental samples analysis by gas chromatography. J. Chromatogr. Sci. 1991;29:8–10. [Google Scholar]
- Nojiri H, Sugimora M, Yamane H, Nishimura Y, Yamada A, Shibuya N, Kodama O, Murofushi N, Omori T. Involvement of jasmonic acid in elicitor-induced phytoalexin production in suspension-cultured rice cells. Plant Physiol. 1996;110:387–392. doi: 10.1104/pp.110.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn AE, Field B. Operons. Cell. Mol. Life Sci. 2009;66:3755–3775. doi: 10.1007/s00018-009-0114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RJ. Uncovering the complex metabolic network underlying diterpenoid phytoalexin biosynthesis in rice and other cereal crop plants. Phytochemistry. 2006;67:2307–2317. doi: 10.1016/j.phytochem.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Prisic S, Xu M, Wilderman PR, Peters RJ. Rice contains two disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol. 2004;136:4228–4236. doi: 10.1104/pp.104.050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura K, Okada A, Okada K, Jikumaru Y, Ko K-W, Toyomasu T, Sassa T, Hasegawa M, Kodama O, Shibuya N, Koga J, Nojiri H, Yamane H. Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 2007;282:34013–34018. doi: 10.1074/jbc.M703344200. [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ. CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell. 2009;21:3315–3325. doi: 10.1105/tpc.108.063677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot NJ. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 2003;57:177–202. doi: 10.1146/annurev.micro.57.030502.090957. [DOI] [PubMed] [Google Scholar]
- Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ. Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 2004;135:2098–2105. doi: 10.1104/pp.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hillwig ML, Prisic S, Coates RM, Peters RJ. Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J. 2004;39:309–318. doi: 10.1111/j.1365-313X.2004.02137.x. [DOI] [PubMed] [Google Scholar]
- Xu M, Wilderman PR, Morrone D, Xu J, Roy A, Margis-Pinheiro M, Upadhyaya N, Coates RM, Peters RJ. Functional characterization of the rice kaurene synthase-like gene family. Phytochemistry. 2007;68:312–326. doi: 10.1016/j.phytochem.2006.10.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.