Abstract
Background
Computer-aided drug design (CADD) often involves virtual screening (VS) of large compound datasets and the availability of such is vital for drug discovery protocols. This paper presents an assessment of the “drug-likeness” and pharmacokinetic profile of > 2,400 compounds of natural origin, currently available in the recently published StreptomeDB database.
Methods
The evaluation of “drug-likeness” was performed on the basis of Lipinski’s “Rule of Five”, while 46 computed physicochemical properties or molecular descriptors were used to predict the absorption, distribution, metabolism, elimination and toxicity (ADMET) of the compounds.
Results
This survey demonstrated that, of the computed molecular descriptors, about 28% of the compounds within the StreptomeDB database were compliant, having properties which fell within the range of ADMET properties of 95% of currently known drugs, while about 44% of the compounds had ≤ 2 violations. Moreover, about 50% of the compounds within the corresponding “drug-like” subset showed compliance, while >83% of the “drug-like” compounds had ≤ 2 violations.
Conclusions
In addition to the previously verified range of measured biological activities, the compounds in the StreptomeDB database show interesting DMPK profiles and hence could represent an important starting point for hit/lead discovery from natural sources. The generated data are available and could be highly useful for natural product lead generation programs.
Electronic supplementary material
The online version of this article (doi:10.1186/2193-1801-2-353) contains supplementary material, which is available to authorized users.
Keywords: 3D structures; Streptomyces, Database, In silico, Natural products, Virtual screening
Background
Many drugs often fail to enter the market as a result of poor pharmacokinetic profiles (Darvas et al. 2002). Thus, it has become imperative nowadays to design lead compounds which can be easily orally absorbed, easily transported to their desired site of action, not easily metabolised into toxic metabolic products before reaching the targeted site of action and easily eliminated from the body before accumulating in sufficient amounts that may produce adverse side effects. The sum of the above mentioned properties is often referred to as ADME (absorption, distribution, metabolism and elimination) properties, or better still ADMET, ADME/T or ADMETox (when considerations are given to toxicity issues). The inclusion of pharmacokinetic considerations at earlier stages of drug discovery programs (Hodgson 2001; Navia and Chaturvedi 1996) using computer-based methods is becoming increasingly popular (Lipinski et al. 1997; Lombardo et al. 2003; Gleeson et al. 2011). The rationale behind in silico approaches are the relatively lower cost and the time factor involved, when compared to standard experimental approaches for ADMET profiling (DiMasi et al. 2003; Darvas et al. 2002). As an example, it only takes a minute in an in silico model to screen 20,000 molecules, but takes 20 weeks in the “wet” laboratory to do the same exercise (Hodgson 2001).
Due to the accumulated ADMET data in the late 1990s, many pharmaceutical companies are now using computational models that, in some cases, are replacing the “wet” screens (Hodgson 2001). This paradigm shift has therefore spurred up the development of several theoretical methods for the prediction of ADMET parameters. A host of these theoretical models have been implemented in a number of software programs currently available for drug discovery protocols ( OCHEM platform; Lhasa 2010; Schrödinger 2011a; Cruciani et al. 2000), even though some of the predictions are often disappointing (Tetko et al. 2006). The software tools currently used to predict the ADMET properties of potential drug candidates often make use of quantitative structure-activity relationships, QSAR (Tetko et al. 2006; Hansch et al. 2004) or knowledge-base methods (Greene et al. 1999; Button et al. 2003; Cronin 2003). A promising lead compound may therefore be defined as one which combines potency with an attractive ADMET profile. As such, compounds with uninteresting predicted ADMET profiles may be completely dismissed from the list of potential drug candidates early enough (even if these prove to be highly potent). Otherwise, the DMPK properties are “fine-tuned” in order to improve their chances of making it to clinical trials (Hou and Wang 2008).
In this paper, we assess the pharmacokinetic profile of the recently published StreptomeDB database (Lucas et al. 2013) using an in silico method. A number of computed molecular descriptors, currently implemented in a wide range of software, have been used as indicators of the pharmacokinetic properties of a large proportion of currently known drugs.
Methods
Data source and initial treatment of chemical structures
The 2,444 3D structures of the compounds in the StreptomeDB database were downloaded from the official webpage of the Pharmaceutical Bioinformatics group of the University of Freiburg (http://www.pharmaceutical-bioinformatics.de/streptomedb/). These were initially treated with LigPrep (Schrödinger 2011b). The implementation was carried out with the graphical user interface (GUI) of the Maestro software package (Schrödinger 2011c), using the OPLS forcefield (Shivakumar et al. 2010; Jorgensen et al. 1996; Jorgensen and Tirado-Rives 1988). Protonation states at biologically relevant pH were correctly assigned (group I metals in simple salts were disconnected, strong acids were deprotonated, strong bases protonated, while topological duplicates and explicit hydrogens were added). All molecular modelling was carried out on a Linux workstation with a 3.5 GHz Intel Core2 Duo processor.
Calculation of ADMET-related descriptors
A set of ADMET-related properties (a total of 46 molecular descriptors) were calculated by using the QikProp program (Schrödinger
2011d) running in normal mode. QikProp generates physically relevant descriptors, and uses them to perform ADMET predictions. An overall ADME-compliance score – drug-likeness parameter (indicated by #stars), was used to assess the pharmacokinetic profiles of the compounds within the StreptomeDB library. The #stars parameter indicates the number of property descriptors computed by QikProp that fall outside the optimum range of values for 95% of known drugs. The methods implemented were developed by Jorgensen and Duffy (Jorgensen and Duffy
2002; Duffy and Jorgensen
2000; Jorgensen and Duffy
2000) and among the calculated descriptors are: the total solvent-accessible molecular surface, Smol in Å2 (probe radius 1.4 Å) (range for 95% of drugs: 300–1000 Å2); the hydrophobic portion of the solvent-accessible molecular surface, Smol,hfob in Å2 (probe radius 1.4 Å) (range for 95% of drugs: 0–750 Å2); the total volume of molecule enclosed by solvent-accessible molecular surface, Vmol in Å3 (probe radius 1.4 Å) (range for 95% of drugs: 500–2000 Å3); the logarithm of aqueous solubility, log Swat (range for 95% of drugs: -6.0 to 0.5) (Jorgensen and Duffy
2002; Jorgensen and Duffy
2000); the logarithm of predicted binding constant to human serum albumin, log KHSA (range for 95% of drugs: -1.5 to 1.2) (Colmenarejo et al.
2001); the logarithm of predicted blood/brain barrier partition coefficient, log B/B (range for 95% of drugs: -3.0 to 1.0) (Luco
1999; Kelder et al.
1999; Ajay et al.
1999); the predicted apparent Caco-2 cell membrane permeability (BIPcaco − 2) in Boehringer–Ingelheim scale, in nm s-1 (range for 95% of drugs: < 5 low, > 100 high) (Yazdanian et al.
1998; Irvine et al.
1999; Stenberg et al.
2001); the predicted apparent Madin-Darby canine kidney (MDCK) cell permeability in nm s-1 (< 25 poor, > 500 great) (Irvine et al.
1999); the index of cohesion interaction in solids, Indcoh, calculated from the number of hydrogen bond acceptors (HBA), donors (HBD) and the surface area accessible to the solvent, SASA (Smol) by the relation
 (0.00 to 0.05 for 95% of drugs) (Jorgensen and Duffy
2000); the globularity descriptor, Glob = (4πr2)/Smol, where r is the radius of the sphere whose volume is equal to the molecular volume (0.75 to 0.95 for 95% of drugs); the predicted polarizability, QPpolrz (13.0 to 70.0 for 95% of drugs); the predicted IC50 value for blockage of HERG K+ channels, log HERG (concern < −5) (Cavalli et al.
2002; De Ponti et al.
2001); the predicted skin permeability, log Kp (−8.0 to −1.0 for 95% of drugs) (Potts and Guy
1992; Potts and Guy
1995); and the number of likely metabolic reactions, #metab (range for 95% of drugs: 1–8).
(0.00 to 0.05 for 95% of drugs) (Jorgensen and Duffy
2000); the globularity descriptor, Glob = (4πr2)/Smol, where r is the radius of the sphere whose volume is equal to the molecular volume (0.75 to 0.95 for 95% of drugs); the predicted polarizability, QPpolrz (13.0 to 70.0 for 95% of drugs); the predicted IC50 value for blockage of HERG K+ channels, log HERG (concern < −5) (Cavalli et al.
2002; De Ponti et al.
2001); the predicted skin permeability, log Kp (−8.0 to −1.0 for 95% of drugs) (Potts and Guy
1992; Potts and Guy
1995); and the number of likely metabolic reactions, #metab (range for 95% of drugs: 1–8).
Results and discussion
Drug-likeness assessment
The “drug-likeness” test was carried out using Lipinski’s “Rule of Five”, ro5 (Lipinski et al. 1997). The distributions of the compound molecular weights (MW), calculated lipophilicity (log P), number of hydrogen bond acceptors (HBA) and number of hydrogen bond donors (HBD) were used to assess the “drug-likeness” of StreptomeDB. It is noteworthy that natural products exhibit a wide range of flexibility, from rigid conformationally constrained molecules to very flexible compounds. Thus, the number of rotatable bonds (NRB) within the StreptomeDB library was used as an additional criterion to test for the favourable drug metabolism and pharmacokinetics (DMPK) outcomes. It was observed that 47.5% of the compounds within StreptomeDB showed no Lipinski violations and 77.3% showed ≤ 2 violations (Figure 1), while the peak of the distribution of the NRB was between 1 and 2 (Figure 2E). Moreover, the analysis of the distributions of MW (truncated at MW = 1000 Da for the sake of clarity), showed a peak value between 301 and 400 Da (Figure 2A), with a curve similar to those previously reported for other “drug-like” NP libraries in the literature (Ntie-Kang et al. 2013; Quinn et al. 2008; Feher and Schmidt 2003) and about 42% of MW > 500 Da. The distribution of the log P values showed a Gaussian shaped curve with a peak centred at 2.5 log P units (Figure 2C). However, some of the compounds had exceptionally large log P values (truncated at 10 log P units for the sake of clarity), which went up to > 19 units. This may be explained by the fact that the training database/algorithm used to calculate log P may not suit the types and combinations of functional groups found in natural products (Quinn et al. 2008). It should however be noted that, inspite of this limitation, 85.3% of the compounds from StreptomeDB had log P values < 5 units. The peaks of the HBA and HBD were respectively at 5 acceptors and 2 donors and both curves fell off rapidly to maximum numbers of 57 and 38 respectively (truncated at 40 HBA and 10 HBD respectively, Figure 2B and 2D). It was also noted that ~40% of the compounds in StreptomeDB had HBA > 10 and only ~24% had HBD > 5. Additionally, the pairwise comparison displaying the mutual relationship between the molecular weight versus the calculated log P, HBA, HBD and NRB are specified in Figure 3A-D, respectively. The plots show that the regions with the highest population densities fall within the “Lipinski region of interest” (MW < 500, -2 < log P < 5, HBA < 10 and HBD < 5), and for which NRB < 5.
Figure 1.

Distributions of violations of Lipinski’s ro5 and Jorgensen’s ro3 within the StreptomeDB database.
Figure 2.
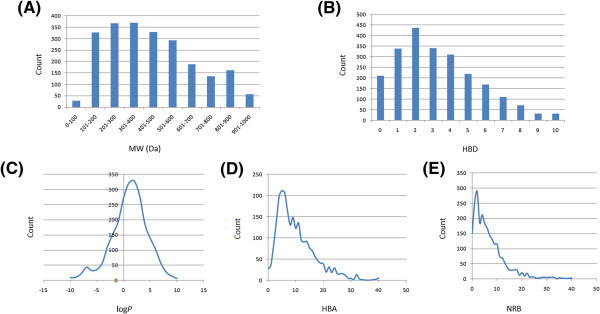
Distributions of features that determine “drug-likeness” in StreptomeDB. (A) MW, (B) HBD, (C) log P, (D) HBA, (E) NRB.
Figure 3.

Scatter diagrams showing pair wise distribution of “drug-likeness” descriptors. (A) MW against log P, (B) MW against HBA, (C) MW against HBD and (D) MW against NRB.
Overall DMPK compliance of the SreptomeDB library
The 24 most relevant molecular descriptors calculated by QikProp are used to determine the #star parameter (Schrödinger 2011d). A plot of the #stars parameter (on x-axis) against the corresponding counts (on y-axis) in the StreptomeDB is plotted within the same set of axes with those of the “drug-like”, “lead-like”, and “fragment-like” standard subsets, as shown in Figure 4. The criteria for the respective standard subsets were defined as (MW < 500; log P < 5; HBD ≤ 5; HBA ≤ 10) (Lipinski et al. 1997), (150 ≤ MW ≤ 350; log P ≤ 4; HBD ≤ 3; HBA ≤ 6) (Teague et al. 1999; Oprea 2002; Schneider 2002) and (MW ≤ 250; -2 ≤ log P ≤ 3; HBD < 3; HBA < 6; NRB < 3) (Verdonk et al. 2003). The ADMET descriptors for some 316 compounds in the total library were not computed by QikProp. This could be due to technical difficulties with running the software, due to errors in the input structures downloaded from the StreptomeDB website. Of the remaining 2,128 compounds, 27.2% showed #star = 0, while 53.8% had #star ≤ 2. Among the 925 compounds of the “drug-like” subset whose pharmacokinetic properties were predicted, 49.7% had pharmacokinetic descriptors within the acceptable range for 95% of known drugs, while 83.5% showed #stars ≤ 2. The “lead-like” and “fragment-like” subsets were respectively 63.5% and 31.5% compliant for all of the 24 most relevant computed descriptors. The average values for 19 selected computed descriptors have been shown in Table 1 for all 4 compound libraries. The average values indicate a high probability of finding drug leads within the StreptomeDB compound library.
Figure 4.

Distribution curves for #stars within the StreptomeDB library, along with the standard “drug-like”, “lead-like” and “fragment-like” subsets. Blue = StreptomeDB library, red = “drug-like” subset, green = “lead-like” subset and violet = “fragment-like” subset.
Table 1.
Summary of average pharmacokinetic property distributions of the total StreptomeDB library in comparison with the various subsets
| Library name | aLib. size | bNo. compl. | cMW (Da) | dLogP | eHBA | fHBD | gNRB |
|---|---|---|---|---|---|---|---|
| StreptomeDB | 2,444 | 577 | 485 | 1.30 | 11.69 | 3.47 | 11.46 |
| Drug-like | 925 | 459 | 262 | 1.19 | 5.27 | 1.92 | 4.78 |
| Lead-like | 326 | 207 | 230 | 1.61 | 3.96 | 1.46 | 3.68 |
| Fragment-like | 127 | 40 | 151 | 0.96 | 2.99 | 1.07 | 1.28 |
| Library name | hLogB/B | iBIPcaco-2(nm s-1) | jSmol(Å2) | kSmol,hfob(Å2) | lVmol(Å3) | mLogSwat(S in mol L-1) | nLogKHSA |
| StreptomeDB | −2.25 | 522 | 748 | 417 | 1426 | −3.20 | −0.48 |
| Drug-like | −1.01 | 734 | 490 | 213 | 840 | −2.42 | −0.37 |
| Lead-like | −0.72 | 986 | 460 | 179 | 768 | −2.50 | −0.29 |
| Fragment-like | −0.31 | 1275 | 343 | 112 | 536 | −1.14 | −0.63 |
| Library name | oMDCK | pIndcoh | qGlob | rQPpolrz(Å3) | sLogHERG | tLogKp |
u# metab |
| StreptomeDB | 368 | 0.028 | 0.82 | 44.53 | −3.90 | −4.76 | 6.62 |
| Drug-like | 545 | 0.015 | 0.88 | 25.44 | −3.33 | −3.93 | 3.48 |
| Lead-like | 638 | 0.010 | 0.88 | 23.56 | −3.59 | −3.28 | 2.75 |
| Fragment-like | 896 | 0.008 | 0.93 | 15.59 | −2.45 | −2.98 | 1.45 |
aSize or number of compounds in library; bNumber of compounds with #star = 0; cMolar weight (range for 95% of drugs: 130–725 Da); dLogarithm of partitioning coefficient between n-octanol and water phases (range for 95% of drugs: -2 to 6); eNumber of hydrogen bonds accepted by the molecule (range for 95% of drugs: 2–20); fNumber of hydrogen bonds donated by the molecule (range for 95% of drugs: 0–6).; gNumber of rotatable bonds (range for 95% of drugs: 0–15); hLogarithm of predicted blood/brain barrier partition coefficient (range for 95% of drugs: -3.0 to 1.0); iPredicted apparent Caco-2 cell membrane permeability in Boehringer–Ingelheim scale, in nm/s (range for 95% of drugs: < 5 low, > 100 high); jTotal solvent-accessible molecular surface, in Å2 (probe radius 1.4 Å) (range for 95% of drugs: 300–1000 Å2); kHydrophobic portion of the solvent-accessible molecular surface, in Å2 (probe radius 1.4 Å) (range for 95% of drugs: 0–750 (Å2); lTotal volume of molecule enclosed by solvent-accessible molecular surface, in Å3 (probe radius 1.4 Å) (range for 95% of drugs: 500–2000 Å3); mLogarithm of aqueous solubility (range for 95% of drugs: -6.0 to 0.5); nLogarithm of predicted binding constant to human serum albumin (range for 95% of drugs: -1.5 to 1.2); oPredicted apparent MDCK cell permeability in nm/sec (< 25 poor, > 500 great); pIndex of cohesion interaction in solids (0.0 to 0.05 for 95% of drugs); qGlobularity descriptor (0.75 to 0.95 for 95% of drugs); rPredicted polarizability (13.0 to 70.0 for 95% of drugs); sPredicted IC50 value for blockage of HERG K+ channels (concern < −5); tPredicted skin permeability (−8.0 to −1.0 for 95% of drugs); uNumber of likely metabolic reactions (range for 95% of drugs: 1–8).
Bioavailability prediction
According to Jorgensen’s ro3, if a compound complies to all or some of the rules (log Swat > −5.7, BIPcaco − 2 > 22 nm/s and # Primary Metabolites < 7), then it is more likely to be orally available (Jorgensen and Duffy 2000; Jorgensen and Duffy 2002; Schrödinger 2011d). The bioavailability of a compound depends on the processes of absorption and liver first-pass metabolism (Van de Waterbeemd and Gifford 2003). Absorption in turn depends on the solubility and permeability of the compound, as well as interactions with transporters and metabolizing enzymes in the gut wall. The computed parameters used to assess oral absorption are the predicted aqueous solubility, log Swat, the conformation-independent predicted aqueous solubility, CI log Swat, the predicted qualitative human oral absorption, the predicted % human oral absorption and compliance to Jorgensen’s famous “Rule of Three” (ro3). The solubility calculation procedure implemented depends on the similarity property space between the given molecule and its most similar analogue within the experimental training set used to develop the model implemented in QikProp, i.e., if the similarity is < 0.9, then the QikProp predicted value is taken, otherwise, the predicted property, Ppred , is adjusted such that:
| 1 |
where S is the similarity, and Pexp and PQP are the respective experimental and QikProp predictions for the most similar molecule within the training set. In equation (1), if S = 1, then the predicted property is equal to the measured experimental property of the training set compound. The distribution curves for two of the three determinants for the ro3 (log Swat and BIPcaco − 2) are shown in Figure 5. In general 37.3% of the StreptomeDB library was compliant to the ro3, while the respective % compliances for the various subsets were 72.7%, 92.3% and 97.6% for the “drug-like”, “lead-like” and “fragment-like” libraries. Among the individual computed parameters, the most remarkable was log Swat, which was met by 85.1% of the compounds within the StreptomeDB library. This property showed a Gaussian distribution for the “drug-like” and “lead-like” subsets. Only 39.9% of the compounds fell within the respected range for the BIPcaco − 2 criterion. The predicted apparent Caco-2 cell permeability, BIPcaco − 2 (in nm s-1), model the permeability of the gut-blood barrier (for non-active transport), even though this parameter is not often correctly predicted computationally (Veber et al. 2002). The histograms of the predicted qualitative human oral absorption parameter (in the scale 1 = low, 2 = medium and 3 = high) are shown in Figure 6. It was observed 26.6% of the compounds in the total StreptomeDB were predicted to have high human oral absorption. The predicted% human oral absorption (on 0 to 100% scale) shows a similar trend, 10.9% of the compounds being predicted to be absorbed at 100% and 15.9% of the compounds predicted to be absorbed at > 90%.
Figure 5.

Distribution curves for compliance to Jorgensen’s “Rule of Three” (A) calculated log Swatagainst count, (B) predicted BIPcaco − 2against count. Colour codes are as defined in Figure 4.
Figure 6.
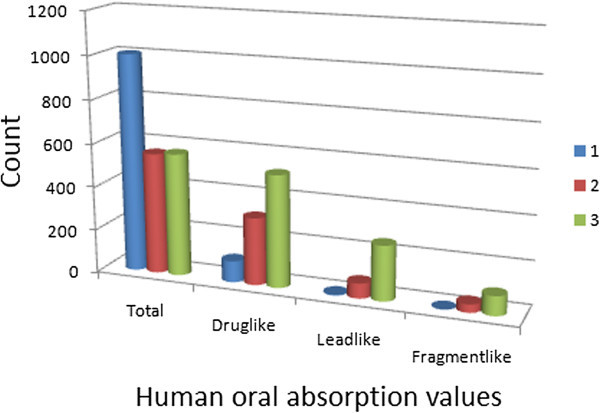
Histograms showing the distribution of human oral absorption predictions.
A molecule’s size, as well as its capacity to make hydrogen bonds, its overall lipophilicity and its shape and flexibility are important properties to consider when determining permeability. Molecular flexibility has been seen as a parameter which is dependent on the number of rotatable bonds (NRB), a property which influences bioavailability in rats (Veber et al. 2002). The results for distribution of the NRB for this dataset revealed that the compounds within the StreptomeDB library show some degree of conformational flexibility, the peak value for the NRB being between 1 and 2, while the average values is 11.46 (Table 1). The large gap between the peak and average value could be explained by the presence of very huge NPs within the dataset, containing as many as 72 rotatable single bonds (truncated at 40 RBs in Figure 2E).
Prediction of blood–brain barrier (BBB) penetration
Too polar drugs do not cross the BBB. The blood/brain partition coefficients (log B/B) were computed and used as a predictor for access to the central nervous system (CNS). The predicted CNS activity was computed on a −2 (inactive) to +2 (active) scale and showed that only 2.47% of the compounds in StreptomeDB could be active in the CNS (predicted CNS activity > 1). A distribution of log B/B (Figure 7) shows a right-slanted Gaussian-shaped curve with a peak at −1.5 log B/B units for the total library, and −0.5 log B/B units for the standard subsets, with 73.7% of the compounds in StreptomeDB falling within the recommended range for the predicted brain/blood partition coefficient (−3.0 to 1.2). Madin-Darby canine kidney (MDCK) monolayers, are widely used to make oral absorption estimates, the reason being that these cells also express transporter proteins, but only express very low levels of metabolizing enzymes (Veber et al. 2002). They are also used as an additional criterion to predict BBB penetration. Thus, our calculated apparent MDCK cell permeability could be considered to be a good mimic for the BBB (for non-active transport). It was estimated that only about 35% of the compounds had apparent MDCK cell permeabilities falling within the recommended range of 25–500 nm s-1 for 95% of known drugs. This situation knew no improvements in the “drug-like” and “lead-like” subsets.
Figure 7.

Plot of the physico-chemical descriptor used to predict BBB penetration. Predicted log B/B against count. The x-axis label is the lower limit of binned data, e.g. 0 is equivalent to 0.0 to 1.0. Colour codes are as defined in Figure 4.
Prediction of dermal penetration
This factor is important for drugs administered through the skin. The distribution of computed skin permeability parameter, log Kp, showed smooth Gaussian-shaped graphs centred at −4.5 log Kp units for the total database, at −3.5 log Kp units for the “drug-like” subset and −2.5 log Kp units for the “lead-like” and “fragment-like” subsets (Figure 8), with ~88% of the compounds in the StreptomeDB database falling within the recommended range for 95% of known drugs. The predicted maximum transdermal transport rates, Jm (in μ cm-2 hr-1), were computed from the aqueous solubility (Swat) and skin permeability (Kp), using the relation (2):
| 2 |
Figure 8.
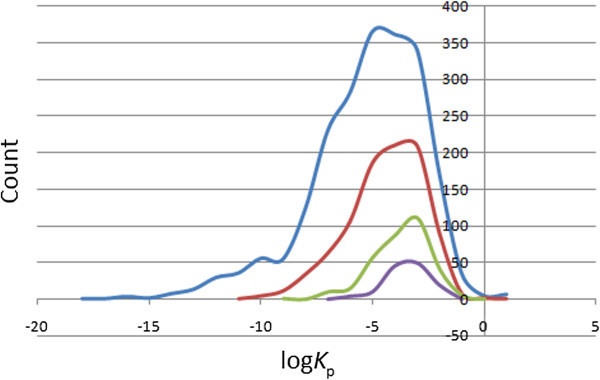
Distribution curves for the predicted skin penetration parameter. Colour codes are as defined in Figure 4.
This parameter showed variations from 0 to about 86,100 μ cm-2 hr-1, with only about 2.7% of the compounds in StreptomeDB having predicted value of Jm > 100 μ cm-2 hr-1.
Prediction of plasma-protein binding
The efficiency of a drug may be affected by the degree to which it binds to the proteins within blood plasma. It is noteworthy that binding of drugs to plasma proteins (like human serum albumin, lipoprotein, glycoprotein, α, β, and γ globulins) greatly reduces the quantity of the drug in general blood circulation and hence the less bound a drug is, the more efficiently it can traverse cell membranes or diffuse. The predicted plasma-protein binding has been estimated by the prediction of binding to human serum albumin; the log KHSA parameter (recommended range is −1.5 to 1.5 for 95% of known drugs). Figure 9 shows the variation of this calculated parameter within the StreptomeDB dataset, as well as for the standard subsets. This equally gave smooth Gaussian-shaped curves centred on −0.5 log KHSA units for all the datasets. In addition, our calculations reveal that > 86% of the compounds within the StreptomeDB library are compliant to this parameter, indicating that a majority of the compounds are likely to circulate freely within the blood stream and hence have access to the target site.
Figure 9.

Distribution curves for predicted plasma-protein binding. Colour codes are as defined in Figure 4.
Metabolism prediction
An estimated number of possible metabolic reactions has also been predicted by QikProp and used to determine whether the molecules can easily gain access to the target site after entering the blood stream. The average estimated number of possible metabolic reactions for the StreptomeDB library is between 5 and 6, while those of the standard subsets are respectively between 6 and 7, between 3 and 4 and between 1 and 2 for the “drug-like”, “lead-like” and “fragment-like” libraries (Table 1). Even though some of the compounds are likely to undergo as many as up to 30 metabolic reactions due to the complexity of some of the secondary metabolites within the database (Figure 8), ~68% of the compounds are predicted to undergo the recommended number of metabolic steps (1 to 8 reactions), with the situation improving to ~92% and almost 100% in the “drug-like” and “lead-like” subsets respectively. From Figure 10, it can be observed that the total, “drug-like”, “lead-like”, and “fragment-like” libraries both show respective peak values at 5, 4, 3 and 2 metabolic steps.
Figure 10.

Graphs showing the distribution of the predicted number of metabolic reactions. Colour codes are as defined in Figure 4.
Prediction of blockage of human ether-a-go-go-related gene potassium (HERG K+) channel
Human ether-a-go-go related gene (HERG) encodes a potassium ion (K+) channel that is implicated in the fatal arrhythmia known as torsade de pointes or the long QT syndrome (Hedley et al. 2009). The HERG K+ channel, which is best known for its contribution to the electrical activity of the heart that coordinates the heart's beating, appears to be the molecular target responsible for the cardiac toxicity of a wide range of therapeutic drugs (Vandenberg et al. 2001). HERG has also been associated with modulating the functions of some cells of the nervous system and with establishing and maintaining cancer-like features in leukemic cells (Chiesa et al. 1997). Thus, HERG K+ channel blockers are potentially toxic and the predicted IC50 values often provide reasonable predictions for cardiac toxicity of drugs in the early stages of drug discovery (Aronov 2005). In this work, the estimated or predicted IC50 values for blockage of this channel have been used to model the process in silico. The recommended range for predicted log IC50 values for blockage of HERG K+ channels (logHERG) is > −5. A distribution curve for the variation of the predicted logHERG is shown in Figure 11, which is a left-slanted Gaussian-shaped curve, with a peak at −5.5 logHERG units for the total library, as well as for the “drug-like” and “lead-like” subsets. It was observed that in general, this parameter is predicted to fall within the recommended range for about 66% of the compounds within the StreptomeDB dataset, ~87% for the “drug-like” subset and ~86% for the “lead-like” subset.
Figure 11.

A plot of predicted logHERG values for StreptomeDB and standard subsets. Colour codes are as defined in Figure 4.
Usefulness of the compound library
The treated 3D structures of the compounds, as well as their physico-chemical properties that were used to evaluate the “drug-likeness” and DMPK profile, can be freely downloaded as additional files accompanying this publication (Additional file 1, Additional file 2, Additional file 3, Additional file 4 and Additional file 5). The computed properties included in the attached files could be a useful guide in compound selection during virtual screening campaigns and hence help users carefully select which compounds to further develop in a drug discovery program which begins with the StreptomeDB database.
Conclusions
Modern drug discovery programs usually involve the search for small molecule leads with attractive pharmacokinetic profiles. The presence of such within the material in the additional files accompanying this publication (for non commercial use) is of major importance and therefore renders the database attractive, in addition to the already known properties – “drug-likeness”, “lead-likeness”, “fragment-likeness” and diversity (Lucas et al. 2013). This is an indication that the 3D structures of naturally occurring compounds within StreptomeDB could be a good starting point for docking, neural networking and pharmacophore-based virtual screening campaigns, thus rendering StreptomeDB a useful asset for the drug discovery community.
Authors’ information
FNK is a computational chemist focused on natural product drug discovery and computer-aided drug design.
Electronic supplementary material
Additional file 1: Prepared 3D structures of compounds currently included in StreptomeDB with calculated “drug-likeness” descriptors. (MDB 2 MB)
Additional file 2: 3D structures of the “drug-like” subset derived from the StreptomeDB library. (MOL2 2 MB)
Additional file 3: 3D structures of the “lead-like” subset derived from the StreptomeDB library. (MOL2 720 KB)
Additional file 4: 3D structures of the “fragment-like” subset derived from the StreptomeDB library. (MOL2 208 KB)
Additional file 5: 3D structures of the total StreptomeDB dataset with calculated pharmacokinetic descriptors. (SDF 13 MB)
Acknowledgements
Financial support is acknowledged from the German Academic Exchange Service (DAAD) to FNK for his stay in Halle, Germany for part of his PhD and from the ICTP via the OEA-AC71 program. Financial support is also acknowledged from a grant from Lhasa Ltd, Leeds, UK, through the Chemical and Bioactivity Information Centre (CBIC), University of Buea, Cameroon The author also acknowledge the academic license generously offered by Schrodinger Inc, for this work.
Abbreviations
- 3D
Three dimensional
- ADME/T
Absorption, distribution, metabolism, elimination, and toxicity
- CADD
Computer-aided drug design
- DMPK
Drug metabolism and pharmacokinetics
- log P
logarithm of the octan-1-ol/water partition coefficient
- MDCK
Madin-Darby canine kidney
- MW
Molar weight
- NP
Natural product
- NRB
Number of rotatable bonds
- RBs
Rotatable bonds
- VS
Virtual screening.
Footnotes
Competing interests
The author declares he has no competing interest.
Authors’ contributions
FNK conceived the project, designed and performed the simulations, analysed the results and wrote the paper as a single author.
References
- Ajay, Bermis GW, Murkco MA. Designing libraries with CNS activity. J Med Chem. 1999;42:4942–4951. doi: 10.1021/jm990017w. [DOI] [PubMed] [Google Scholar]
- Aronov AM. Predictive in silico modeling for hERG channel blockers. Drug Discov Today. 2005;10:149–155. doi: 10.1016/S1359-6446(04)03278-7. [DOI] [PubMed] [Google Scholar]
- Button WG, Judson PN, Long A, Vessey JD. Using absolute and relative reasoning in the prediction of the potential metabolism of xenobiotics. J Chem Inf Comput Sci. 2003;43:1371–1377. doi: 10.1021/ci0202739. [DOI] [PubMed] [Google Scholar]
- Cavalli A, Poluzzi E, De Ponti F, Recanatini M. Toward a pharmacophore for drugs inducing the long QT syndrome: Insights from a CoMFA Study of HERG K+ channel blockers. J Med Chem. 2002;45:3844–3853. doi: 10.1021/jm0208875. [DOI] [PubMed] [Google Scholar]
- Chiesa N, Rosati B, Arcangeli A, Olivotto M, Wanke E. A novel role for HERG K+ channels: spike-frequency adaptation. J Physiol. 1997;501:313–318. doi: 10.1111/j.1469-7793.1997.313bn.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenarejo G, Alvarez-Pedraglio A, Lavandera J-L. Cheminformatic models to predict binding affinities to human serum albumin. J Med Chem. 2001;44:4370–4378. doi: 10.1021/jm010960b. [DOI] [PubMed] [Google Scholar]
- Cronin MTD. Computer-assisted prediction of drug toxicity and metabolism in Modern Methods of Drug Discovery. In: Hillisch A, Hilgenfeld R, editors. Modern methods of drug discovery. Basel: Birkhäuser; 2003. [Google Scholar]
- Cruciani C, Crivori P, Carrupt PA, Testa B. Molecular fields in quantitative structure-permeation relationships: the VolSurf approach. J Mol Struc-Theochem. 2000;503:17–30. doi: 10.1016/S0166-1280(99)00360-7. [DOI] [Google Scholar]
- Darvas F, Keseru G, Papp A, Dormán G, Urge L, Krajcsi P. In Silico and Ex silico ADME approaches for drug discovery. Top Med Chem. 2002;2:1287–1304. doi: 10.2174/1568026023392841. [DOI] [PubMed] [Google Scholar]
- De Ponti F, Poluzzi E, Montanaro N. Organising evidence on QT prolongation and occurrence of Torsades de Pointes with non-antiarrhythmic drugs: a call for consensus. Eur J Clin Pharmacol. 2001;57:185–209. doi: 10.1007/s002280100290. [DOI] [PubMed] [Google Scholar]
- DiMasi JA, Hansen RW, Grabowsk HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003;22:151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- Duffy EM, Jorgensen WL. Prediction of properties from simulations: free energies of solvation in hexadecane, octanol, and water. J Am Chem Soc. 2000;122:2878–2888. doi: 10.1021/ja993663t. [DOI] [Google Scholar]
- Feher M, Schmidt JM. Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. J Chem Inf Comput Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- Gleeson MP, Hersey A, Hannongbua S. In-silico ADME models: a general assessment of their utility in drug discovery applications. Curr Top Med Chem. 2011;11(4):358–381. doi: 10.2174/156802611794480927. [DOI] [PubMed] [Google Scholar]
- Greene N, Judson PN, Langowski JJ. Knowledge-based expert systems for toxicity and metabolism prediction: DEREK, StAR and METEOR. SAR QSAR Environ Res. 1999;10:299–314. doi: 10.1080/10629369908039182. [DOI] [PubMed] [Google Scholar]
- Hansch C, Leo A, Mekapatia SB, Kurup A. QSAR and ADME. Bioorg Med Chem. 2004;12:3391–3400. doi: 10.1016/j.bmc.2003.11.037. [DOI] [PubMed] [Google Scholar]
- Hedley PL, Jørgensen P, Schlamowitz S, Wangari R, Moolman-Smook J, Brink PA, Kanters JK, Corfield VA, Christiansen M. The genetic basis of long QT and short QT syndromes: A mutation update. Human Mutation. 2009;30:1486–1511. doi: 10.1002/humu.21106. [DOI] [PubMed] [Google Scholar]
- Hodgson J. ADMET – turning chemicals into drugs. Nature Biotechnology. 2001;19:722–726. doi: 10.1038/90761. [DOI] [PubMed] [Google Scholar]
- Hou T, Wang J. Structure-ADME relationship: still a long way to go? Expert Opin Drug Metab Toxicol. 2008;4(6):759–770. doi: 10.1517/17425255.4.6.759. [DOI] [PubMed] [Google Scholar]
- Irvine JD, Takahashi L, Lockhart K, Cheong J, Tolan JW, Selick HE, Grove JR. MDCK (Madin-Darby canine kidney) cells: a tool for membrane permeability screening. J Pharm Sci. 1999;88:28–33. doi: 10.1021/js9803205. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Duffy EM. Prediction of drug solubility from Monte Carlo simulations. Bioorg Med Chem Lett. 2000;10:1155–1158. doi: 10.1016/S0960-894X(00)00172-4. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Duffy EM. Prediction of drug solubility from structure. Adv Drug Deliv Rev. 2002;54:355–366. doi: 10.1016/S0169-409X(02)00008-X. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110(6):1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118(45):11225–11236. doi: 10.1021/ja9621760. [DOI] [Google Scholar]
- Kelder J, Grootenhuis PD, Bayada DM, Delbresine LP, Ploemen JP. Polar molecular surface as a dominating determinant for oral absorption and brain penetration of drugs. Pharm Res. 1999;16:1514–1519. doi: 10.1023/A:1015040217741. [DOI] [PubMed] [Google Scholar]
- Lhasa L. Meteor, version 13.0.0. Leeds, UK: Lhasa; 2010. [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Delivery Rev. 1997;23:3–25. doi: 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Lombardo F, Gifford E, Shalaeva MY. In silico ADME prediction: data, models, facts and myths. Mini Rev Med Chem. 2003;3:861–875. doi: 10.2174/1389557033487629. [DOI] [PubMed] [Google Scholar]
- Lucas X, Senger C, Erxleben A, Grüning BA, Döring K, Mosch J, Flemming S, Günther S. StreptomeDB: a resource for natural compounds isolated from Streptomyces species. Nucleic Acids Res. 2013;47:D1130–D1136. doi: 10.1093/nar/gks1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco JM. Prediction of brain–blood distribution of a large set of drugs from structurally derived descriptors using partial least squares (PLS) modelling. J Chem Inf Comput Sci. 1999;39:396–404. doi: 10.1021/ci980411n. [DOI] [PubMed] [Google Scholar]
- Navia MA, Chaturvedi PR. Design principles for orally bioavailable drugs. Drug Dev Today. 1996;1:179–189. doi: 10.1016/1359-6446(96)10020-9. [DOI] [Google Scholar]
- Ntie-Kang F, Mbah JA, Mbaze LM, Lifongo LL, Scharfe M, Ngo Hanna J, Cho-Ngwa F, Amoa Onguéné P, Owono Owono LC, Megnassan E, Sippl W, Efange SMN. CamMedNP: Building the Cameroonian 3D structural natural products database for virtual screening. BMC Complement Altern Med. 2013;13:88. doi: 10.1186/1472-6882-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OCHEM: A platform for the creation of in silico ADME /Tox prediction models. http://www.eadmet.com/en/ochem.php
- Oprea TI. Current trends in lead discovery: are we looking for the appropriate properties? J Comput-Aided Mol Des. 2002;16:325–334. doi: 10.1023/A:1020877402759. [DOI] [PubMed] [Google Scholar]
- Potts RO, Guy RH. Skin permeability. Pharm Res. 1992;9:663–669. doi: 10.1023/A:1015810312465. [DOI] [PubMed] [Google Scholar]
- Potts RO, Guy RH. A predictive algorithm for skin permeability: The effects of molecular size and hydrogen bond activity. Pharm Res. 1995;12:1628–1633. doi: 10.1023/A:1016236932339. [DOI] [PubMed] [Google Scholar]
- Quinn RJ, Carroll AR, Pham MB, Baron P, Palframan ME, Suraweera L, Pierens GK, Muresan S. Developing a drug-like natural product library. J Nat Prod. 2008;71:464–468. doi: 10.1021/np070526y. [DOI] [PubMed] [Google Scholar]
- Schneider G. Trends in virtual computational library design. Curr Med Chem. 2002;9:2095–2102. doi: 10.2174/0929867023368755. [DOI] [PubMed] [Google Scholar]
- QikProp, version 3.4. New York, NY: LLC; 2011. [Google Scholar]
- LigPrep software, version 2.5. New York, NY: LLC; 2011. [Google Scholar]
- Maestro, version 9.2. New York, NY: LLC; 2011. [Google Scholar]
- QikProp 3.4 User Manual. New York, NY: LLC; 2011. [Google Scholar]
- Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput. 2010;6:1509–1519. doi: 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- Stenberg P, Norinder U, Luthman K, Artursson P. Experimental and computational screening models for the prediction of intestinal drug absorption. J Med Chem. 2001;44:1927–1937. doi: 10.1021/jm001101a. [DOI] [PubMed] [Google Scholar]
- Teague SJ, Davis AM, Leeson PD, Opea TI. The design of leadlike combinatorial libraries. Angew Chem, Int Ed. 1999;38:3743–3748. doi: 10.1002/(SICI)1521-3773(19991216)38:24<3743::AID-ANIE3743>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Tetko IV, Bruneau P, Mewes H-W, Rohrer DC, Poda GI. Can we estimate the accuracy of ADMET predictions? Drug Discov Today. 2006;11:700–707. doi: 10.1016/j.drudis.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Van de Waterbeemd H, Gifford E. ADMET in silico modelling: towards prediction paradise? Nat Rev Drug Discov. 2003;2:192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- Vandenberg JI, Walker BD, Campbell TJ. HERG K+ channels: Friend or foe. Trends Pharmacol Sci. 2001;22:240–246. doi: 10.1016/S0165-6147(00)01662-X. [DOI] [PubMed] [Google Scholar]
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Verdonk ML, Cole JC, Hartshorn ML, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- Yazdanian M, Glynn SL, Wright JL, Hawi A. Correlating partitioning and caco-2 cell permeability of structurally diverse small molecular weight compounds. Pharm Res. 1998;15:1490–1494. doi: 10.1023/A:1011930411574. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Prepared 3D structures of compounds currently included in StreptomeDB with calculated “drug-likeness” descriptors. (MDB 2 MB)
Additional file 2: 3D structures of the “drug-like” subset derived from the StreptomeDB library. (MOL2 2 MB)
Additional file 3: 3D structures of the “lead-like” subset derived from the StreptomeDB library. (MOL2 720 KB)
Additional file 4: 3D structures of the “fragment-like” subset derived from the StreptomeDB library. (MOL2 208 KB)
Additional file 5: 3D structures of the total StreptomeDB dataset with calculated pharmacokinetic descriptors. (SDF 13 MB)


