EMBO reports(2013) 14 8, 733–740 doi:; DOI: 10.1038/embor.2013.86
Intracellular bacterial pathogens exploit the compartmentalization of the eukaryotic cell to create an environment that supports their own survival and growth. Among cellular organelles, the nucleus has long been considered to be mostly ‘safe’ from direct bacterial attacks. However, a growing number of molecules secreted by bacteria—termed ‘nucleomodulins’—target this central organelle to subvert the host defences of plants and animals [1].
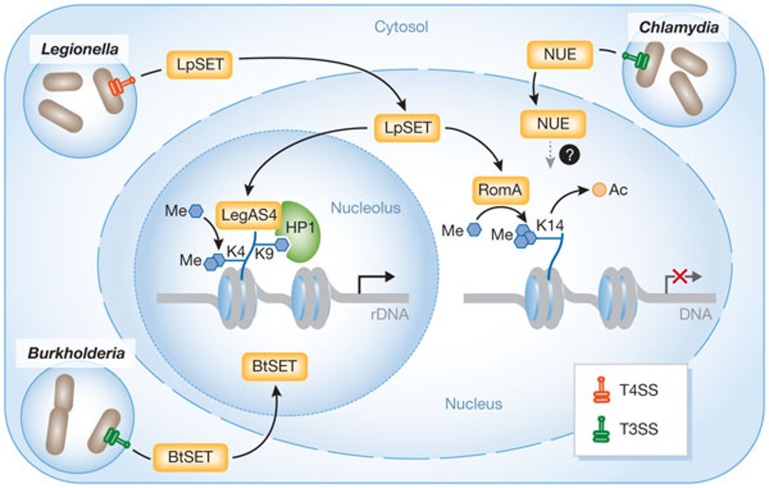
The nucleus itself is a spatially organized compartment, with distinct chromosome territories and many non-membrane-delineated nuclear bodies. The most prominent of such subnuclear structures is the nucleolus, which is the site of both ribosomal RNA (rRNA) synthesis and the assembly of ribosomal subunits. In this issue of EMBO reports, new research by Ting Li, Feng Shao and colleagues shows that bacterial pathogens, such as Legionella and Burkholderia spp., can hijack the nucleolus by secreting SET-domain effectors that target and modify rDNA chromatin (Fig 1; [2]). This activates rDNA transcription and promotes bacterial intracellular survival. SET-related proteins exist in diverse bacterial species, which suggests that controlling rDNA transcription in the nucleolus might be a general virulence strategy.
Figure 1.
Bacterial histone methyltransferases control host gene expression in the nucleus and nucleolus. Legionella, Burkholderia and Chlamydia bacteria residing in cytosolic vacuoles secrete SET-domain-containing effector proteins in the cytosol through type 3 (T3SS; Burkholderia and Chlamydia) or type 4 (T4SS; Legionella) secretion systems. C. trachomatis secretes the nuclear effector (NUE), which targets the nucleus at unknown loci [4]. L. pneumophila secretes a methyltransferase (LpSET) that has been characterized in two ways: (i) in the nucleus, LpSET termed ‘RomA’ trimethylates histone H3 at Lys 14, causing a switch from acetylated to methylated H3K14 at specific gene promoters and thus transcriptional repression [9]; (ii) in the nucleolus, LpSET termed ‘LegAS4’ is proposed to bind to HP1 at rDNA silent promoters and activate transcription by stimulating H3K4 methylation [2]. B. thailandensis secretes a LegAS4-like protein (BtSET) that also activates rDNA transcription in the nucleolus [2]. These mechanisms promote bacterial replication.
In eukaryotic cells, the state of chromatin compaction has a major role in nuclear processes requiring access to DNA. Compaction is regulated by a plethora of histone post-translational modifications (PTMs), and catalysed by a wide range of enzymes termed ‘writers’, such as histone methyltransferases (HMTases). Histone marks created by these enzymes can either activate (for example, H3K4me) or repress (for example, H3K9me and H3K27me) transcription. Histone PTMs serve as signalling platforms that recruit regulatory proteins, termed ‘readers’, which themselves recruit or stabilize other chromatin components. Readers are docked to histone PTMs through chromatin-binding modules. One such reader is heterochromatin protein 1 (HP1), which binds to H3K9me2/3 by its chromodomain and contributes mainly to heterochromatin formation and gene silencing.
Bacterial pathogens can alter host chromatin structure, and thus gene expression, by interfering with signalling pathways that regulate histone-modifying enzymes or by producing mimics of these enzymes [3]. The first example of a bacterial HMTase mimic was discovered in the human pathogen Chlamydia trachomatis by finding a secreted protein, nuclear effector (NUE), that has sequence similarities with eukaryotic SET-domain-containing lysine-specific methyltransferases. NUE enters the nucleus and associates with chromatin (Fig 1; [4]). However, although NUE methylates mammalian histones in vitro, its target genes in the host infected cell remain unknown.
Li and colleagues have now identified a SET-domain-containing protein in the opportunistic bacterial pathogen Legionella pneumophila, which causes a life-threatening pneumonia called Legionnaires’ disease. On phagocytosis by macrophages, Legionella uses a Dot/Icm type IV secretion system (T4SS) to deliver bacterial effector proteins that transform the Legionella-containing vacuole (LCV) into a protective compartment [5]. Li and colleagues describe a secreted effector, LegAS4, acting in the nucleolus, far away from the LCV [2]. Human cells contain several hundred rDNA genes organized in tandem repeats that are clustered into nucleolar organizer regions. LegAS4 binds to rDNA at promoter and intergenic-spacer regions, which results in increased 45S pre-RNA synthesis. LegAS4 harbours a SET-domain-sharing 35% sequence identity with eukaryotic NSD2/3 Lys HMTases of the SET2 family. In vitro studies on histone H3 substrate, using methylation-specific H3 antibodies, suggest that LegAS4 catalyses dimethylation of histone H3 on Lys 4 (H3K4me2). Consistently, ectopic expression of LegAS4 in human cells is associated with increased levels of H3K4me2 at rDNA promoters and the activation of the transcription of these genes.
Epigenetics and the chromatin structure of rRNA genes have a fundamental role in regulating transcription of rDNA loci [6]. In each cell, a fraction of rRNA genes is transcriptionally active, while the rest remain silent. In this regard, the identification of a specific interaction between LegAS4 and the chromatin reader HP1 (HP1α/γ) provides a molecular rationale for the specific recruitment of this bacterial effector to rDNA (Fig 1; [2]). HP1 bound to H3K9me2/3 is enriched in rDNA heterochromatic regions. Docking of LegAS4 to these regions through binding to HP1, and subsequent methylation of H3K4, might convert the epigenetically silent state of rDNA genes to an active state (Fig 1). It is worth mentioning that HP1 not only localizes to silent rRNA genes, but also marks active genes, after H3K9 methylation by the eukaryotic HMTase G9a/KMT1C [7]. Therefore, future investigations are required to dissect the precise order of events that result in LegAS4-mediated rDNA expression.
Interestingly, LegAS4-like proteins are present in various pathogenic bacterial species of animals and plants [2]. In particular, the Burkholderia thailandensis T3SS effector BtSET also promotes H3K4 methylation of rDNA chromatin and transcription (Fig 1). Thus, epigenetic modulation of host rDNA expression might be a pathogenic mechanism shared by various bacteria, as stimulation of rDNA expression seems to contribute to bacterial replication, although the mechanism at play is not yet understood.
Hijacking the nucleolus
The primary role of the nucleolus is rRNA synthesis and ribosome biogenesis. One might speculate that bacterial pathogens induce rDNA transcription to exploit host ribosome activity for their own survival. In line with this idea, Legionella-containing vacuoles are studded with an increasing number of ribosomes during the first 8 h after bacterial internalization, after which the bacteria start to replicate in the vacuole [5]. The nucleolus is also involved in other important functions, such as regulation of mitosis and cell proliferation. Stimulation of host rDNA transcription by bacteria might also promote the proliferation of infected cells, hence providing a better niche for bacterial replication.
Not surprisingly, various plant and animal viruses also target nucleolar functions as part of their infection strategy [8]. Many virally encoded proteins target the nucleolus in various ways, and it will be of interest to see whether bacterial SET effectors use any of these mechanisms.
Bacterial HMTase with dual functions
While the Shao group has characterized LegAS4 from the L. pneumophila strain Philadelphia, Monica Rolando, Carmen Buchrieser and colleagues have studied the equivalent effector from the L. pneumophila strain Paris, which they have named RomA (regulator of methylation A) [9]. RomA has a major role in bacterial replication in THP1 macrophages. Although both studies have found that Legionella SET effectors act as HMTases, their conclusions on the function of these homologous enzymes are different. RomA has a genome-wide function; instead of catalysing H3K4me2, it trimethylates Lys 14 of H3 (H3K14me3), a histone mark not previously described in mammals. By promoting a burst of H3K14me3, RomA decreases H3K14 acetylation, which is an activating mark, thus leading to repression of host gene expression (Fig 1; [9]). In addition, ChIP-seq analysis by Rolando et al identified 4,870 H3K14 methylated promoter regions, including at innate immune genes, during Legionella infection.
Discrepancies between the two papers are puzzling, but not contradictory. It is possible that the Legionella SET effectors behave similarly to eukaryotic HMTases in their ability to have different substrates and functions. For instance, the H3K9 HMTase G9a/KMT1C predominantly represses genes at euchromatic regions but acts as a positive activator of rDNA transcription [7]. It is interesting to note that LegAS4/RomA has identity with NSD2/3 HMTases, for which it has been shown that the residues they modify on histone H3 depend on the nature of the substrate provided [10]. The modification catalysed by RomA was identified by mass spectrometry on H3 and by using a large panel of antibodies on histone octamers [9], while LegAS4-induced modification was less extensively investigated [2]. In future studies, it will be important to determine whether LegAS4 can also catalyse H4K14me3 in the nucleolus. Also of interest is the potential role of the LegAS4 host partner HP1 in RomA-mediated genome-wide repression.
These studies illustrate the fascinating ability of intracellular pathogens to use molecular mimicry to manipulate host processes and fine-tune host gene expression. But this raises many questions. For instance, how are the target specificities of LegAS4/RomA regulated? Is there any link with other epigenetic marks? Do these methyltransferases have non-histone substrates? What are the roles of regions other than SET domains in the bacterial HMTase functions? What is the biological significance of a nuclear/nucleolar tropism? The crosstalk between cellular microbiology and epigenetics, in a new discipline that we might call ‘nuclear microbiology’, should help to shed light on the function of these bacterial effectors in reprogramming the host response to infection.
Footnotes
The author declares that she has no conflict of interest.
References
- Bierne H, Cossart P (2012) Cell Microbiol 14: 622–633 [DOI] [PubMed] [Google Scholar]
- Li T et al. (2013) EMBO Rep (In the press) [Google Scholar]
- Bierne H et al. (2012) Cold Spring Harb Perspect Med 2: a010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penini ME et al. (2010) PLoS Pathog 6: e1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubber A, Roy CR (2010) Annu Rev Cell Dev Biol 26: 261–283 [DOI] [PubMed] [Google Scholar]
- Guetg C, Santoro R (2012) Epigenetics 7: 811–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X et al. (2007) Mol Cell 27: 585–595 [DOI] [PubMed] [Google Scholar]
- Ni L, Wang S, Zhen C (2012) J Med Microbiol 61: 1637–1643 [DOI] [PubMed] [Google Scholar]
- Rolando M et al. (2013) Cell Host Microbe 13: 395–405 [DOI] [PubMed] [Google Scholar]
- Li Y et al. (2009) J Biol Chem 284: 34283–34295 [DOI] [PMC free article] [PubMed] [Google Scholar]