Abstract
Constitutive heterochromatin is crucial for the integrity of chromosomes and genomic stability. Here, we show that the chromatin remodelling complex NoRC, known to silence a fraction of rRNA genes, also establishes a repressive heterochromatic structure at centromeres and telomeres, preserving the structural integrity of these repetitive loci. Knockdown of NoRC leads to relaxation of centromeric and telomeric heterochromatin, abnormalities in mitotic spindle assembly, impaired chromosome segregation and enhanced chromosomal instability. The results demonstrate that NoRC safeguards genomic stability by coordinating enzymatic activities that establish features of repressive chromatin at centromeric and telomeric regions, and this heterochromatic structure is required for sustaining genomic integrity.
Keywords: NoRC, heterochromatin, telomeres, centromeres, genome stability
Introduction
Constitutive heterochromatin exerts a crucial function in maintaining genomic stability by repressing recombination and mutagenic transposition of repetitive elements. In eukaryotes, heterochromatin is concentrated in pericentromeric and telomeric regions, is enriched for repetitive sequences and has a relatively low gene density. As repetitive sequences present a serious challenge for genomic stability, mechanisms must exist that suppress homologous recombination and protect the structural integrity of these tandem repeats by maintaining them in a compact, transcriptionally refractory state. For this, distinct proteins need to be guided to such sequences to establish a heterochromatic structure. Previous studies have demonstrated that a fraction of ribosomal RNA genes is constitutively maintained in a silent, heterochromatic state that safeguards rDNA stability by preventing aberrant homologous recombination. The heterochromatic, silent state of rRNA genes is established by NoRC (nucleolar remodelling complex), a member of SNF2h-containing chromatin remodelling complexes [1]. TIP5, the large subunit of NoRC, targets histone-modifying enzymes to rDNA to generate a repressive heterochromatic structure [2, 3].
Likewise, repetitive pericentromeric regions have a repressive, heterochromatic structure [4]. Centromeric regions are embedded in pericentromeric repeats, providing a platform for kinetochore assembly and mitotic spindle attachment [5, 6]. In human and mice, the centromere-specific histone H3 variant CENP-A replaces a fraction of canonical histone H3 [7]. Cells deficient in CENP-A show severe defects in chromosome segregation [8], suggesting that specific histone modifications determine the structure of centromeric and pericentromeric chromatin, which in turn is relevant for the integrity and function of kinetochores [9].
Moreover, nucleosomes at telomeres and subtelomeres are enriched in heterochromatic marks, for example, H3K9me3, which is responsible for the recruitment of HP1 proteins and trimethylation of histone H4 at lysine 20 (H4K20me3) [10]. Sequestration of telomeres into protective nucleoprotein caps masks chromosome ends from exposure to the DNA-damage-response machinery, thus protecting chromosome integrity. Loss of heterochromatic histone modifications and transient decondensation of chromatin at chromosome ends impairs telomere homeostasis, leading to uncontrolled telomere elongation, enhanced recombination and genomic instability, underscoring the importance of a proper heterochromatic conformation for telomere function [11, 12].
Although our understanding of the function of heterochromatin has tremendously increased, the mechanisms underlying heterochromatin formation at centromeric, pericentromeric and telomeric repeats remained elusive. Considering the repetitive nature and heterochromatic features of rRNA genes, centromeres and telomeres, we reasoned that the chromatin remodelling complex NoRC that silences rRNA genes might also mediate heterochromatin formation at other genomic regions. Here, we show that NoRC also establishes a repressive chromatin environment at pericentromeres, subtelomeres and telomeres, and NoRC-dependent heterochromatin formation at these repetitive gene loci is crucial for genomic integrity.
Results and discussion
NoRC establishes pericentromeric heterochromatin
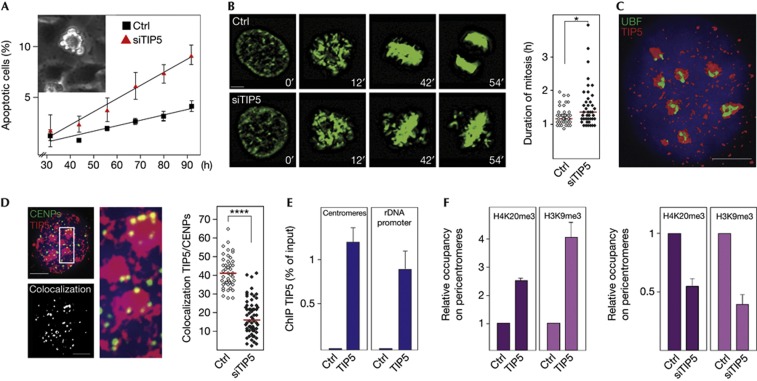
As NoRC promotes heterochromatin formation and silencing of rRNA genes, we hypothesized that this chromatin remodelling complex might have a general role in higher-order chromatin formation at repetitive sequences and serve a function in cell viability and genome stability. Indeed, a moderate but reproducible delay in cell proliferation was observed after knockdown of TIP5, the large subunit of NoRC (supplementary Fig S1A–D online). To unravel the mechanism causing growth defects in TIP5-deficient cells, we monitored cell cycle progression by live cell microscopy of HeLa/Kyoto cells that express green fluorescent protein-tagged histone H2B. After knockdown of TIP5, an increase in the number of apoptotic cells was observed (Fig 1A; supplementary Movie 1 online). Fluorescent images captured during time-lapse microscopy showed impaired metaphase plate formation that was accompanied by slightly prolonged mitosis in TIP5-depleted cells, suggesting that spindle organization has been altered (Fig 1B; supplementary Movie 2 online). Analysis of TIP5 localization by high-resolution deconvolution microscopy and three-dimensional reconstruction revealed that most of TIP5 localized within nucleoli (Fig 1C). However, TIP5 was also found at distinct foci outside the nucleolus. In accord with a previous study showing that NoRC affects perinucleolar heterochromatin [13], a significant fraction of TIP5 co-stained with kinetochore proteins (CENPs, Fig 1D). Chromatin immunoprecipitation (ChIP) assays confirmed TIP5 association with pericentromeric regions, the amount of TIP5 associated with pericentromeric chromatin being similar to TIP5 bound to rDNA (Fig 1E). TIP5 was not associated with the pre-rRNA-coding region, repetitive SINE elements or the X-chromosome, underscoring the specificity of the ChIP (supplementary Fig S2 online). Consistent with NoRC promoting heterochromatin formation, short interfering RNA-mediated depletion of TIP5 decreased the amount of H3K9me3 and K4K20me3 at major satellite repeats (Fig 1F, right). Conversely, overexpression of TIP5 increased the amount of heterochromatic histone marks, supporting the role of NoRC in centromeric chromatin structure (Fig 1F, left).
Figure 1.
NoRC affects centromere function. (A) Depletion of TIP5 induces apoptosis. Percentage of apoptotic HeLa/Kyoto cells transfected with control siRNA (ctrl) or siRNA against TIP5 (siTIP5). Data represent mean values and error bars indicate s.e.m. (n=3). A representative apoptotic cell is shown in the inset. (B) Knockdown of TIP5 delays metaphase plate formation and prolongs mitosis. Representative images of mitotic HeLa/Kyoto cells 72 h after transfection with control (ctrl) and TIP5-specific siRNA (siTIP5). The graph indicates the length of mitosis in a representative experiment (N=3), each dot represents one cell and red lines indicate the median duration of mitosis (Mann–Whitney test, *P=0.028, n=43). Scale bar, 5 μm. (C) NoRC is localized in nucleoli and the nucleoplasm. Three-dimensional reconstruction of deconvolved image of an interphase U2OS cell nucleus immunostained with α-TIP5 (red) and α-UBF (green) antibodies. DNA was counterstained with DAPI. Scale bar, 5 μm. (D) TIP5 localizes at centromeres. Left: Z-projection of a representative nucleus from U2OS cells immunostained with α-TIP5 (red) and α-CENPs antibodies (green). The same nucleus showing colocalization of TIP5 with CENPs (white spots) is shown below. Middle: magnification of an indicated section showing colocalization of TIP5 and CENPs (yellow). Scale bar, 5 μm. Right: quantification of TIP5-positive spots colocalizating with CENPs in a representative experiment (N=4), each dot represents one cell (unpaired t-test, P-value ****<0.0001, n=45). (E) ChIP comparing TIP5 occupancy at major satellite repeats and the rDNA promoter in NIH3T3 cells. Error bars represent s.e.m. (n=3). (F) NoRC triggers heterochromatin formation at pericentromeres. H4K20me3 and H3K9me3 occupancy at pericentromeres of NIH3T3 cells overexpressing TIP5 (left) and in TIP5-depleted cells (right). ChIPs from mock-transfected cells were used as reference (ctrl=1). Error bars represent s.e.m. (n=3). ChIP, chromatin immunoprecipitation; DAPI, 4,6-diamidino-2-phenylindole; NoRC, nucleolar remodelling complex; siRNA, short interfering RNA.
NoRC affects spindle assembly and genome stability
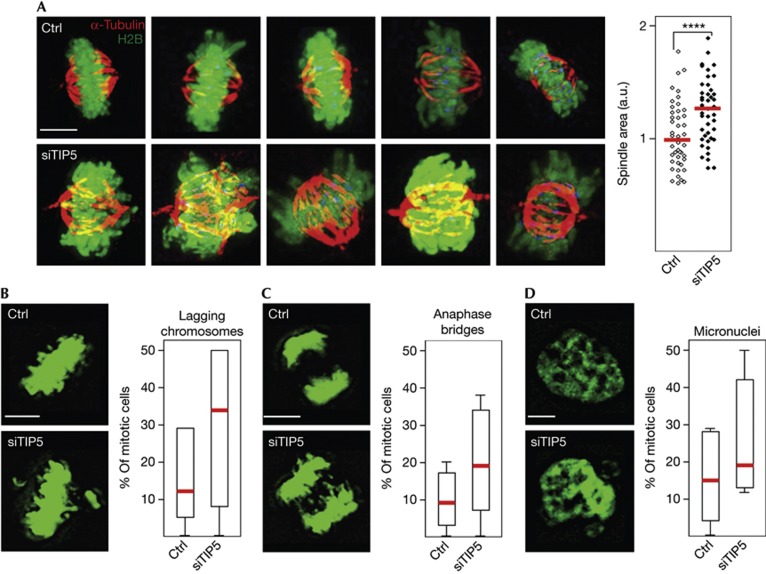
Incorporation of CENP-A, the centromere-specific histone H3 variant, is essential for kinetochore assembly and mitotic spindle formation [7, 9]. Consistent with previous studies [13], TIP5 interacts with CENP-A and with HP1α, suggesting a protein-based mechanism that recruits NoRC to centromeres and adjacent regions (supplementary Fig S3A–B online). The finding that TIP5 depletion impaired cell proliferation suggested that compromised heterochromatin formation has affected mitotic spindle organization. In HeLa/Kyoto cells, mitotic spindles appeared as compact, well-tailored structures. In contrast, abnormally large spindles (30% increase in size) appeared in TIP5-deficient cells, emphasizing the relevance of NoRC for proper kinetochore structure (Fig 2A). Consistent with TIP5 depletion having disturbed centromere structure and function, the number of anaphase bridges and metaphase plates with lagging chromosomes increased. Moreover, micronuclei were frequently observed (Fig 2B–D). These results support the notion that NoRC-dependent heterochromatin formation at centromeric and pericentromeric regions is essential for proper attachment to the mitotic spindle and the fidelity of chromosome segregation.
Figure 2.
NoRC is required for mitotic spindle organization and genome stability. Mitotic spindle is disorganized in TIP5-deficient cells. (A) Mitotic spindles of control (ctrl) and siTIP5-treated HeLa/Kyoto cells stained with α-tubulin antibodies (red) and for GFP-tagged histone H2B with α-GFP (green). Right: quantification of the spindle area in TIP5-depleted compared with control cells equalized to 1, each dot represents one cell (representative experiment, N=3, unpaired t-test, P-value ****<0.0001, n=45). Red bars indicate medians. Scale bar, 5 μm. (B–D). Knockdown of TIP5 impairs genomic stability. Images from live cell microscopy of control (ctrl) and TIP5-deficient (siTIP5) HeLa/Kyoto cells show an increased fraction of mitotic cells with anaphase bridges (B), misaligned chromosomes (C) and interphase nuclei containing micronuclei (D). The box plots represent the abundance of chromosomal mitosis in control and siTIP5-treated cells in a representative experiment (N=2). The overall increase in all types of abnormal mitosis was determined by two-way ANOVA test, P-value=0.0145, n=84. Red bars indicate mean values. Scale bars, 5 μm. ANOVA, analysis of variance; GFP, green fluorescent protein; NoRC, nucleolar remodelling complex.
NoRC controls telomeric chromatin structure
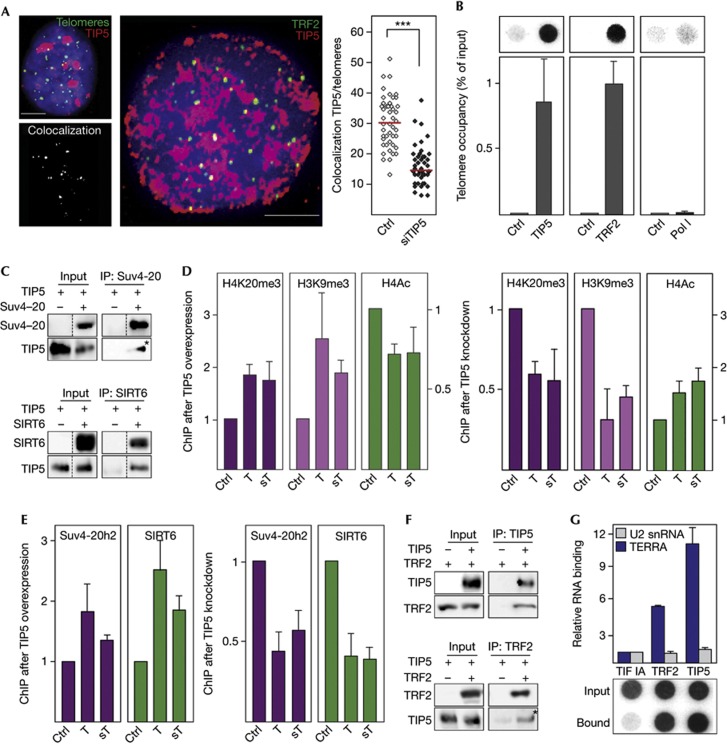
Next, we examined whether NoRC is also involved in the establishment of higher-order chromatin structure at telomeres. Immunofluorescence in situ hybridization (FISH) with a telomere-specific PNA probe as well as co-immunostaining of TRF2 (telomere repeat-binding factor 2) and TIP5 revealed binding of TIP5 to a significant fraction of telomeres (Fig 3A). Moreover, ChIP experiments show TIP5 occupancy at telomeric repeats comparable to that of the telomere repeat-binding factor TRF2 (Fig 3B). Further, NoRC has been shown to specifically interact with several chromatin-modifying enzymes, including the histone deacetylases HDAC1&2, SIRT1 and the methyltransferase SET-DB1 but not with HDAC4 and CHD4/Mi-2 [2, 3, 14]. Here, we demonstrate by co-immunoprecipitation experiments that TIP5 also interacts with the histone methyltransferases Suv3-9 and Suv4-20h2 that deposit H3K9me3 and H4K20me3 at telomeres and subtelomeres [11, 12], with SIRT6 that deacetylates H3K9 [15], and with HP1 proteins that promote spreading of heterochromatin (Fig 3C; supplementary Fig S3B–D online). In accord with NoRC recruiting repressive chromatin modifiers to chromosome ends, overexpression of TIP5 increased the levels of H3K9me3 and H4K20me3 and decreased acetylation of histone H4 (H4ac) at telomeres and subtelomeres, whereas knockdown of TIP5 led to opposite effects (Fig 3D). Consistently, the association of Suv4-20h2 and SIRT6 was increased by overexpression and decreased by depletion of TIP5 (Fig 3E). Given that NoRC is recruited to rDNA by interaction with chromatin-bound proteins, NoRC might be targeted to telomeres by the shelterin complex. In support of this hypothesis, TIP5 co-immunoprecipitated with TRF2 suggesting that TRF2 might recruit NoRC to telomeres (Fig 3F).
Figure 3.
NoRC regulates telomeric heterochromatin. (A) TIP5 localizes at telomeres. Left: Z-projection of a deconvolved image of a representative U2OS cell stained with α-TIP5 antibody (red) followed by Q-FISH with a telomere-specific PNA probe (green). The same nucleus depicting colocalization of TIP5 with telomeres (white spots) is shown below. Middle: a representative U2OS cell immunostained with α-TIP5 (red) and α-TRF2 (green) antibodies is shown. Colocalization of TIP5 and TRF2 is depicted in yellow. Right: quantification of telomeres colocalizing with TIP5 in a representative experiment (N=4), each dot on the plot represents one cell (unpaired t-test, P-value ***<0.001, n=45). Red bars indicate mean values. Scale bars, 5 μm. (B) TIP5 is associated with telomeres. ChIPs in U2OS cells monitoring TIP5, TRF2 and Pol I at telomeres. Telomere DNA was assayed by dot hybridization with a telomere-specific riboprobe. Data are normalized to input values. Error bars represent s.e.m. (n=3). (C) TIP5 interacts with chromatin modifiers. Co-IP experiments showing association of Flag-tagged TIP5 with GFP-tagged Suv4-20h2 and SIRT6. Non-adjacent lanes in the original blots are indicated by dotted lines. Co-IP lanes exposed longer than the input lanes are marked with an asterisk. (D) TIP5 triggers heterochromatic histone marks at subtelomeres and telomeres. ChIP of H4K20me3, H3K9me3 and H4Ac at T and sT after overexpression of TIP5 (n=5, left panels) and after knockdown of TIP5 (n=3, right panels) in U2OS cells. ChIPs from cells transfected with empty vector or control siRNA were used as a reference (ctrl=1). Data are normalized to input values. Error bars represent s.e.m. (E) TIP5 recruits histone-modifying enzymes to telomeres and subtelomeres. ChIPs showing binding of ectopic Suv4-20h2 and SIRT6 to telomeres and subtelomeres after overexpression (left panels) and knockdown of TIP5 (right panels) in NIH3T3/ER-Suv4-20h2 cells. ChIPs from cells transfected with empty vector or control siRNA, respectively, were used as a reference (ctrl=1). Data are normalized to input values. Error bars represent s.e.m. (n=3). (F) TIP5 interacts with TRF2. Co-IP experiment showing association of Flag-tagged TIP5 with GFP-tagged TRF2. Co-IP lanes exposed longer than the input lanes are marked with an asterisk. (G) NoRC interacts with TERRA in vivo. GFP-tagged proteins were immunoprecipitated from HEK293T cells and associated RNAs were quantified by dot blot (TERRA) or qPCR (U2 snRNA). Data represent fold excess of precipitated RNA (normalized to input) in cells expressing TIP5 over control cells. Error bars represent s.e.m. (n=3). ChIP, chromatin immunoprecipitation; Co-IP, co-immunoprecipitation; GFP, green fluorescent protein; NoRC, nucleolar remodelling complex; Q-FISH, quantitative fluorescence in situ hybridization; siRNA, short interfering RNA; snRNA, small nuclear RNA; sT, subtelomeres; T, telomeres; TERRA, telomeric repeat-containing RNA.
The establishment of higher-order chromatin structure at rDNA, telomeres and centromeres has been shown to depend on specific noncoding RNAs, such as pRNA [16, 17], telomeric repeat-containing RNA (TERRA) [18, 19] and RNAs originating from satellite repeats [4, 20]. Notably, overexpression of TIP5 increased the levels of TERRA and centromeric satellite transcripts (supplementary Fig S4A online), whereas these ncRNAs were downregulated after TIP5 knockdown (supplementary Fig S4B online). The fact that overexpression or depletion of TIP5 led to corresponding changes of telomeric and centromeric RNA suggests that NoRC cooperates with important components of telomeric and centromeric silencing networks that preserve the structural and functional integrity of these loci. As NoRC requires associated RNA (pRNA) to be targeted to the rDNA promoter [16, 17], we reasoned that TERRA might have a similar role in guiding NoRC to telomeres. To test this, we performed RNA immunoprecipitation experiments to monitor the association of TERRA with TRF2 and TIP5. Dot blot hybridization revealed a strong association of TIP5 with TERRA (Fig 3G). No binding of TERRA to the Pol I-specific transcription factor TIF-IA was observed. Moreover, U2 small nuclear RNA, an abundant control RNA, was not associated with either protein, underscoring the specificity of the RNA immunoprecipitation assay. Together, these results indicate that TERRA might serve a guiding function in NoRC-dependent heterochromatin formation at telomeres.
NoRC suppresses telomeric recombination
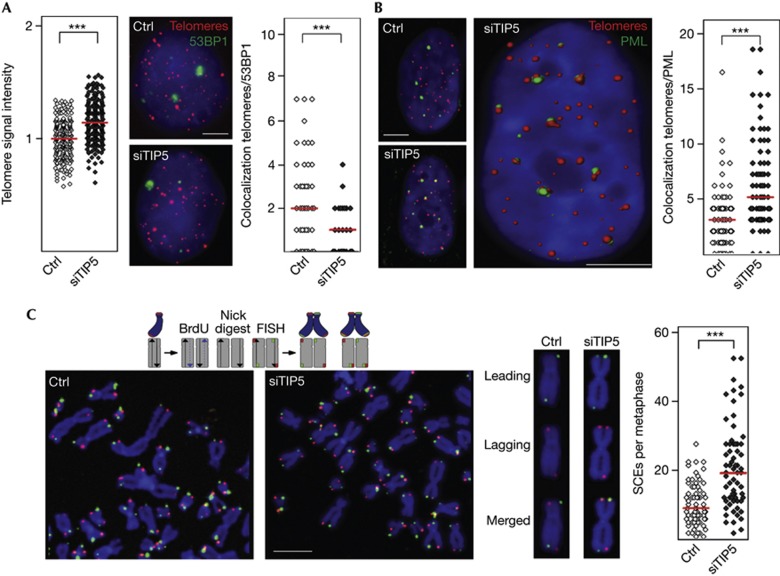
As loss of telomeric heterochromatin correlates with telomere elongation [11, 12], depletion of NoRC should lead to changes in telomere homeostasis. Indeed, interphase Q-FISH (quantitative FISH) and dot blot hybridization with telomeric probes revealed enhanced signal intensities in TIP5-deficient cells (Fig 4A; supplementary Fig S5A online), indicating that disruption of silent chromatin has compromised telomere-length control. However, in contrast to inactivation of TRF2 known to induce uncapped dysfunctional telomeres [21], depletion of TIP5 did not increase the number of telomere dysfunction-induced foci, monitored by telomeric immuno-FISH of p53BP1 or γ-H2AX (Fig 4A; supplementary Fig S5B online). Furthermore, no obvious telomere aberrations, such as loss of telomeres or telomere duplications, were observed in TIP5-deficient cells (supplementary Fig S5C online). Noteworthy, depletion of TIP5 increased the association of telomeres with promyelocytic leukaemia nuclear bodies (PML-NBs, Fig 4B), a phenotype known to coincide with the loss of telomeric heterochromatin [11]. As the interaction with PML-NBs indicates increased telomeric recombination, we assayed the frequency of telomeric sister chromatid exchange on metaphase spreads of control and TIP5-deficient cells by CO-FISH (chromosome orientation-FISH) using differently labelled strand-specific telomeric probes. Quantification of telomeric sister chromatid exchange events revealed a twofold increase in homologous recombination in TIP5-depleted cells as compared with control cells (Fig 4C), reinforcing that NoRC suppresses telomeric recombination. Moreover, multiplex-FISH (M-FISH) on metaphase spreads showed a >2-fold increase in telomere translocations as early as 32 h after TIP5 knockdown, emphasizing the impact of NoRC on telomere and chromosome stability (supplementary Table S2 online).
Figure 4.
NoRC suppresses recombination between telomeric repeats. (A) Left: quantification of telomeric Q-FISH in U2OS cells transfected with control siRNA (ctrl) or siRNA against TIP5 (siTIP5). Telomeric signals in a representative experiment (N=3) were quantified and compared by unpaired t-test (P-value ***<0.001, n=240). Red bars indicate mean values. Middle: U2OS cells transfected with control siRNA or siTIP5 were immunostained for 53BP1 protein (green) followed by Q-FISH with a telomere-specific PNA probe (red). Scale bar, 5 μm. Right: quantification of telomeres colocalizing with 53BP1 in a representative experiment (N=5, unpaired t-test, P-value ***<0.001, n=42). (B) Enhanced association of telomeres with PML bodies in TIP5-deficient U2OS cells. Left: U2OS cells transfected with control siRNA or siTIP5 were immunostained for PML protein (green) followed by Q-FISH with a telomere-specific PNA probe (red). Middle: reconstructed three-dimensional image of the same nucleus from TIP5-depleted cells. Right: colocalization of PML bodies and telomeres was measured in control and siTIP5-treated cells in a representative experiment (N=4) and compared by Mann–Whitney test (P-value ***<0.001, n=77). Red bars indicate medians. Scale bars, 5 μm. (C) T-SCE is increased in TIP5-depleted U2OS cells. Left: images of metaphase spreads from control (ctrl) and TIP5-deficient cells (siTIP5). A scheme illustrating the CO-FISH is shown above. Middle: chromosome images from control and siTIP5-depleted cells. The leading telomeric strand is labelled in green, the lagging strand in red; yellow signals indicate recombination between telomeres. Right: quantification of T-SCE events in control and siTIP5-treated cells in a representative experiment (N=3, unpaired t-test, P-value ***<0.001, n=68). Red bars indicate medians. Scale bar, 5 μm. CO-FISH, chromosome orientation fluorescence in situ hybridization; NoRC, nucleolar remodelling complex; PML, promyelocytic leukaemia body; Q-FISH, quantitative FISH; siRNA, short interfering RNA; T-SCE, telomeric sister chromatid exchange.
Together, our results demonstrate that NoRC function is not restricted to silencing of rRNA genes but is also required for heterochromatin formation at telomeres and centromeres. In the absence of TIP5, enhanced recombination between telomeric repeats and mitotic defects were observed, underscoring the significance of NoRC-dependent heterochromatic structure in sustaining the integrity of telomeres and centromeres. TIP5 interacts with HP1α and with histone-modifying enzymes, for example, HDAC1&2 [2, 3], SIRT6, SET-DB1 [14], Suv3-9 and Suv4-20, all of which mediate higher-order chromatin structure at telomeres and centromeres. Regarding the mechanism of NoRC-directed heterochromatin formation, our results reveal that NoRC is recruited to distinct genome loci by specific chromatin-associated proteins, for example, to rDNA by TTF-I, to centromeres by CENP-A and to telomeres by the shelterin complex. The establishment of higher-order chromatin structure at repetitive elements has been shown to depend on specific noncoding RNAs, including pRNA [16, 17], TERRA [18, 19] and major satellite RNA [4, 20]. In accord with previous studies demonstrating compromised heterochromatin structure at telomeres on depletion of TERRA [19], the levels of TERRA and satellite RNA correlate with levels of TIP5 (supplementary Fig S4 online). Moreover, the observation that TIP5 is associated with these ncRNAs suggest that protein- and RNA-based mechanisms guide NoRC to specific genomic loci to preserve their structural and functional integrity.
Our finding that depletion of TIP5 impairs cell cycle progression and leads to mitotic aberrations is in apparent contradiction to previous work reporting increased proliferation after TIP5 depletion [13]. However, while the former study has been performed with a TIP5-deficient cell line that exhibited pronounced alterations in morphology and might have been transformed during long-term cultivation, we have now monitored functional consequences of TIP5 depletion early (60 h) after transfection and at the single-cell level. The results demonstrate that depletion of TIP5 leads to abnormalities in chromosome segregation, mitotic defects and growth retardation, reinforcing that NoRC safeguards genomic stability. Although the molecular details underlying NoRC function at telomeres and centromeres need further analysis, we propose that NoRC, surely among other factors as well, triggers the establishment or maintenance of compact chromatin structure at other genomic sequences with wide-ranging implications for genome structure and function.
Methods
Description of plasmids, antibodies, transfection, knockdown of RNA, ChIPs and co-immunoprecipitations can be found in Supplemental Methods online.
Immunofluorescence and telomere FISH assays. Cells were permeabilized in 20 mM Tris–HCl (pH 7.4), 25% glycerol, 0.04% Triton X-100, 0.5 mM EDTA, 5 mM MgCl2 (3 min, room temperature), fixed in methanol (6 min, −20 °C) and acetone (1 min), incubated with primary antibodies, stained with fluorophore-coupled secondary antibodies, hybridized with a Cy3- or FITC-labelled telomere-specific PNA probe (DAKO, Denmark) and analysed by deconvolution microscopy. Telomeric FISH, deconvolution microscopy, three-dimensional reconstruction and image processing were performed as described [22]. For multiplex FISH (M-FISH), cells were arrested with vinblastin (80 ng/ml, 60 min) with or without calyculin A (0.013 μg/ml, 30 min), fixed and spread metaphases were hybridized using the multicolour probe kit 24XCyte (Metasystems, Germany), as described [23].
Chromosomal recombination analysis. For CO-FISH, cells grown in the presence of 7.5 mM 5′-BrdU and 2.5 mM 5′-BrdC for 20 h were arrested by colcemid (0.05 μg/ml, 3 h). Fixed mitotic cells were spread on coverslips, exposed to UV light, treated with exonuclease III and single-stranded DNA was hybridized with Cy3-OO-(TTAGGG)3 followed by FITC-OO-(CCCTAA)3 telomeric PNA probe (Panagene Inc., [24]).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank M. Evyilmerz and Y. Odunsi for help in the cytogenetic analyses and acquisition of microscopic images. I.G.’s work has been supported by the DFG, the BMBF (0315502A ‘EpiSys’) and the ERC (232645). Work in P.B.’s group was supported by the Deutsche Krebshilfe e.V. (Tumorstammzell Verbund) and the BMBF (02NUK003).
Author contributions: A.P.-I., and D.K. designed and carried out most experiments, N.S. performed experiments, K.M.G.-B. performed M-FISH. I.G. and P.B. guided the project and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Längst G, Grummt I (2001) NoRC—a novel member of mammalian ISWI chromatin remodeling machines. EMBO J 20: 4892–4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32: 393–396 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Santoro R, Grummt I (2002) The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J 21: 4632–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C, Bailly D, AHFM Peters, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G (2002) Higher order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet 30: 329–334 [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenatri M, Bailly D, Maison C, Almouzni G (2004) Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol 166: 493–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Bassett EA (2008) The histone variant CENP-A and centromere specification. Curr Opin Cell Biol 20: 91–100 [DOI] [PubMed] [Google Scholar]
- Regnier V, Vagnarelli P, Fukagawa T, Zerjal T, Burns E, Trouche D, Earnshaw W, Brown W (2005) CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol Cell Biol 25: 3967–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco HD, Pidoux AL, Urano T, Allshire RC (2008) Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science 319: 94–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA (2007) The epigenetic regulation of mammalian telomeres. Nat Rev Genet 8: 299–309 [DOI] [PubMed] [Google Scholar]
- Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA (2007) Suv4-20 deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol 178: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA (2004) Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferase. Nat Genet 36: 94–99 [DOI] [PubMed] [Google Scholar]
- Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottinger M, Fussenegger M, Santoro R (2010) The NoRC complex mediates heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J 29: 2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Feng W, Imhof A, Grummt I, Zhou Y (2007) Activation of RNA Polymerase I transcription by Cockayne Syndrome Group B protein and histone methyltransferase G9a. Mol Cell 27: 585–595 [DOI] [PubMed] [Google Scholar]
- Michishita E et al. (2008) SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452: 492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Santoro R, Grummt I (2006) Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22: 351–361 [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24: 2264–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoefter S, Blasco M (2007) Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol 10: 228–236 [DOI] [PubMed] [Google Scholar]
- Deng Z, Norsen J, Wiedmer A, Riethman H, Lieberman PM (2009) TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35: 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M (2002) Coordinated methyl and RNA binding is required fro heterochromatin localization of mammalian HP1 alpha. EMBO Rep 3: 975–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, de Lange T (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448: 1068–1071 [DOI] [PubMed] [Google Scholar]
- Krunic D et al. (2009) Tissue context-activated telomerase in human epidermis correlates with little age-dependent telomere loss. Biochim Biophys Acta 1792: 297–308 [DOI] [PubMed] [Google Scholar]
- Greulich KM, Kreja L, Heinze B, Brucker M, Fuchs P, Rhein AP, Molls M (2000) M-FISH detects stable and unstable radiation induced chromosomal aberrations. Mut Res 425: 73–81 [DOI] [PubMed] [Google Scholar]
- Celli GB, Denchi EL, de Lange T (2006) Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol 8: 885–890 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.