Abstract
Cancers of various organs have been categorized into distinct subtypes after increasingly sophisticated taxonomies. Additionally, within a seemingly homogeneous subclass, individual cancers contain diverse tumour cell populations that vary in important cancer-specific traits such as clonogenicity and invasive potential. Differences that exist between and within a given tumour type have hampered significantly both the proper selection of patients that might benefit from therapy, as well as the development of new targeted agents. In this review, we discuss the differences associated with organ-specific cancer subtypes and the factors that contribute to intra-tumour heterogeneity. It is of utmost importance to understand the biological causes that distinguish tumours as well as distinct tumour cell populations within malignancies, as these will ultimately point the way to more rational anti-cancer treatments. EMBO reports advance online publication 12 July 2013; doi:10.1038/embor.2013.92
Keywords: clonal evolution, cancer stem cells, cancer subtypes, therapy resistance
See the Glossary for abbreviations used in this article.
Glossary.
- ABL
abelson murine leukemia viral oncogene
- APC
Adenomatous polyposis coli
- BCR
B-cell receptor
- BRAF
v-raf murine sarcoma viral oncogene homologue B1
- c-KIT
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homologue
- c-MET
hepatocyte growth factor receptor
- DLL4
delta-like ligand 4
- EGFR
epidermal growth factor receptor
- EMT
epithelial–mesenchymal transition
- HER2
human epidermal growth factor receptor 2
- HGF
hepatocyte growth factor
- KRAS
kirsten rat sarcoma viral oncogene homologue
- MLH1
MutL homologue 1
- MMTV
mouse mammary tumor virus
- NFκB
nuclear factor kappa-light-chain-enhancer of activated B cells
- PDGFR-α
platelet-derived growth factor receptor alpha
- PyMT
polyoma middle T antigen
- TGF
transforming growth factor
- TGFBR2
TGF beta receptor 2
- TP53
tumor protein p53
Introduction
The first pathological reports made it clear that a single neoplasm presents with tremendous variability among individual tumour cells. This so-called intra-tumour heterogeneity poses an important challenge for predicting tumour behaviour and clinical outcome. Differences exist not only within a single tumour, but also across individual patients presenting with cancers originating in the same organ. Increasing knowledge of this inter-tumour heterogeneity has led to an exhaustive categorization of tumour subsets according to staging, differentiation grade, cellular morphology and marker expression. Intra- and inter-tumour heterogeneity and how they relate to each other are the main focus of this review. The emergence of advanced molecular and biochemical technologies has led to a better understanding of the numerous mechanisms by which both faces of tumour heterogeneity are driven. Indeed, gene expression arrays and next-generation sequencing have paved the way for a more comprehensive indexing of a broad variety of cancer types, such as breast and brain tumours [1,2,3], allowing their separation into robust subtypes with markedly different molecular and clinical qualities. In addition, scrutiny within individual tumours has reached tremendous sensitivity, and many genetic and epigenetic modifications, as well as micro-environmental interactions, can be dissected at the single-cell level [4]. Despite all these efforts, we are only starting to appreciate the complexity of tumours and our limited understanding of the molecular mechanisms by which individual cancers are driven has precluded the development of curative targeted therapies. In more advanced disease for instance, tumours relapse in most cases even after a seemingly successful therapy. This distressing fact has often been attributed to the presence of heterogeneity within tumours, and two main conceptual frameworks have been elaborated to conceptualize the link between intra-tumour heterogeneity and therapy resistance. The first, and most well-established idea, rests on classical evolutionary concepts and postulates that selective pressure acting on tumour cells ultimately leads to (epi-)genetically distinct, resistant clones [5,6]. The second, and more recent theory is based on the realization that subsets of tumour cells inherently have a dissimilar ability to drive tumour growth and metastasis [7,8]. In this scenario, tumours are perceived as hierarchically organized tissues and differences between tumour cells are mainly explained by a gradient of differentiation that exists between cancer stem cells (CSCs) residing at the top of the hierarchy, and their more differentiated progeny. As we discuss below, these two theories coexist perfectly and are complementary rather than mutually exclusive [9]. In addition, tumour heterogeneity is also influenced by the cell of origin and involves the presence of non-neoplastic cells that establish a tumour microenvironment. Here, we provide an overview of several aspects that contribute crucially to tumour heterogeneity (Fig 1) and discuss their implications in a clinical context. Moreover, we highlight the connections that exist between intra- and inter-tumoral heterogeneity in an effort to draw parallels that might contribute to our basic understanding of these two sides of the same coin.
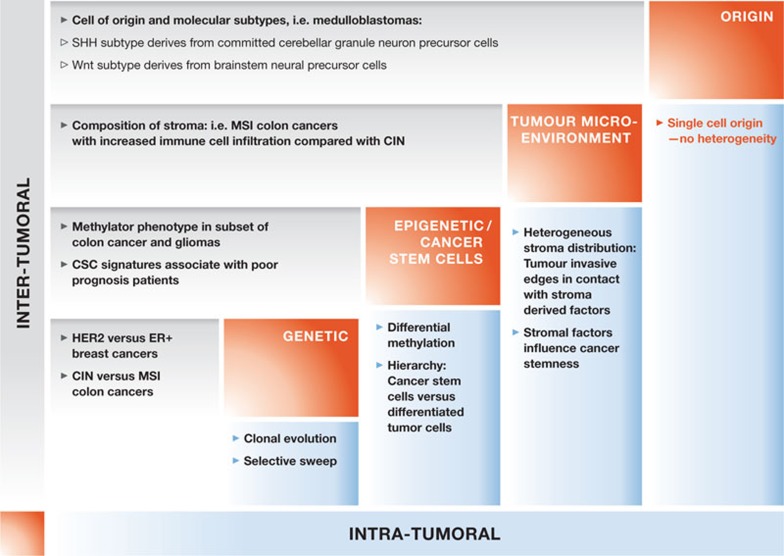
Figure 1.
Schematic representation of the various factors (diagonal, red boxes) that affect intra- (x-axis, light blue) and inter- (y-axis, light grey) tumour heterogeneity. Several examples are depicted for each factor and for both forms of tumour heterogeneity. CIN, chromosomal instability; CSC, cancer stem cell; MSI, microsatellite instability; SHH, sonic hedgehog signalling.
The actors of tumour heterogeneity
The genetic contribution. Genetic variation is undoubtedly the most established foundation of both intra- and inter-tumour heterogeneity. The stepwise acquisition of mutations during cancer evolution was first advocated by Nowell in 1976 [10], and later on substantiated by a large body of experimental work. As tumours progress, multiple and changing environmental pressures operate on the cancer cell population and select for clones that are best endowed to survive and respond to these changes (Fig 2A). Adaptation can be enabled by genetic mutations that confer a selective advantage to a particular clone. This process is highly dynamic and usually selects for clones that will grow and dominate the tumour through so-called ‘selective sweeps’. This is reiterated as long as the tumour ecosystem evolves, leading to the formation of complex clonal patterns [6]. In their landmark paper, Fearon and Vogelstein crystallized this concept by looking at different mutations in a series of patients comparing normal epithelium with cancer precursor lesions and cancer tissue. They reported increasing amounts of mutations with more advanced disease, but importantly, also proposed a preferential sequence of events by inferring the prevalence of certain mutations at each stage [11]. Today the picture has expanded considerably and a full genomic landscape has been drawn for a large variety of malignancies [12,13,14], only to strengthen the notion that clonal evolution is crucial to tumorigenesis. Individual cancers can harbour from a handful [15,16] to hundreds of mutations [17]. This startling diversity has urged efforts to define patterns and sets of mutations that actively contribute to disease initiation and progression, also dubbed ‘driver’ mutations as compared with bystander or ‘passenger’ mutations, which are not causally implicated in oncogenesis but are carried along and accumulate as cancers develop. Identifying the biological nature of driver mutations in tumours, such as those conferring a growth advantage, is relatively straightforward, but the actual benefit they provide for tumour cells remains poorly investigated. Indeed, few studies have attempted to quantify the evolutionary advantage that mutations confer to a clone [18]. Models in which clone size can be easily measured longitudinally [19] are needed for this purpose (Sidebar A), such as the normal colonic crypt, which provides a unique architecture to study stem cell homeostasis. Colonic crypts become monoclonal over time, following a pattern of neutral drift dynamics in which functionally equivalent stem cells either expand or disappear stochastically until they either take over the crypt or are lost [20,21]. Therefore, a mutation specifically initiated in stem cells can affect both neutral drift and the speed at which an affected crypt reaches monoclonality. This can subsequently be used to infer the evolutionary benefit of that particular mutation (L.V., unpublished work). In contrast to the lack of quantitative measurements on genetic hits, it is more apparent that distinct mutations have different impacts on tumour heterogeneity. For instance, inactivation of the tumour suppressor MLH1 in colorectal cancer (CRC) impairs the DNA damage response and leads to the formation of tumours with a high mutation rate and a so-called ‘hypermutator’ phenotype [22]. Finally, assessing clonal heterogeneity with existing techniques requires the evaluation of topologically distinct regions. This is beautifully illustrated in renal carcinoma in which whole-exome sequencing of multiple distinct tumour regions has revealed considerable genetic disparities [23].
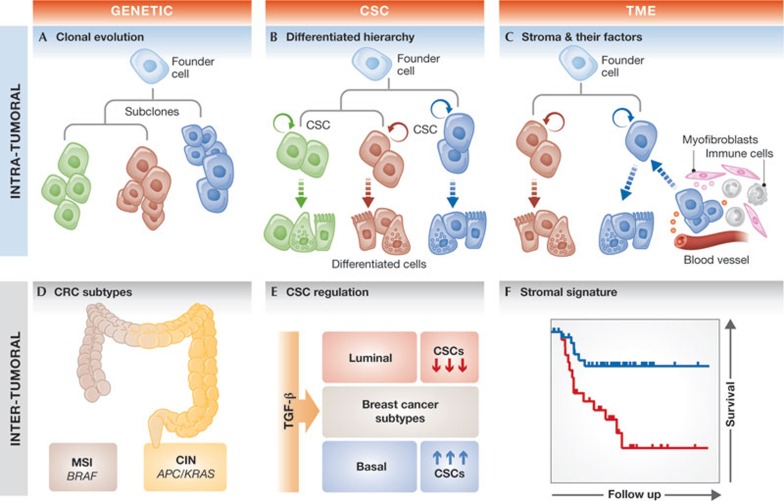
Figure 2.
Cartoon representation of several factors affecting tumour heterogeneity. (A–C) Intra-tumour heterogeneity . (A) From a common ancestor (blue founder cell), different subclones (represented by different colours) emerge due to selection and distinct mutations. (B) In the CSC model, clonal evolution still takes place but only acts on the CSCs. These cells can self-renew and give rise to the various cell lineages present in a tumour (differentiated cells in respective colours). (C) The TME affects intra-tumour heterogeneity by its composition (myofibroblasts in pink and immune cells in grey) as well as their derived factors that can induce the reversion of differentiated cells to CSCs. (D–F) Inter-tumour heterogeneity. (D) The MSI or CIN colon cancer subtypes are distinct subtypes that associate with BRAF or APC and KRAS mutations, respectively. (E) TGF-β pathway activation results in an increase or decrease of CSC phenotype when triggered in the basal or luminal breast cancer subtype, respectively. (F) SDPPs can be used to identify breast cancer patients that differ in their survival probability. APC, Adenomatous polyposis coli; BRAF, v-raf murine sarcoma viral oncogene homologue B1; CIN, chromosomal instability; CSC, cancer stem cell; KRAS, kirsten rat sarcoma viral oncogene homologue; MSI, microsatellite instability; SDPP, stroma-derived prognostic profile; TGF-β, transforming growth factor beta; TME, tumour microenvironment.
Sidebar A | In need of answers.
What is the evolutionary benefit of a single or combination of genetic hits on normal stem cells? Models in which clone size can be measured after alteration of specific genes are needed for this purpose.
Is the cancer stem cell state a fixed quality? What is the rate of de-differentiation in a tumour and what are the factors (tumour microenvironment factors or stochasticity) that promote it?
What are the best models to recapitulate most of the heterogeneity found in primary tumours and which can also be easily used for pre-clinical drug testing?
What are the common biological backgrounds between patients from a given subtype?
Are cancer subtypes suitably evaluated during clinical trials?
Could we approach clonal variation sequentially in the clinic, targeting one subclone at a time and following their evolution?
Genomic differences do not exist solely within an individual cancer but also, and arguably most prominently, among cancers. It is evident that the range of mutations between seemingly identical cancers can vary substantially between patients. Even the type of driver mutations found in each cancer can differ highly. This has often resulted in the identification of molecular subsets of cancers that are made up of diverse mutational spectra (Fig 2D). In the case of CRC, many previous reports have attempted to classify this disease in different categories according to its respective genetic defects [24,25]. For instance, activating mutations of the BRAF oncogene are associated with a particular type of CRC that displays instability in microsatellite (MSI) DNA repeats. Another category shows chromosomal instability and associates with mutations in the TP53 and KRAS genes. Although certain mutation patterns associate with a subset of cancers, the link between the genetic make-up of a tumour and its phenotype is fairly weak. For example, on the most abstract level, similar mutations can result in a vastly different phenotype, whilst the same phenotype can result from different mutations. CRC patients with BRAF mutations illustrate the former, and have markedly different biology and clinical outcome whether or not they present with MSI [24]. Conversely, gastrointestinal stromal tumours (GISTs) that are induced by either c-KIT or PDGFR-α mutations, present with remarkably similar histology and therapy response [26]. Furthermore, our group has shown that unbiased determination of the most biologically distinct CRCs by using gene expression arrays results in the identification of robust subsets that have crucial differences in prognosis, despite a significant overlap in their mutation spectra [27]. This further demonstrates that determining mutations per se is insufficient to appreciate fully biological and, as a consequence, clinical behaviour.
The contribution of epigenetics to tumour heterogeneity. Besides genetic diversity, there is increasing recognition that epigenetic changes, such as DNA methylation and histone deacetylation, can occur throughout tumorigenesis [28]. These changes, and the subsequent biological processes they affect, are inherited during cell division without altering the DNA sequence. DNA methylation is instrumental for crucial biological processes during embryonic development and cellular differentiation [29], and is mediated by DNA methyltransferases that transfer methyl groups to cytosine residues. This process occurs mostly at CpG dinucleotides, which are distributed with high frequency at the promoter region of genes [30]. Aberrant DNA methylation is a characteristic detected in many cancer types that present often with global loss of DNA methylation [31]. More specific alterations are also frequently seen, such as those that target p16 [32], which is a prime example of methylation-mediated inactivation of tumour suppressor genes. In addition to contributing to tumorigenesis, epigenetic alterations are also linked to tumour heterogeneity in numerous ways. Within an individual tumour, DNA methylation patterns are particularly polymorphic, even at single DNA promoter regions that are frequently methylated, which suggests the presence of heterogeneous cell populations with regards to DNA methylation [33]. In breast cancers, subsets of tumour cells differentially methylate the TGFBR2 promoter, which results in differential TGF pathway activity [34]. Furthermore, methylation-mediated silencing of key gatekeeper genes, such as MLH1, might fuel clonal evolution as they promote a hypermutation phenotype [22]. Interestingly, this particular case is directly related to inter-tumour heterogeneity as MLH1 methylation is a defining feature of the MSI–CRC subtype [25]. Moreover, methylation of a subset of Wnt target genes has been shown to identify colon cancers with poor prognosis [35]. Finally, certain subtypes of CRCs and gliomas have even been defined by a high level of methylation at the CpG-rich promoter regions of a subset of genes [36,37]. The reversible nature of epigenetic modifications has attracted much research efforts to develop agents that target the enzymes responsible for these changes. Several clinical examples illustrate the promise of epigenetic targeting, translated to the bedside by the US Food and Drug Administration approval of drugs that inhibit DNA methylation, and histone deacetylation for the treatment of myelodysplastic syndrome and cutaneous T-cell lymphoma, respectively [30].
The role of cancer stem cells in tumour heterogeneity. Normal tissue homeostasis is orchestrated by complex interactions between cells and their microenvironment that maintain a crucial balance between proliferation and differentiation. Importantly, all these processes occur in an ecosystem that is genetically identical and relies on a gradient of differentiation between stem cells and their differentiated progeny. An equivalent paradigm is not new in cancer [38], but only recent advances in technologies have made it possible to investigate critically the role of cancer cells endowed with stem cell properties. Initially described in leukaemia, the concept of CSCs has been rapidly embraced by the scientific community and led to their identification in virtually all tumour types [7,8,9]. The principle generally relies on the use of markers that enrich for cells able to propagate the malignancy in serial xenotransplantation assays. The exact identity of CSCs in many tumours is still disputed [39], especially due to the imperfect nature of the methodologies available to assess cancer stemness. Although more recent reports lend support for the existence of CSCs in endogenously growing neoplasia [40,41,42], they also have their own caveats. For instance, both intestinal adenoma and skin papillomas used in these studies are only benign precursor lesions that progress infrequently to carcinomas. The CSC concept nevertheless provides an additional source of heterogeneity in cancer; heterogeneous cell populations are defined by their differentiation state, non-differentiated (or CSCs) as compared with differentiated cells, as well as the various differentiation lineages that can be adopted (Fig 2B). Importantly, the genetic and CSC models are not mutually exclusive but rather complementary in fuelling tumour heterogeneity [9]. For example, CSCs are—by definition—the only cells with self-renewal capacity and as such would serve as a repository for clonal evolution to take place. Indeed, genetically diverse CSC populations within individual tumours have been demonstrated in both haematological and solid malignancies [34,43,44]. Reciprocally, genetic mutations such as inactivation of TP53 can influence the CSC content [45]. The molecular attributes that maintain the CSC state remain largely elusive but we have gained an increased insight into the molecular determinants of cancer stemness. For example, colon CSCs were shown to have relatively higher levels of Wnt pathway activity as compared with their differentiated progeny, despite the presence of Wnt-activating mutations [46]. Although deregulated in all colon carcinomas, Wnt does not necessarily define colon CSCs in every tumour. There might be tumours with CSCs that rely on alternative signal transduction cascades or that do not even follow a hierarchical organization [39,47], as illustrated in CRC or breast cancer cell lines [48,49].
The role of CSCs in regulating inter-tumour heterogeneity is more elusive, although subtype-specific regulation of CSC traits has also been noted. First, experimental mouse models of lung cancer with distinct genetic backgrounds were shown to have a crucial impact on the identity of the CSC population [50]. Second, an elegant study by Caldas et al reported that TGF-β potently induces a stem cell phenotype in basal—and low-claudin subtypes, but has a markedly different and even opposing role in other breast subtypes, such as luminal (Fig 2E; [51]). Finally, besides xenotransplantation assays, analyses of phylogenic trees of tumour populations have also resolved that CSC fractions vary among cancers [52]. These subtype-specific features of CSCs might explain some long-lasting inconsistencies in the field, such as the discrepancies associated with markers commonly used to identify CSC populations [39]. Regardless, these results point towards the CSC phenotype as an important contributor to heterogeneity across cancer subtypes. The implications of the CSC model for tumour growth are further illustrated by mathematical modelling of tumours that follow this model, which has revealed that hierarchical organization promotes the acquisition of new genetic and epigenetic traits and, thereafter, increased heterogeneity [53]. It should be noted however that differentiation hierarchy is not the only non-genetic explanation of tumour heterogeneity. In some instances, more stochastic processes seem to dominate the scene [48,54].
The impact of the tumour microenvironment on heterogeneity. Environmental cues have a major role in tumour development and progression, but also determine the heterogeneity observed within and across tumours. The tumour microenvironment (TME) comprises a wide variety of non-neoplastic cells, coopted by tumour cells, that influence the various steps of cancer development. The TME includes cancer-associated fibroblasts, endothelial cells and a variety of infiltrating immune cells, and promotes tumour heterogeneity in various ways. The mere presence of TME components is, by itself, a source of heterogeneity, and these cell types can be distributed in different tumour microenvironments, such as the invasive compared with the non-invasive edge [55]. Extrinsic factors produced by stromal cells also affect tumour heterogeneity by mediating changes in clonal evolution rate or stem cell content. For example, chronic inflammation can lead to oxidative stress and increased production of reactive oxygen species, which are potent inducers of DNA damage [56]. Consequently, this might influence intra-tumour heterogeneity in tumour regions with localized inflammation, but also affect cancer subtypes that associate strongly with chronic inflammation.
In the context of CSCs, the TME regulates tumour hierarchy and stem cell traits. Normal stem cell homeostasis is ensured by the integration of signals that fine-tune the balance between self-renewal and lineage commitment. Part of these signals emanate from the microenvironment, commonly referred to as the ‘stem cell niche’ [57]. An equivalent entity has been shown to have similar albeit aberrant functions in cancer. For instance, tumour-associated myofibroblasts produce HGF that binds to c-MET and activates Wnt activity to support colon CSCs [46]. The Notch ligands DLL4 and more recently Jagged1 were shown to have similar effects [58,59]. In glioma, endothelial cells co-localize to and are required for glioma stem cell maintenance [60]. Altogether, these and other studies [55] have demonstrated the impact of the TME on the heterogeneity of tumours. Perhaps even more importantly, factors provided by tumour-associated cells not only maintain stem cell attributes but also induce the phenotype in more differentiated cells (Fig 2C). For instance, HGF was shown to induce differentiated colon cancer cells to revert to a CSC state [46], and similarly TGF-β triggers an EMT–CSC programme in mammary epithelial cells [61]. This extrinsic control of tumour cell flexibility poses an obvious challenge in the design of therapies, especially those aimed at targeting the clonogenic core of tumours [62], and suggests that alternative therapeutical options have to be considered [63]. The composition of the TME is not only important in the regulation of tumour cell properties, but it also can be used as a defining peculiarity among different cancer subtypes. For example, activation of CD8+ T cells in specific tumour regions predicts tumour recurrence in colon cancer [55]. Furthermore, stromal-derived profiles from breast cancer have been associated with cancer subtypes suffering with a worse outcome (Fig 2F; [64]). Interestingly, the impact of the TME might also be specific to certain tumour subtypes. Mammary tumour metastasis in the MMTV-PyMT breast cancer model depends crucially on the recruitment of CD4+ T cells [65] unlike MMTV-NeuT-derived tumours [66]. The recognition of the TME as a defining feature of cancer subtypes is crucial when differences in mutations or stem cell content cannot explain the apparent diversity. For example, similar mutations might accumulate in a different sequential order, leading to a distinct requirement towards the environment. Alternatively, analogous mutations following the same sequence of events might give rise to divergent molecular subtypes depending on the inflammatory or environmental response.
The effect of the cell of origin on tumour heterogeneity. Cancer develops from a single founding cell, the identity of which is still highly debated [67,68]. In most adult tissues, self-renewal capability is restricted to stem cells and therefore these are probably the prime target for cellular transformation. Adult somatic stem cells are usually long-lived, which is a prerequisite to accumulate sufficient transforming events. Mouse models of intestinal cancer have initially confirmed the latter hypothesis as adenomas were shown to develop preferentially when loss of APC is induced in the stem cell compartment [69]. However, more recent work has challenged this view and has demonstrated that dedifferentiation of intestinal enterocytes can occur and lead to intestinal tumours [68], provided they have the right conditions. The Verma group reported another example in which they used a model of glioma and showed that terminally differentiated cells are also potent inducers of cancer, given that the right combination of genetic aberrations is induced. More specifically, the authors [67] deleted the tumour suppressor p53 together with the activation of the KRAS oncogene in differentiated neurons. They observed the induction of tumours that progressed rapidly towards invasive gliomas. Although these examples suggest that there is no restriction on the cell type that is initially targeted, the presence or acquisition of stem cell traits could still represent a limiting step to transformation. In the intestine, enforced activity of both Wnt signalling and NFκB in enterocytes is required to initiate adenomas from these cells and is accompanied by the re-expression of genes present in the intestinal stem cell signature. Similarly, acute myeloid leukaemia can develop either from haematopoietic stem cells (HSCs) or more committed progenitor cells on translocation of the mixed lineage leukaemia (MLL) gene. When occurring in committed granulocyte–macrophage progenitors, MLL-AF9 activates a self-renewal signature normally restricted to HSCs and generates tumours with high leukaemic stem cell content [70].
Moreover, the cell of origin can be a source for inter-tumour heterogeneity, and mapping the susceptible cells of different cancer subtypes is intensely investigated. A striking example was reported in medulloblastomas, one of the most frequent paediatric brain tumours originally thought to arise predominantly in the cerebellum. At least two main subtypes have been described—one driven by aberrant sonic hedgehog signalling (SHH) and the other seemingly dependent on Wnt activation—which have a dismal and good prognosis, respectively. By comparing the transcriptomes of these subtypes to those of distinct precursor cells, Gibson et al revealed that tumours from the SHH subtype were closely associated with committed cerebellar granule neuron precursor cells and developed from the cerebellum [71]. By contrast, the Wnt-driven subtype is associated with a gene expression profile of neural precursor cells located in the embryonic dorsal brainstem. Several additional examples have been described [72], and it is expected that many molecularly distinct subtypes from various tumours will be quickly trailed back to their origin. Importantly, in some cases, transcriptional programmes alone might be insufficient or even misleading in inferring the cell of origin of cancer subtypes. Basal cell carcinomas (BCCs) develop through activation of the hedgehog pathway, and during progression these tumours often show molecular markers normally expressed by hair follicles. Although this would suggest these tumours originate from hair follicle stem cells, conditional activation of the hedgehog pathway in various skin lineages has demonstrated that most BCCs develop preferentially from stem or progenitor cells found in the interfollicular epidermis [73].
In summary, we have reviewed two forms of tumour heterogeneity, namely intra- and inter-tumour heterogeneity, and discussed various sources that can promote them (Fig 1). First, we described the genetic contribution to heterogeneity resulting from an accumulation of genetic mutations in tumour cells and subsequent clonal selection; the process of clonal selection acts mostly on the CSCs as they are uniquely endowed with the capacity to self-renew. Moreover, we have discussed how CSCs add another layer of heterogeneity as these cells can spin off different tumour lineages of differentiated tumour cells. Finally, we concluded this first part by presenting the role of the tumour microenvironment and the cell of origin.
Implications and crosstalk of both forms of heterogeneity
Clinical implications. The mere presence of heterogeneity, regardless of the underlying foundation that promotes it, has a profound clinical impact. In the case of inter-tumour heterogeneity, the presence of oncogenic mutations is often used to guide treatment decisions. Two main situations can be described: on the one hand, the presence of a mutation could directly predict a lack of response to a particular treatment. This is the case in metastatic CRC, for which anti-EGFR therapy is relatively effective in KRAS wild-type cancer subtypes, but is not effective in KRAS-mutant tumours [74]. On the other hand, certain tumours are ‘addicted’ to oncogenic aberrations and targeting these mutations has proven clinically useful in those tumours. For instance, melanomas with a BRAF mutation are eligible and sensitive to treatment with the BRAF inhibitor vemurafenib as opposed to BRAF wild-type tumours [75]. As mentioned previously, mutation-based categorization incompletely recapitulates the complexity and diversity of cancer subtypes, and sub-classification of tumours based on gene expression profiles potentially improves clinical decisions. In this respect, unbiased identification of CRCs on the basis of gene expression has revealed the presence of a resistant subset to anti-EGFR treatment, independent of KRAS mutation status [27]. Similarly, different pancreatic adenocarcinoma subtypes show differential response to anti-EGFR treatment despite the presence of KRAS mutations [76]. The use of genomic technologies has resulted in an extensive characterization of breast cancers into distinct molecular subtypes and has yielded a major clinical benefit for some of these subsets. For example, the use of tamoxifen (an oestrogen receptor antagonist; [77]) and trastuzumab (an anti-HER2 monoclonal antibody; [78]) has crucially improved the survival of patients belonging to the oestrogen receptor or HER2 subtype, respectively.
Intra-tumour heterogeneity also has an impact on clinical outcome. In diagnostic terms, the choice of a therapeutic intervention is constrained by topological heterogeneity, as sampling procedures do not yield a fully representative picture of a tumour [23]. Furthermore, diagnosis is generally derived from the primary tumour whereas treatment often aims at eradicating metastatic disease presenting with phenotypic attributes that have progressed beyond the primary tumour. Finally, much of our knowledge on tumour heterogeneity is derived from ensemble measurements, which reflect changes present in most cells. Although emerging, the genetic make-up of single cells within a tumour [4] remains a formidable challenge, but will result in a better interpretation of clonal genotypes. The question remains, however, of whether these diverse issues are rate-limiting steps that need to be resolved to improve clinical outcome for patients? In other words, is it essential to obtain multi-regional sampling to have a correct diagnosis and successful therapy? For instance, some approaches might be available to filter through tumour complexity. One of them relies on the use of xenotransplantation as xenografts of human breast primary tumours display an enrichment of mutations present in the metastatic lesion counterpart. This suggests that this method could be used as an extra step to enrich for the most aggressive clones [79], from which diagnosis and therapy design should be made. Furthermore, the clinical benefit of obtaining genetic information on single tumour cells remains uncertain as it will only provide a complex architectural view of the clonal genotype without further revealing the dependence of tumour cells on specific growth factors or signalling pathways. Finally, the relevance of single-cell-derived diagnosis is unclear, as data derived from these cells does not provide any direct information on the remnant tumour cell population, and other more accessible methods can be used to infer clonal relations [5,80].
Therapy response is also affected by heterogeneity as targeted drugs often suffer from their highly selective nature towards specific gene alterations. When a selective pressure is applied, such as therapy, the fittest—most resistant—subclones are invariably selected to survive and expand to manifest clinically as a tumour relapse [5,6]. Mutations can directly affect the target itself, as in the case of chronic myelogenous leukaemia (CML) when treated with imatinib [81]. Alternatively, mutations can act synergistically to reactivate signalling pathways that are targeted. As mentioned above, relapses of metastatic CRC treated with anti-EGFR are frequently accompanied with mutations in KRAS, a downstream effector of EGFR signalling [74]. Alternatively, feedback mechanisms are often activated to confer resistance. One such example has been described in CRC in which resistance to BRAF inhibition is bypassed by EGFR overexpression [82]. In most cases, therapy resistance is accompanied by the outgrowth—selective sweep—of genetic variants that existed before the introduction of the treatment. In CML, a variant of the ABL kinase, a crucial target of the drug imatinib can be detected before therapy in patients that will develop resistance to that drug [83]. Similarly, the emergence of KRAS-mutant clones might be noticed months before radiographic progression in metastatic CRC patients treated with anti-EGFR therapy [84]. More recently, longitudinal assessment of genomic alterations before and after treatment in a panel of chronic lymphocytic leukaemias has revealed that an adverse clinical outcome was related to the expansion of subclones that were already present before treatment [80].
Importantly, in the context of CSCs, therapy seems to select for pre-existing, highly resistant tumour cells. It is generally assumed that these cells are more resistant to a variety of treatments [62,85]. Evidently this has a major clinical impact, as eradication of this subpopulation should be the priority when designing new therapeutic interventions. For example, leukaemic stem cells (LSCs) identified in CML are reportedly more resistant to the drug imatinib than their differentiated progeny. This is often attributed to the quiescent nature of LSCs [86,87] as imatinib preferentially targets dividing cells. Similarly, the quiescent state of a fraction of colon CSCs has also been associated with chemotherapy resistance [54], and altogether these studies highlight the potential therapeutic benefit of reverting quiescence [87]. Additional experimental evidence comes from glioma in which CSC marker-expressing cells repair DNA damage more efficiently [88], and the ability of breast CSCs to maintain low levels of reactive oxygen species, which protects them against radiation [89]. More clinical examples have also been reported, for instance in GISTs, whereby patients showing complete disease remission under imatinib treatment relapsed quickly after imatinib withdrawal [90]. This suggests that a fraction of cancer cells remain untouched during the treatment and cause a relapse.
The number of CSCs might relate to metastatic potential and, therefore, it is not too surprising that many efforts have been made to better identify tumours at a higher risk of relapse, with the assumption that they would present with a higher CSC content. The identification of patients with poor prognosis in breast cancers [91], colon cancers [35,92] and leukaemia [93], based on their association with CSC signatures, has indeed supported the relevance of CSCs in a clinical setting. Frequently, the underlying biological association of the tumour transcriptome to a CSC signature has been thought to reflect the fraction of CSCs present in a tumour. Although these associations might exist in malignancies that have a high CSC content, it is unlikely to be a general feature, as most cancers are believed to harbour only a minute number of CSCs. Consequently, several important conclusions can be drawn from the prognostic power of CSC signatures. First, CSC-associated expression profiles are clinically relevant as elements of stemness influence the clinical outcome of various cancers. Second, the association of CSC-derived profiles with prognosis should be interpreted with caution. This is well-illustrated for CRC in which a mouse intestinal stem cell (ISC)-derived signature was shown to associate strongly with patients at a high risk of relapse [92], suggesting that stem cell content is important. However, our data show that the presence of ISC-related genes in both pure ISC or colon CSC-derived gene signatures is not a crucial determinant of the prognostic power of these signatures [35]. Importantly, in the illustrated case, ISC and CSC signatures pointed to a distinct, more immature and poorly differentiated subset, and thereby reflected a clonal trait of the malignant tissue, rather than CSC number.
Connecting inter- and intra-tumour heterogeneity. As described above, both forms of heterogeneity influence clinical outcome in many ways. The mere presence of intra-tumoral heterogeneity impinges on adequate diagnosis and results frequently in therapy resistance. Moreover, the realization that distinct molecular subsets exist requires a shift in cancer drug development, from a ‘one-size fits all’ chemotherapy to a more personalized and group-based drug design, or even an approach directed towards specific tumour clones [44]. Unfortunately, our crude understanding of the molecular mechanisms by which intra-tumour heterogeneity is driven, coupled with uncertainty about the diversity of existing molecular subtypes, might preclude the development of significantly improved therapies, particularly for more advanced metastatic disease.
Can one find analogy between these two forms of heterogeneity that could be exploited to improve rational therapy design? In other words, could a better appreciation of the biological foundation of distinct subtypes be used to infer the behaviour of intra-tumour heterogeneity? A study of acquired resistance to EGFR inhibitors in lung cancer revealed that recurrent tumours in a fraction of patients undergo a subtype shift from non-small cell to small cell carcinoma [94]. The acquisition of the small cell lineage is marked by the acquisition of traits specific for this particular lineage, including sensitivity to conventional chemotherapy. Although the authors conclude that a lineage switch occurred by an unknown mechanism, it is equally feasible that robust subsets of lung cancer were pre-existing within these tumours and contracted or expanded during the therapy. Although this observation remains anecdotal, the presence of a heterogeneous tumour cell population in a tumour could be a reflection of the diverse molecular subtypes that belong to a malignancy (Fig 3).
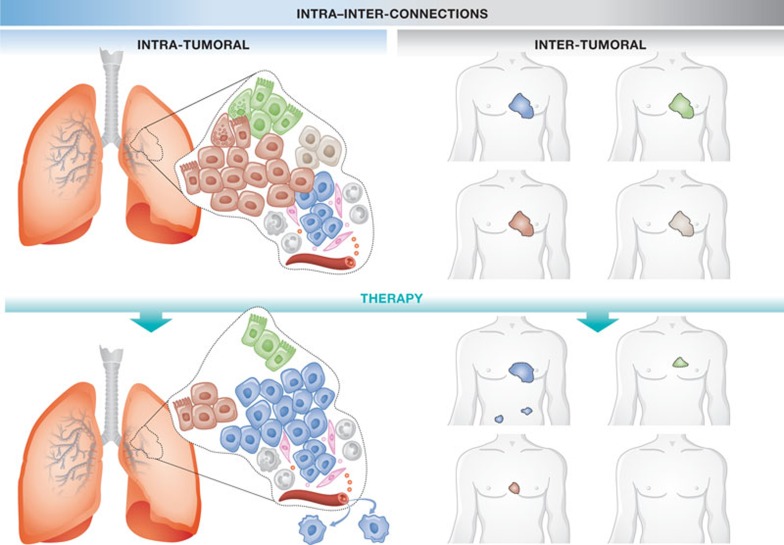
Figure 3.
Connecting intra- and inter-tumour heterogeneity. (Top) Multiple subclones exist in an individual lung tumour each being represented by a different subtype of lung cancer. (Bottom) Some subtypes of lung cancer are more sensitive to therapy than others. This is reflected in an individual tumour for which a therapy selects the most aggressive subclone, such as the subclone resembling the most aggressive cancer subtype.
Conclusion and outlook
Tumours are recognised as heterogeneous entities but the crucial question remains of how to use that knowledge and turn it into a clinical benefit. For instance, heterogeneity in Barrett oesophagus, measured by several indexes initially developed for ecological studies, is an important predictor of progression to oesophageal carcinoma; a similar study performed in breast tissue suggests that the degree of heterogeneity could be a general predictor for disease progression [95]. Whether the presence of more clones is crucial for progression or simply the reflection of genomically unstable tumours remains to be established. Ingeniously designed, targeted therapeutic agents are tested in preclinical models at tremendous speed but at a high cost as well. Tumour heterogeneity reflects the biggest challenge for drug development—single agents that target known aberrations to which cancer cells might be addicted—such as BCR–ABL in CML—might simply fail due to the presence of distinct non-targeted clones. Moreover, a better appreciation of inter-tumour heterogeneity and how this might affect tumour response is needed when drug responses are being evaluated. For example, ‘drug x’ might be discarded because it does not produce clinical benefit in the overall population, although it might be potent in a small subset of the population given that the latter is represented adequately. This is not straightforward as the identification of subtypes is generally achieved on primary tumour-derived material, whereas phase II clinical trials are usually performed on late-stage cancers that might not necessarily encompass the same diversity of subtypes as those present in early stages. Assuming the latter is true, how do we progress towards more effective treatments (Sidebar A)? One of the limiting steps here is having appropriate models to use to tackle these questions. Mouse models have been cleverly engineered to recapitulate crucial aspects of human tumour progression but, in particular, fail to reflect the heterogeneity that is present in human cancers, especially inter-tumorally. Human xenograft models are superior in that respect as they are derived directly from patients and can be maintained almost indefinitely in immuno-compromised mice. The main criticism here rests on the lack of interaction with the microenvironment and the selection of tumour cells and clones that are more apt to survive in the mouse environment. Moreover, drug screening in this setting is cumbersome. A potential alternative is to fall back on established and well-characterized cancer cell lines. Studies have begun to elucidate drug response in large panels of cell lines [96], but these lack sufficient insight into the extent to which these represent the cancer population. Based on the unbiased identification of distinct subtypes of primary colon cancers, we have identified subtypes of colon cancer cell lines that have important biological attributes, such as invasive properties and therapy resistance, corresponding with primary cancers [27]. Research in this area will be facilitated by a wealth of data such as genotype, drug response and expression profiles that are available for a wide range of cell lines from various lineages. Although clearly imperfect, these simple in vitro models might be the first crucial step to understanding the different biological backgrounds that are hard-wired into distinct cancer entities.
Acknowledgments
We thank Maarten Bijlsma for critically reading the manuscript and for useful comments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Perou CM et al. (2000) Molecular portraits of human breast tumours. Nature 406: 747–752 [DOI] [PubMed] [Google Scholar]
- Stephens PJ et al. (2012) The landscape of cancer genes and mutational processes in breast cancer. Nature 486: 400–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG et al. (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navin N et al. (2011) Tumour evolution inferred by single-cell sequencing. Nature 472: 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Caldas C (2013) The implications of clonal genome evolution for cancer medicine. N Engl J Med 368: 842–851 [DOI] [PubMed] [Google Scholar]
- Greaves M, Maley CC (2012) Clonal evolution in cancer. Nature 481: 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM (2006) Cancer stem cells—perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res 66: 9339–9344 [DOI] [PubMed] [Google Scholar]
- Dick JE (2008) Stem cell concepts renew cancer research. Blood 112: 4793–4807 [DOI] [PubMed] [Google Scholar]
- Vermeulen L, Sprick MR, Kemper K, Stassi G, Medema JP (2008) Cancer stem cells—old concepts, new insights. Cell Death Differ 15: 947–958 [DOI] [PubMed] [Google Scholar]
- Nowell PC (1976) The clonal evolution of tumor cell populations. Science 194: 23–28 [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B (1990) A genetic model for colorectal tumorigenesis. Cell 61: 759–767 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321: 1807–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LD et al. (2007) The genomic landscapes of human breast and colorectal cancers. Science 318: 1108–1113 [DOI] [PubMed] [Google Scholar]
- Molenaar JJ et al. (2012) Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483: 589–593 [DOI] [PubMed] [Google Scholar]
- Parsons DW et al. (2011) The genetic landscape of the childhood cancer medulloblastoma. Science 331: 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L et al. (2008) Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic I et al. (2010) Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci USA 107: 18545–18550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaza M et al. (2013) Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497: 108–112 [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia C, Klein AM, Simons BD, Winton DJ (2010) Intestinal stem cell replacement follows a pattern of neutral drift. Science 330: 822–825 [DOI] [PubMed] [Google Scholar]
- Snippert HJ et al. (2010) Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143: 134–144 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M et al. (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366: 883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovici V et al. (2012) Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J Clin Oncol 30: 1288–1295 [DOI] [PubMed] [Google Scholar]
- Shen L et al. (2007) Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci USA 104: 18654–18659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway KA et al. (2011) Defects in succinate dehydrogenase in gastrointestinal stromal tumors lacking KIT and PDGFRA mutations. Proc Natl Acad Sci USA 108: 314–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa E Melo et al. (2013) Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med 19: 614–618 [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA (2011) A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 11: 726–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Verbrugge I, Johnstone RW, Bots M (2011) Promises and challenges of anticancer drugs that target the epigenome. Epigenomics 3: 547–565 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, Perucho M (2006) Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell 9: 199–207 [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG (1998) Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA 95: 11891–11896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landan G et al. (2012) Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat Genet 44: 1207–1214 [DOI] [PubMed] [Google Scholar]
- Shipitsin M et al. (2007) Molecular definition of breast tumor heterogeneity. Cancer Cell 11: 259–273 [DOI] [PubMed] [Google Scholar]
- de Sousa EMF et al. (2011) Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 9: 476–485 [DOI] [PubMed] [Google Scholar]
- Noushmehr H et al. (2010) Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP (1999) CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 96: 8681–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema JP, Vermeulen L (2011) Microenvironmental regulation of stem cells in intestinal homeostasis and cancer. Nature 474: 318–326 [DOI] [PubMed] [Google Scholar]
- Medema JP (2013) Cancer stem cells: The challenges ahead. Nat Cell Biol 15: 338–344 [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF (2012) A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 488: 522–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C (2012) Defining the mode of tumour growth by clonal analysis. Nature 488: 527–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H (2012) Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337: 730–735 [DOI] [PubMed] [Google Scholar]
- Anderson K et al. (2011) Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature 469: 356–361 [DOI] [PubMed] [Google Scholar]
- Notta F et al. (2011) Evolution of human BCR–ABL1 lymphoblastic leukaemia-initiating cells. Nature 469: 362–367 [DOI] [PubMed] [Google Scholar]
- Cicalese A et al. (2009) The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138: 1083–1095 [DOI] [PubMed] [Google Scholar]
- Vermeulen L et al. (2010) Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol 12: 468–476 [DOI] [PubMed] [Google Scholar]
- Horst D, Chen J, Morikawa T, Ogino S, Kirchner T, Shivdasani RA (2012) Differential WNT activity in colorectal cancer confers limited tumorigenic potential and is regulated by MAPK signaling. Cancer Res 72: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES (2011) Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146: 633–644 [DOI] [PubMed] [Google Scholar]
- Yeung TM, Gandhi SC, Wilding JL, Muschel R, Bodmer WF (2010) Cancer stem cells from colorectal cancer-derived cell lines. Proc Natl Acad Sci USA 107: 3722–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis SJ et al. (2010) Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell 7: 127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruna A et al. (2012) TGFβ induces the formation of tumour-initiating cells in claudinlow breast cancer. Nat Commun 3: 1055. [DOI] [PubMed] [Google Scholar]
- Sottoriva A, Spiteri I, Shibata D, Curtis C, Tavare S (2013) Single-molecule genomic data delineate patient-specific tumor profiles and cancer stem cell organization. Cancer Res 73: 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A, Verhoeff JJ, Borovski T, McWeeney SK, Naumov L, Medema JP, Sloot PM, Vermeulen L (2010) Cancer stem cell tumor model reveals invasive morphology and increased phenotypical heterogeneity. Cancer Res 70: 46–56 [DOI] [PubMed] [Google Scholar]
- Kreso A et al. (2013) Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 339: 543–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagès F et al. (2009) In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol 27: 5944–5951 [DOI] [PubMed] [Google Scholar]
- Meira LB et al. (2008) DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J Clin Invest 118: 2516–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden DT (2006) The stem-cell niche as an entity of action. Nature 441: 1075–1079 [DOI] [PubMed] [Google Scholar]
- Hoey T et al. (2009) DLL4 blockade inhibits tumor growth and reduces tumor-initiating cell frequency. Cell Stem Cell 5: 168–177 [DOI] [PubMed] [Google Scholar]
- Lu J et al. (2013) Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell 23: 171–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese C et al. (2007) A perivascular niche for brain tumor stem cells. Cancer Cell 11: 69–82 [DOI] [PubMed] [Google Scholar]
- Scheel C et al. (2011) Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 145: 926–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L, de Sousa e Melo F, Richel DJ, Medema JP (2012) The developing cancer stem-cell model: clinical challenges and opportunities. Lancet Oncol 13: e83–e89 [DOI] [PubMed] [Google Scholar]
- Pienta KJ, McGregor N, Axelrod R, Axelrod DE (2008) Ecological therapy for cancer: defining tumors using an ecosystem paradigm suggests new opportunities for novel cancer treatments. Transl Oncol 1: 158–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finak G et al. (2008) Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 14: 518–527 [DOI] [PubMed] [Google Scholar]
- DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, Coussens LM (2009) CD4+ T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16: 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampricotti M, Vrijland K, Hau CS, Pemovska T, Doornebal CW, Speksnijder EN, Wartha K, Jonkers J, de Visser KE (2011) Development of metastatic HER2+ breast cancer is independent of the adaptive immune system. J Pathol 224: 56–66 [DOI] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM (2012) Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science 338: 1080–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla S et al. (2013) Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152: 25–38 [DOI] [PubMed] [Google Scholar]
- Barker N et al. (2009) Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457: 608–611 [DOI] [PubMed] [Google Scholar]
- Krivtsov AV et al. (2006) Transformation from committed progenitor to leukaemia stem cell initiated by MLL–AF9. Nature 442: 818–822 [DOI] [PubMed] [Google Scholar]
- Gibson P et al. (2010) Subtypes of medulloblastoma have distinct developmental origins. Nature 468: 1095–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C (2013) Tracing the cellular origin of cancer. Nat Cell Biol 15: 126–134 [DOI] [PubMed] [Google Scholar]
- Youssef KK, Van Keymeulen A, Lapouge G, Beck B, Michaux C, Achouri Y, Sotiropoulou PA, Blanpain C (2010) Identification of the cell lineage at the origin of basal cell carcinoma. Nat Cell Biol 12: 299–305 [DOI] [PubMed] [Google Scholar]
- Khambata-Ford S et al. (2007) Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol 25: 3230–3237 [DOI] [PubMed] [Google Scholar]
- Flaherty KT et al. (2010) Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 363: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson EA et al. (2011) Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 17: 500–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher B et al. (1989) A randomized clinical trial evaluating tamoxifen in the treatment of patients with node-negative breast cancer who have estrogen-receptor-positive tumors. N Engl J Med 320: 479–484 [DOI] [PubMed] [Google Scholar]
- Vogel CL et al. (2002) Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 20: 719–726 [DOI] [PubMed] [Google Scholar]
- Ding L et al. (2010) Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464: 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA et al. (2013) Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 152: 714–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche-Lestienne C, Laï JL, Darré S, Facon T, Preudhomme C (2003) A mutation conferring resistance to imatinib at the time of diagnosis of chronic myelogenous leukemia. N Engl J Med 348: 2265–2266 [DOI] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R (2012) Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483: 100–103 [DOI] [PubMed] [Google Scholar]
- Roche-Lestienne C, Soenen-Cornu V, Grardel-Duflos N, Laï JL, Philippe N, Facon T, Fenaux P, Preudhomme C (2002) Several types of mutations of the Abl gene can be found in chronic myeloid leukemia patients resistant to STI571, and they can pre-exist to the onset of treatment. Blood 100: 1014–1018 [DOI] [PubMed] [Google Scholar]
- Misale S et al. (2012) Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES (2009) Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell 138: 645–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntly BJ, Gilliland DG (2005) Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer 5: 311–321 [DOI] [PubMed] [Google Scholar]
- Takeishi S, Matsumoto A, Onoyama I, Naka K, Hirao A, Nakayama KI (2013) Ablation of Fbxw7 eliminates leukemia-initiating cells by preventing quiescence. Cancer Cell 23: 347–361 [DOI] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN (2006) Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444: 756–760 [DOI] [PubMed] [Google Scholar]
- Diehn M et al. (2009) Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458: 780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cesne A et al. (2010) Discontinuation of imatinib in patients with advanced gastrointestinal stromal tumours after 3 years of treatment: an open-label multicentre randomised phase 3 trial. Lancet Oncol 11: 942–949 [DOI] [PubMed] [Google Scholar]
- Liu R et al. (2007) The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med 356: 217–226 [DOI] [PubMed] [Google Scholar]
- Merlos-Suarez A et al. (2011) The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 8: 511–524 [DOI] [PubMed] [Google Scholar]
- Eppert K et al. (2011) Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med 17: 1086–1093 [DOI] [PubMed] [Google Scholar]
- Sequist LV et al. (2011) Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 3: 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo LM, Shah NA, Li X, Blount PL, Vaughan TL, Reid BJ, Maley CC (2010) A comprehensive survey of clonal diversity measures in Barrett's esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev Res (Phila) 3: 1388–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]