Cell (2013) 153 6, 1266–1280 doi: ; DOI: 10.1016/j.cell.2013.05.023
DNA double-stranded breaks (DSBs) are highly cytotoxic lesions that can induce genome rearrangements if not accurately repaired. DSBs can be repaired either through homologous recombination (HR) or non-homologous end-joining (NHEJ). HR is the preferred repair pathway during the S and G2 cell cycle phases because a sister chromatid provides a perfect template for ‘error-free’ repair. During G1, when HR is suppressed to prevent recombination with homologues, repair is achieved primarily by NHEJ. Molecularly, DSB repair pathway choice is largely regulated at the level of 5′ to 3′ DNA end resection, that is, the formation of the 3′ end single-stranded DNA overhangs that are used to initiate HR. End resection inhibits NHEJ and promotes HR.
In the June issue of Cell, Nussenzweig and colleagues identified the protein PTIP (also known as PAXIP) as a new component of the regulatory network that controls DSB repair pathway choice [1]. This work has important implications for our understanding of the mechanisms by which genomic integrity is underpinned, and is especially germane to those interested in the genesis of breast and ovarian cancer caused by a defective BRCA1 protein, which is crucial for DSB repair by HR.
53BP1 (also known as TP53BP1) is a key determinant of DSB repair pathway choice [2]. In response to DSBs, 53BP1 binds to chromatin at damaged sites, where it promotes NHEJ by blocking end resection. 53BP1 has a crucial role during class switch recombination (CSR) in B cells and the fusion of dysfunctional telomeres. An even more striking phenotype was observed in mice in which loss of 53BP1 reversed most of the phenotypes associated with BRCA1 deficiency, including cell and embryonic lethality as well as tumorigenesis [2]. These findings suggest that 53BP1 and BRCA1 battle each other to influence DSB repair pathway choice.
Molecularly, 53BP1 is responsible for the defective HR seen in BRCA1-deficient cells. Furthermore, in those cells, 53BP1 promotes the formation of characteristic radial chromosomes that are caused by toxic NHEJ events, presumably during S phase. Understanding exactly how 53BP1 carries out its many functions has been a major challenge to the field as 53BP1 does not harbour any enzymatic activity. However, it has been shown that 53BP1 must accumulate on chromatin to be functional. In addition, a mutant 53BP1 allele in which all 28 ataxia telangiectasia-mutated (ATM) phosphorylation sites were changed to alanine (53BP128A) failed to rescue 53BP1 deficiency, suggesting that 53BP1 acts through phosphorylation-dependent protein interactions to promote NHEJ [2].
RIF1 was identified as the first effector of 53BP1 in DSB repair [3,4,5,6,7]. RIF1 accumulates at DSB sites by binding to phosphorylated 53BP1 but, intriguingly, the loss of RIF1 has a milder effect than the loss of 53BP1 with respect to the fusion of dysfunctional telomeres [3], and RIF1 deficiency does not fully restore HR in BRCA1-deficient cells [7]. As the 53BP128A mutant is nearly as defective as the complete loss of 53BP1 for these activities, these observations indicate that additional 53BP1 effector proteins contribute to some of the 53BP1 functions.
Nussenzweig and colleagues provide compelling evidence that the BRCT domain-containing protein PTIP is the missing 53BP1 effector protein [1]. The authors identified a separation-of-function mutation in 53BP1 that disrupted the first eight amino-terminal ATM sites (53BP18A). The 53BP18A mutant behaved the same as the wild-type protein with respect to CSR—a physiological process dependent on NHEJ—but failed to promote genome instability (radial chromosome formation) in BRCA1-deficient cells after treatment with a PARP inhibitor. Since RIF1-deficient cells have impaired CSR and RIF1 can localize to break sites in cells expressing the 53BP18A mutant, this suggests that a protein other than RIF1 binds to the N-terminal region of 53BP1 to inhibit HR.
The newly identified 53BP1 effector protein PTIP is a multifunctional DNA repair factor that interacts with phosphorylated Ser 25 of 53BP1 through its tandem BRCT domains [8]—a site that was mutated in the 53BP18A allele. PTIP is also part of the MLL3/MLL4 histone H3 Lys 4 methyltransferase complexes but this function seems to be unrelated to its role as a 53BP1 co-factor.
Nussenzweig and co-workers found that PTIP-deficient cells are sensitive to ionizing radiation but tolerant of DNA damaging agents that are toxic to HR-deficient cells, which suggests a role for PTIP in NHEJ. In agreement with this, the fusion frequency of uncapped telomeres was reduced in PTIP-deficient cells. Interestingly, as in the case of the 53BP18A allele, PTIP-deficient B cells were proficient in switching their immunoglobulin locus, although this switching event is impaired in RIF1−/− B cells. This suggests that PTIP might participate selectively in pathological NHEJ.
Nussenzweig and colleagues next generated a conditional BRCA1−/− PTIP−/− mouse to investigate the contribution of PTIP to the genome instability of BRCA1-deficient B cells. Loss of PTIP restored normal growth kinetics and genome stability to BRCA1-deficient cells treated with a PARP inhibitor. In addition, RAD51 IR-induced focus formation was restored in BRCA1−/− PTIP−/− cells. As the primary defect of BRCA1-deficient cells with respect to HR seems to be at the level of resection, the accumulation of the single-stranded DNA-binding protein RPA into IR-induced foci was then analysed. The finding that PTIP-deficient cells have an increased number of RPA foci per cell supports a role for PTIP in blocking resection. Together, this suggests that PTIP opposes DNA end resection and mutagenic DSB repair in BRCA1-deficient cells.
These results were surprising as they revealed that the 53BP1 activities relating to physiological NHEJ (during CSR) and mutagenic NHEJ (after PARP inhibition) can be separated, and that they are carried out by two distinct proteins that ‘read’ ATM-dependent 53BP1 phosphorylation. The relationship between 53BP1, RIF1 and PTIP is probably complex, as suggested by the possible competition between RIF1 and PTIP, and the observation that both proteins contribute in an additive manner to the fusion of dysfunctional telomeres, downstream from 53BP1.
According to these findings, multiple phosphorylation events in 53BP1 seem to integrate ATM activity to control distinct aspects of DSB repair pathway choice (Fig 1). Establishing exactly how an increase of ATM activity at break sites is translated into the coordination of 53BP1 phosphorylation, with RIF1 and PTIP binding, will be an important milestone towards understanding 53BP1 function. Indeed, multi-site phosphorylation and its recognition by binding proteins can be used to develop switch-like responses that might be important for organizing the chromatin at DSB sites.
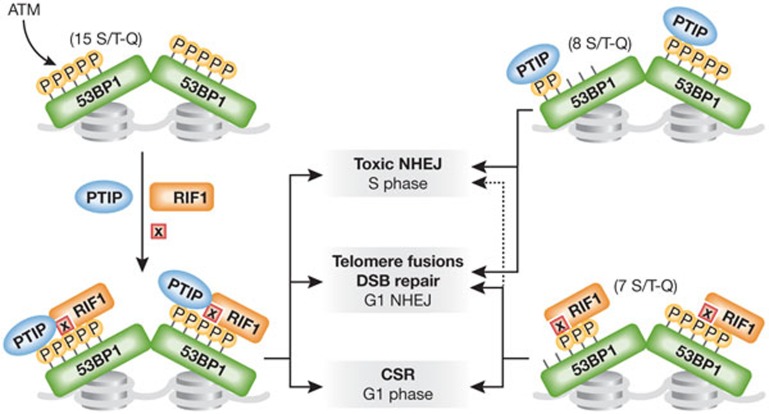
Figure 1.
53BP1 phospho-dependent interactions involved in DSB repair. PTIP and RIF1 interact with chromatin-bound and ATM-phosphorylated 53BP1 at DSB sites. PTIP binds directly to 53BP1 phosphorylated on Ser 14;25 (within the first eight Ser/Thr-Q sites). RIF1 binds to phosphorylated 53BP1 either directly or through an intermediate factor (X). The carboxy-terminal seven Ser/Thr-Q sites (9–15 Ser/Thr-Q sites) are involved in the interaction of RIF1–53BP1, although the amino-terminal eight Ser/Thr-Q sites might stabilize the binding. It is unknown whether PTIP and RIF1 can associate simultaneously with 53BP1 (left side of the figure), or if the binding is exclusive, due to either differential phosphorylation of the Ser/Thr-Q sites or steric hindrance (right side of the figure). 53BP1, PTIP and RIF1 block DNA end-resection and promote NHEJ repair. Although both PTIP and RIF1 contribute to dysfunctional telomere fusions, they also have distinct functions downstream from 53BP1. While RIF1 is essential for CSR and has a milder effect on toxic NHEJ events, PTIP is dispensable for CSR and has a more prominent role in toxic NHEJ events that lead to genome instability in BRCA1-deficient cells. ATM, ataxia telangiectasia-mutated; CSR, class switch recombination; DSB, double-stranded break; NHEJ, non-homologous end-joining.
The identification of PTIP as a new 53BP1 effector also deepens the mystery of DSB repair pathway choice regulation by 53BP1. Future studies are needed to elucidate how 53BP1 and its effector proteins block resection. Are PTIP and RIF1 blocking specific nucleases? Do they act in a temporally distinct fashion or are they distributed in distinct subdomains of the chromatin flanking DSB sites? What is the function of PTIP in relation to the cell cycle? Testing whether RIF1 binds directly to 53BP1, and if so to which phosphorylated site, might answer some of the above questions. The identification of a RIF1 mutation that selectively disrupts 53BP1 binding would enable surgical manipulation of the 53BP1–RIF1–PTIP circuit at DSB sites.
Another unresolved issue is whether 53BP1 acts solely by recruiting RIF1 and PTIP, or whether 53BP1 has a more active role in blocking resection. We have shown that 53BP1 localizes to the chromatin flanking the DSBs by binding to methylated and ubiquitinated nucleosomes, in a wheel clamp-like manner [9]. This suggests that 53BP1 might modify the nucleosomal array structure in a way that makes it refractory to the resection machinery. Recognizing how nucleosomes modified by 53BP1 cooperate with RIF1 and PTIP might provide clues to the role of these two proteins in end protection.
It is important to note that in human cells, PTIP might not be recruited to DSB sites in a 53BP1- and ATM-dependent manner [8]. Furthermore, in the avian B-cell line DT40, PTIP promotes HR instead of inhibiting it [10]. It will be important to revisit these studies to tease out whether these differences are due to context-, experiment- or species-specific effects.
The identification of PTIP as a candidate genetic modifier of BRCA1-deficient tumours is an important finding. As noted by the authors, disabling the PTIP–53BP1 interaction pharmacologically might selectively restore HR in BRCA1-deficient cells, which might be useful in certain contexts, for example as a chemopreventive strategy.
Footnotes
The authors declare that they have no conflict of interest.
References
- Callen E et al. (2013) Cell 153: 1266–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR et al. (2012) Mol Cell 47: 497–510 [DOI] [PubMed] [Google Scholar]
- Zimmermann M et al. (2013) Science 339: 700–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Diaz C et al. (2013) Mol Cell 49: 872–883 [DOI] [PubMed] [Google Scholar]
- Di Virgilio M et al. (2013) Science 339: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR et al. (2013) Mol Cell 49: 858–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L et al. (2013) J Biol Chem 288: 11135–11143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM et al. (2007) Nucleic Acids Res 35: 5312–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet-Turcotte A et al. (2013) Nature [Epub ahead of print] doi:; DOI: 10.1038/nature12318 [DOI] [Google Scholar]
- Wang X et al. (2010) Genes Cells 15: 243–254 [DOI] [PubMed] [Google Scholar]