Abstract
Poly(A) tail elongation after export of an messenger RNA (mRNA) to the cytoplasm is called cytoplasmic polyadenylation. It was first discovered in oocytes and embryos, where it has roles in meiosis and development. In recent years, however, has been implicated in many other processes, including synaptic plasticity and mitosis. This review aims to introduce cytoplasmic polyadenylation with an emphasis on the factors and elements mediating this process for different mRNAs and in different animal species. We will discuss the RNA sequence elements mediating cytoplasmic polyadenylation in the 3′ untranslated regions of mRNAs, including the CPE, MBE, TCS, eCPE, and C-CPE. In addition to describing the role of general polyadenylation factors, we discuss the specific RNA binding protein families associated with cytoplasmic polyadenylation elements, including CPEB (CPEB1, CPEB2, CPEB3, and CPEB4), Pumilio (PUM2), Musashi (MSI1, MSI2), zygote arrest (ZAR2), ELAV like proteins (ELAVL1, HuR), poly(C) binding proteins (PCBP2, αCP2, hnRNP-E2), and Bicaudal C (BICC1). Some emerging themes in cytoplasmic polyadenylation will be highlighted. To facilitate understanding for those working in different organisms and fields, particularly those who are analyzing high throughput data, HUGO gene nomenclature for the human orthologs is used throughout. Where human orthologs have not been clearly identified, reference is made to protein families identified in man. © 2013 John Wiley & Sons, Ltd.
THE POLY(A) TAIL, INITIAL SYNTHESIS, AND FUNCTION
We shall start with a brief overview of nuclear polyadenylation so that we can provide context for discussing cytoplasmic polyadenylation. For more detail, the reader is referred to other reviews that discuss nuclear polyadenylation specifically.1–6 The nuclear polyadenylation of eukaryotic messenger RNAs was discovered in the 1970s and soon was shown to be a nearly universal characteristic of this RNA species.1 In the following decades, polyadenylation was shown to follow cleavage of the newly transcribed messenger RNA (mRNA) and to be tightly coupled to transcription and to splicing of the 3′ intron.1–3 Cleavage of the nascent mRNA is now known to be required for normal transcription termination as well as for polyadenylation.4
The target selectivity of cleavage and polyadenylation in mammals is mediated by four sequence elements flanking the cleavage site, as depicted in Figure 1.5–8 Subunit 1 of the cleavage and polyadenylation specificity complex, CPSF1 (CPSF160), binds the well-known poly(A) signal (AAUAAA or AUU AAA), normally located 15–30 nucleotides upstream of the cleavage site. The CPSF complex also contains CPSF3 (CPSF73) the endonuclease responsible for cleavage,9 and FIP1L, the factor that recruits the poly(A) polymerase together with CPSF1.10,11 The cleavage stimulating factor (CSTF) complex binds to a GU or U rich sequence downstream of the cleavage site called the downstream element (DSE). The association of the CSTF and CPSF complexes with the RNA is thought to be the most important for the selection of the cleavage site, and the two complexes are connected by the 3′ processing scaffold protein Symplekin (SYMPK).12,13 The Cleavage Factor I (CFIm) is a complex of NUDT21 (CFIm25, CPSF5) and CPSF6 (CFIm68), or CPSF7 (CFIm59). NUDT21 binds upstream of the poly(A) signal to the upstream element (USE), which contains U(G/A)UA sequences that enhance recognition of the cleavage site.5,14 A fourth sequence element contributing to cleavage site recognition is a G rich sequence downstream of the DSE, for which the binding factor has not yet been determined.5 If the poly(A) signal diverges from AAUAAA or AUUAAA, the sequence of the other three sequences become more important for correct cleavage. Alternative poly(A) site selection has recently been shown to be regulated for many mRNAs and is likely to play an important role in post-transcriptional gene regulation by generating mRNAs with different 3′ untranslated regions (UTRs).15,16
FIGURE 1.
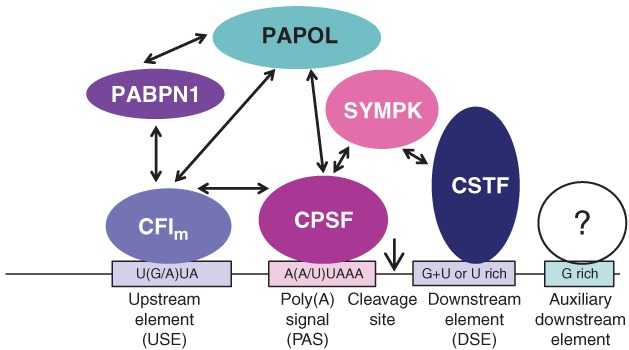
The nuclear cleavage and polyadenylation complex in vertebrates. The Cleavage and Polyadenylation Specificity Factor (CPSF) protein complex binds to the poly(A) signal PAS, it contains CPSF1-4 and the associated factors FIP1L and Symplekin (SYMPK).The CPSF3 subunit is the endonuclease acting at the cleavage site. The Cleavage Factor 1 complex (CFIm) recognizes the upstream element (USE), it is composed of NUDT21, CPSF6, and CPSF7. The CSTF complex recognizes the GU- or U-rich downstream element (DSE). CPSF, CSTF, SYMPK, and CFIm interact at the protein level, stabilizing the RNA binding, thus promoting correct cleavage and polyadenylation site recognition and recruitment of the poly(A) polymerase (PAPOLA or PAPOLG). The nuclear poly(A) binding protein (PABPN1) interacts with CFIm and PAPOLA and contributes to the efficiency of polyadenylation.17–19 Double arrows indicate interactions. The cleavage site is indicated by a bold single headed arrow. Some of these interactions have been inferred from work in yeast. For further details, see two recent reviews.5,8
After cleavage, the poly(A) tail is added by a nuclear poly(A) polymerase. The canonical enzyme that is thought to mediate this in most cells and for most mRNAs is poly(A) polymerase α (PAPOLA), which was originally isolated from calf thymus.20 It is required for both the polyadenylation and the cleavage reaction. However, another canonical mammalian poly(A) polymerase γ (PAPOLG) has been found in the nuclei of many tumor cells and cell lines21 and was the only poly(A) polymerase detected in a proteomic study of polyadenylation complexes.22 In addition, the noncanonical poly(A) polymerase TUT1 (Star-PAP, PAPD2) has been shown to mediate the nuclear polyadenylation of specific mRNAs.23–28
During cleavage and polyadenylation, a number of proteins are deposited on the mRNA which appear to limit the poly(A) tail size to 200–250 nt and play a role in mRNA export. These include nucleophosmin (NPM1), the nuclear poly(A) binding proteins PABPN1 and NAB2, as well as export factors.29–32 The removal of the cleavage and polyadenylation complex from the mRNA is also part of the export process.33
In the cytoplasm, the poly(A) tail enhances translational efficiency and protects the mRNA from degradation.34,35 These functions are mediated by the cytoplasmic poly(A) binding proteins (PABPs), the most well characterized of which is PABPC1 (PABP1 or PAB1). Cytoplasmic PABPs first bind to the poly(A) tail in the nucleus and are exported with the newly made mRNA, where additional PABP molecules may bind.36–38 During translation, PABPs mediate the formation of the ‘closed loop complex’ (Figure 2) in which interactions between PABP and eIF4G, a subunit of the cap binding translation initiation complex, connect the ends of the mRNA. The cap binding complex recruits the ribosome through interactions with eIF3 and contains the eIF4A helicase, which unwinds the 5′ untranslated region of the mRNA. In addition, PABP can recruit further eIF4A and eIF3 through its interaction with PAIP1.39–41 In plants, a further PABPC1 interaction with the eIF4A binding factor eIF4B has been established42,43 and a similar complex may also exist in human cells.44 Thus, the poly(A) tail stimulates translation by remodeling of the mRNP (through eIF4A) and loading of ribosomes (through eIF3), all mediated by interactions with PABPCs.
FIGURE 2.
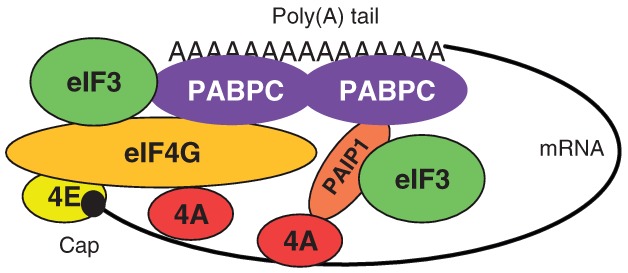
The closed loop complex enhances translation by increasing eIF4A and eIF3 recruitment. Cytoplasmic poly(A) binding proteins (e.g., PABPC1 or PABPC1L) stabilize the cap binding translation initiation complex consisting of eIF4E (4E), eIF4G, and eIF4A (4A), forming the closed loop complex. In addition, PABPC proteins can bind poly(A) binding protein interacting protein 1 (PAIP1), which, like eIF4G can bind eIF4A and the ribosome recruiting complex eIF3. This leads to enhanced translation mediated by the poly(A) tail. For references, see text.
The poly(A) tail also protects the mRNA from degradation, a process in which PABPs play a complex role, by both preventing and stimulating removal of the poly(A) tail, depending on the presence of different deadenylation complexes.34 Most mRNA degradation in eukaryotes is thought to occur by the deadenylation-dependent decapping pathway, in which the poly(A) tail must be shortened to 10–15 nt before the mRNA is decapped and degraded by exonucleases.45–47 The sequences regulating mRNA stability are usually found in the 3′ UTR of the mRNA, although elements in the 5′ UTR and the coding region can also participate. Deadenylation, the removal of the poly(A) tail by specific 3′ exonucleases, is a frequently regulated step in the determination of mRNA stability. Destabilizing 3′ UTR elements include AU and GU rich destabilizing elements that recruit proteins binding to deadenylating enzymes.45,48,49 However, mRNAs with short poly(A) tails are not rare in tissue culture cells50 and many 3′ UTRs can bind ELAVL1 (HuR) through U rich elements, which prevents the degradation of mRNAs with short tails.51–53 The poly(A) tail of an mRNA is therefore involved in its function throughout its lifetime, functioning in mRNA export, translation, and stability, and is the target of extensive regulation.
THE ROLE OF CYTOPLASMIC POLYADENYLATION IN THE REGULATION OF GENE EXPRESSION
Cytoplasmic polyadenylation is the elongation of the poly(A) tail of an mRNA after it has been exported into the cytoplasm. Typically, this process increases protein expression from specific mRNAs by the translational activation of stored mRNAs with short poly(A) tails.54,55 In general mRNAs are thought to be synthesized with long poly(A) tails and the default state is for an mRNA to be translated upon exit from the nucleus. Therefore, regulation by cytoplasmic polyadenylation requires that the mRNA recruits factors that deadenylate and translationally repress the newly exported mRNA, as well as respond to a specific signal to induce expression by cytoplasmic polyadenylation
So far, the defined repressive elements are all in the 3′ untranslated regions of the mRNA and the same elements that convey activation by cytoplasmic polyadenylation sometimes can also mediate translational repression and mRNA deadenylation.56,57 In addition, these mRNAs often contain separate regions that recruit other deadenylation and translational repression factors.58–64 Especially widespread in early development are mRNAs with GU-rich elements, which bind the CELF family of deadenylation factors and mediate deadenylation in the Xenopus embryo, counteracting cytoplasmic polyadenylation activated in the oocyte.49,65–68 Although a shorter poly(A) tail generally reduces translational efficiency, the lack of a poly(A) tail alone is not sufficient for efficient inhibition of translation, as many translational repressors can reduce the expression from nonadenylated mRNAs significantly in cells.56,69 Consequently, in addition to recruiting deadenylating enzymes, mRNAs that are regulated by cytoplasmic polyadenylation also rely on other mechanisms to reduce translation efficiency, predominantly through preventing the association of the cap-binding translation initiation complex,55,64,70 although repression by interference with the elongation phase of protein synthesis has also been implicated recently.71 In addition, several deadenylation factors are able to repress translation directly, independently of their deadenylation activity.70,72 In microRNA-mediated translational repression, deadenylation is known to be secondary to efficient translational repression.73–77 It is not known if deadenylation follows or precedes translational repression in mRNAs repressed by other deadenylation factors.
The activation of translation by cytoplasmic polyadenylation is presumably mediated by the increased recruitment of poly(A) binding proteins to the elongated poly(A) tail and enhanced formation of the closed loop complex.34,35 However, a few studies indicate that active cytoplasmic polyadenylation or other remodeling of the mRNP is required for translational activation in addition to the presence of a poly(A) tail,78–80 as was reviewed previously in more detail.70 In theory, cytoplasmic polyadenylation should also be able to stabilize mRNAs that are dependent on deadenylation for their degradation.45–47 Interestingly, two such potential cases have been reported. In zebrafish embryos, the binding of a protein that appears to mediate cytoplasmic polyadenylation counteracts the destabilization of germline mRNAs by a microRNA.81 In addition, in the nematode Caenorhabditis elegans, mutation of the cytoplasmic polyadenylation machinery leads to a reduction in the levels of its target mRNAs.82
Regulation of translation by cytoplasmic polyadenylation allows nuclear export of mRNAs to be separated from the synthesis of the encoded proteins in time and space. Many of these mRNAs are indeed localized to specific positions in the cell, for instance Bicoid mRNA in early Drosophila embryos, CAMK2A mRNA in the dendrites of neurons and mRNAs encoding cell cycle regulators on the spindle of mitotic or meiotic cells.83–86 In several cases, the sequence elements and RNA binding proteins that mediate the control of poly(A) tail size also contribute to the intracellular localization of mRNAs. Cytoplasmic polyadenylation itself is therefore only one function of a family of RNP complexes that can also mediate translational repression, deadenylation, and mRNA localization.54,55,70,87 In the rest of this review we focus on the components of these complexes that determine which mRNAs are targeted by cytoplasmic polyadenylation, the cytoplasmic polyadenylation specificity factors (CyPSFs).
CYTOPLASMIC POLYADENYLATION OF mRNAS IN GERM CELLS AND EMBRYOS
Cytoplasmic polyadenylation was first described in the 1970s in sea urchin embryos.88 Since then it has been observed in the oocytes and early embryos of many animal species including bivalves (Spisula), insects (Drosophila), amphibians (e.g., Xenopus), fish (e.g., zebrafish), and mammalians (e.g., mouse).86,89–94 In these developmental stages transcription is absent and posttranscriptional regulation is therefore particularly important and more readily detectable. The factors mediating cytoplasmic polyadenylation of mRNAs have predominantly been characterized in Xenopus oocytes and early embryos.94–98 The large size of Xenopus oocytes and embryos enables injection of radioactive RNA substrates, making it relatively easy to demonstrate that cytoplasmic polyadenylation occurs and to map the elements. Most of the mRNAs targeted by cytoplasmic polyadenylation in oocytes and early embryos are directly involved in meiosis and/or mitosis.99–102 In all cases in which this was examined, cytoplasmic polyadenylation in Xenopus requires the poly(A) signal that is also used for nuclear polyadenylation.59,103–106 In addition, specific sequence elements in the 3′ untranslated region are required. Five different sequence elements mediating cytoplasmic polyadenylation have been characterized in this system. Only one of these elements and its associated factors, the cytoplasmic polyadenylation element (CPE), has so far been studied in detail. Table 1 gives a summary of the evidence for the mRNA elements and specificity factors that may mediate cytoplasmic polyadenylation, as discussed further below.
TABLE 1.
Summary of Potential Specificity Factors for Cytoplasmic Polyadenylation with Human Orthologs. For Further Details, See Text
| Factor | Synonyms/Orthologs | Binding Site | Evidence for Cytoplasmic Polyadenylation |
|---|---|---|---|
| CPEB1 | CPEB, Orb | CPE Strong: UUUUAU, UUUUAAU; Weak: UUUUACU,UUUUAAGU, UUUCAU99 | CPE confirmed by polyadenylation in the absence of transcription and cytoplasmic injection of RNA in Xenopus and mouse oocytes, e.g.107–110 CPEB1 function shown by depletion from extracts and dominant mutants in Xenopus.96,111Good correlation in mammalian neurons and mitotic cells.112,110 |
| CPEB4 | Orb2 | CPE Similar to CPEB1.100,112 Or higher affinity for U rich sequence with secondary structure.113 | Injection into Xenopus oocytes late in maturation for CPE and knockdown for CPEB4. Correlation between element and protein function in mitotic cells100,112 |
| MSI1 | Musashi | MBE (G/A) U1-3 AGU114 | Injection of RNA for the MBE and depletion as well as dominant negative mutants in Xenopus oocytes.103,115–117 |
| ZAR2 | Zygote Arrest 2 | TCS (A/U)UU(A/G)UCU59 | Injection of RNA demonstrated existence of TCS mediated polyadenylation in Xenopus oocytes.58,59,63 ZAR2 binds to the element but no other evidence for a function of this protein.63 |
| PCBP2 | αCP2, hnRNP-E2 | C-CPE C-rich, e.g. CCCUC CCUCCUCCCC105 | Injection of RNA into Xenopus embryos proved existence of the element and PCBP2 binds to this element, as well as to polyadenylation factors105 |
| ELAVL1 | HuR | eCPE U rich, U1279,106,118 | eCPE function shown by RNA injection and by polyadenylation of endogenous mRNA in the absence of transcription in Xenopus embryos.79,106,118 ELAVL1 binds this element.119 |
| PUM2 | Pumilio, FBF1, FBF2 | PBE UGUAU(A/U)UAU 120 | In Xenopus, PUM2-CPEB1 interactions contribute to CPE mediated cytoplasmic polyadenylation in oocytes.99,121 In C. elegans, upregulates translation in association with PAPD4122 |
| BICC1 | Bicaudal C, GLD-3 | Unknown | Interacts physically and functionally with PAPD4 and PAPD5 to activate target mRNAs in C. elegans.123,124 |
| DAZL | Deleted in Azoospermia-like | GUU triplet125 | No cytoplasmic polyadenylation observed in Xenopus oocytes, despite clear role in translational activation.126,127 In zebrafish embryos, DAZL binding negates microRNA mediated mRNA destabilization, and increases poly(A) tail size.81 Interacts with Pumilio.128 |
| RBFOX2 | RBM9, FOX-2 | UGCAUG and others characterized in splicing substrates129,130 | Binds to PAPD4 in the cytoplasm of Xenopus oocytes.131 No evidence that binding site confers cytoplasmic polyadenylation. |
| FMRP | Fragile X Mental Retardation Protein | Large number of widely different elements proposed e.g.132–135 | Found to interact and co-localize with PAPD4 in Drosophila extracts and neurons.136 Functional evidence supports a role in translational repression rather than activation in oocytes.137 |
The Cytoplasmic Poly(A) Polymerases in Oocytes and Embryos
Cytoplasmic polyadenylation must be mediated by poly(A) polymerases that are presumably recruited to specific mRNAs by CyPSFs and activated at the appropriate time. The best characterized cytoplasmic poly(A) polymerase, PAPD4 (Gld-2) was first discovered in C. elegans and its enzymatic activity was confirmed in mammalian orthologs.91,138 This enzyme is now thought to be the main polymerase associated with the CyPSF cytoplasmic polyadenylation element binding protein (CPEB) in full grown Xenopus oocytes,95 while in Drosophila oogenesis the CPEB homolog Orb first mediates cytoplasmic polyadenylation with the classical nuclear poly(A) polymerase (PAPOLA) before switching to a PAPD4 ortholog.139 In vitro studies indicated that Xenopus CPEB can also recruit PAPOLA to mediate polyadenylation and a cytoplasmic variant has been characterized, so it is possible that this poly(A) polymerase plays a role in cytoplasmic polyadenylation in vertebrate oocytes as well.140,141 However, a different study has reported that such truncated PAPOLA variants lack polyadenylation activity in vitro, so more evidence for a cytoplasmic role for this enzyme in oocytes is required.142 Although early studies indicated that PAPD4 is also the poly(A) polymerase involved in CPE mediated polyadenylation in mouse oocytes,143 knockout mice do not have changes in cytoplasmic polyadenylation in their oocytes.144 However, an ortholog of a third conserved poly(A) polymerase, PAPD5 (Gld-4) can compensate for PAPD4 (Gld-2) loss in oogenesis in C. elegans, and this protein can mediate CPEB1 dependent cytoplasmic polyadenylation in human fibroblasts, so perhaps such a compensation functions in mouse oocytes as well.123,145 A testis specific cytoplasmic poly(A) polymerase with high homology to PAPOLA, PAPOLB (TPAP), is required for sperm development in mouse, but it is unknown which CyPSF this enzyme is partnered with.146–150 This data therefore indicate that four poly(A) polymerases could participate in cytoplasmic polyadenylation in germ cell development in animals, and further research is required to determine which polymerases are involved in cytoplasmic polyadenylation in each stage of vertebrate oogenesis and development.
The CPE and Its Binding Proteins
The CPE is a sequence that was originally characterized as conferring cytoplasmic polyadenylation during Xenopus oocyte maturation.107 Its first specific binding factor, now called CPEB1, was isolated from oocyte extract and shown to convey cytoplasmic polyadenylation.96 The four CPEB proteins conserved in vertebrates contain two RNA recognition motifs (RRMs), followed by a zinc finger domain located in the C terminus of the proteins.151,152 For CPEB1, all three of these domains are required for RNA binding.153
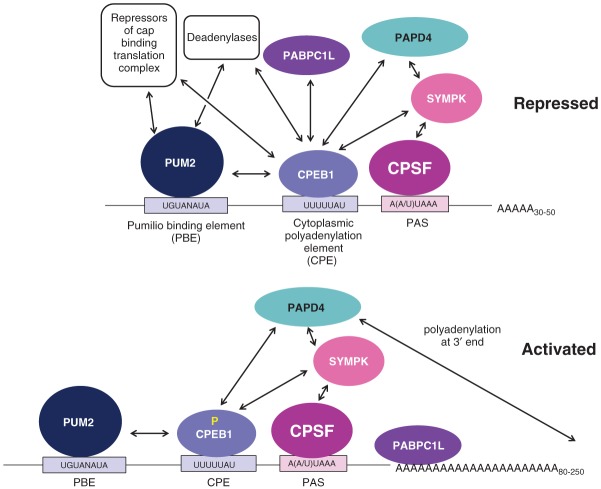
CPEB1 and CPEB4 are required for normal Xenopus oocyte maturation and CPEB1 has been implicated in regulating the cell cycle in early embryos.85,100,154,155 CPEB1 knockout mice also have defects in oocyte and sperm development.156 In addition, CPEB homologs in Drosophila and Caenorhabditis have been shown to mediate translational activation and play a role in germ cell development and early embryogenesis, indicating that these functions are evolutionary conserved.139,156–161 Figure 3 summarizes the regulation of translation by CPEB1 as elucidated in Xenopus oocytes and discussed in detail below.
FIGURE 3.
Cytoplasmic polyadenylation during Xenopus oocyte maturation. The cytoplasmic polyadenylation element (CPE) recruits its binding protein CPEB1.96 This binding can be stabilized, especially on noncanonical CPEs, by the recruitment of Pumilio (PUM2) to the Pumilio binding element (PBE) and protein–protein interactions between CPEB1 and PUM2.99,121 CPEB1 in turn interacts with the cytoplasmic CPSF complex, consisting of CPSF1,2 and 4 and the associated factor SYMPK (the endonuclease CPSF3 is absent and FIP1L has not been tested).162 CPSF1 recognizes the poly(A) signal (PAS). Before oocyte maturation translation is Repressed: CPEB1 and PUM2 mediate mRNA deadenylation and translational repression by recruiting deadenylases (CPEB1 recruits PARN,163 PUM2 can bind the CNOT complex164) and disrupting closed loop complex formation (PUM2 by direct cap interaction,64 CPEB1 by recruiting a choice of eIF4E binding or cap binding proteins70). In addition, CPEB1 binds the embryonic poly(A) binding protein PABPCL1.165 The poly(A) polymerase PAPD4 is secured in the complex by interactions with CPEB1 and CPSF, but its activity is repressed and/or masked by active deadenylation. Upon the stimulation of meiotic maturation, translation is Activated: CPEB1 is phosphorylated (yellow P).166 Deadenylases and translational repression complexes are ejected from the complex and/or inactivated,163,167 PAPD4 mediates elongation of the poly(A) tail95 and PABPC1L dissociates from CPEB1 and transferred to the poly(A) tail, where it can stimulate the formation of the closed loop complex and activate translation.168,169 More details and references can be found in the text.
The CPE is a U-rich element containing a stretch of at least four U residues with a canonical CPE being UUUUAU or UUUUAAU, which binds CPEB1.96,104,108 All the variants that were used in a bioinformatic screen and correlate well with cytoplasmic polyadenylation in Xenopus oocytes can be found in Table 1.99 The RNA binding domains of mouse CPEB3 and CPEB4 have been reported to select a distinct structured recognition element containing a U rich bulge.113 However, Xenopus CPEB4 appears to target the same mRNAs as CPEB1 in oocytes and mediates cytoplasmic polyadenylation of the same mRNAs, although at a later stage of meiosis.100 Mammalian CPEB2 binds to sequences containing CPEs and binding can be competed with poly(U), indicating its binding site is also U-rich.170,171 So far, no structures of the RNA binding domains of CPEB family proteins in complex with RNA have been published, so the relationship between the structure of the protein and the RNA elements it binds remains unclear.
In addition to the CPE, the poly(A) signal that confers nuclear cleavage and polyadenylation (AAUAAA or AUUAAA) is required for CPE-mediated cytoplasmic polyadenylation in Xenopus oocytes.104 The protein complex recognizing the poly(A) signal is thought to be a form of CPSF, with the 160 kDa subunit (CPSF1) recognizing the RNA specifically. The 100 and 30 kDa subunits of CPSF (CPSF2 and CPSF4) are also present in the cytoplasm of Xenopus oocytes, while the 73 kDa endonuclease subunit (CPSF3) is absent.172 Because the poly(A) signal is present in nearly all native mRNAs, mRNA specific regulation must be mediated by binding sites for CyPSFs such as the CPE, but the regulation of cytoplasmic polyadenylation could in principle be influenced by factors associated with the poly(A) signal.
Interaction Between the Complexes Associated with the CPE and the Poly(A) Signal
The spatial arrangement of CPEs in the 3′ untranslated region (3′ UTR) of an mRNA is important for their function.99 Two CPEs in close proximity (less than 50 nucleotides apart), mediate translational repression in immature oocytes regardless of their position in the 3′ UTR. In order to mediate robust cytoplasmic polyadenylation, a CPE normally has to be within 100 nucleotides of the poly(A) signal and ideally within 25 nucleotides. Cytoplasmic polyadenylation mediated by weak CPEs benefits from the presence of a second CPE in close proximity and/or the presence of the RNA recognition element UGUANUAU.99 This represents a binding site for the Pumilio (PUM2) RNA binding protein, which has a conserved interactions with CPEB proteins through protein–protein contacts and binds in their proximity on many mRNAs.173,121,120 Pumilio is thought to stabilize the binding of CPEB in this context, although it is also a deadenylation factor, a translational repressor and a putative CyPSF in its own right (discussed below).64,174,175,122,164 CPEB, in turn, associates with the CPSF complex bound to the poly(A) signal.172,166 CPEB also recruits the scaffold protein SYMPK, which itself can interact with CPSF.95,176 The cytoplasmic poly(A) polymerase PAPD4 is recruited by this complex, although the direct interactions are not yet clear. Cytoplasmic polyadenylation sequence recognition in mRNAs therefore has a multipartite character, with multiple elements recruiting proteins which also interact at the protein level. This is reminiscent of the determination of specificity in nuclear polyadenylation as depicted in Figures 1 and 3.
Regulation of CPEB-Mediated Translational Activation in Oocytes
Activation of CPEB1-mediated cytoplasmic polyadenylation in Xenopus oocytes is through phosphorylation in its N terminal domains.166,111 This phosphorylation can be mediated by the Aurora kinase A (AURKA) and this leads to increased binding of CPSF and expulsion of the deadenylase PARN from the complex, shifting the balance to poly(A) tail elongation.166,163 The activation of CPEB1 mediated cytoplasmic polyadenylation by Aurora kinase phosphorylation is conserved in mice.177,178 However, several studies indicate that the earliest activation of CPEB1 in the Xenopus oocyte is mediated by a different kinase, with CDK1 activated by its noncyclin partner Speedy/Ringo (SPDY) being a strong candidate.179–183 For a more extensive discussion of the regulation of CPEB1 phosphorylation during Xenopus oocyte maturation, we refer to a previous review.70 The N-termini of the CPEB proteins are widely divergent,151,152 and the kinase activating other CPEB homologs is likely to be different. The Drosophila CPEB homolog Orb was recently shown to be activated through phosphorylation by casein kinase 2.184 In addition, the PAN GU kinase, which resembles the NimA (Never in mitosis A, NEK) family of kinases, regulates polyadenylation of cyclin A mRNA during meiosis in Drosophila,185 and is therefore a potential candidate for a regulator of cytoplasmic polyadenylation.
In addition to regulation by phosphorylation, the levels of CPEB proteins and their associated factors can also be regulated during oogenesis by increased synthesis or degradation. Increases are through translational activation, usually involving cytoplasmic polyadenylation. mRNAs regulated in this manner include CPEB4, Aurora kinase A (Eg2), and the cytoplasmic poly(A) polymerase PAPD4 (Gld-2) in Xenopus and Orb in Drosophila.100,186–189 CPEB1 is degraded during the completion of meiosis I through a mechanism involving phosporylation by a polo like kinase (PLK1) and the cyclin dependent kinase CDK1 (cdc2) and ubiquitination by the E3 ligase BTRC (SCFβ-TrCP).101,190,191 This degradation has been proposed to be predominantly on nonRNA bound CPEB1 which is sequestered in the form of a dimer.192 SYMPK-bound CPEB1 levels, presumably representing RNA bound CPEB1 in polyadenylation competent complexes, remain unchanged during oogenesis.95
The timing of translational activation of mRNAs during the oocyte maturation is critical for the correct completion of meiosis. The Mos kinase is an early player in this signal transduction cascade193 and its synthesis is regulated by early cytoplasmic polyadenylation, while activation of cyclin B1 mRNA plays a later role, even though both these mRNAs contain CPEs and bind CPEB1.80,84,108,194,195 CPEs that do not overlap with the poly(A) signal have been reported to mediate earlier cytoplasmic polyadenylation. In contrast, the presence of a CPE overlapping with the poly(A) signal correlates with activation of cytoplasmic polyadenylation late in oocyte maturation, after the breakdown of the nuclear envelope in meioisis I.99,195 The reduction in CPEB1 levels during late oocyte maturation is thought to lead to the availability of the poly(A) signal in these mRNAs. The addition of AU-rich elements that bind deadenylation factors can also affect the timing of cytoplasmic polyadenylation.196 CPEB1-mediated polyadenylation is thought to be required for mouse oocyte maturation as well, and CPEB1 knockout mice fail to translate specific mRNAs very early in oogenesis, long before the full grown oocyte stages usually studied in Xenopus.177,197,109,198
Other CPEB-Associated Factors that Influence Cytoplasmic Polyadenylation
A number of other proteins have been found associated with CPEB1 and are implicated in cytoplasmic polyadenylation in Xenopus oocytes. The poly(A) binding proteins PABPC1 (PAB) and PABPC1L (ePAB) both can bind to CPEB1. Embryonic poly(A) binding protein (PABPC1L) is the predominant poly(A) binding protein in vertebrate oocytes and embryos199,200 and is required for oogenesis in mice and oocyte maturation in Xenopus.165,201 PABPC1L phosphorylation is required for cytoplasmic polyadenylation in Xenopus oocytes and the protein transfers from CPEB1 to the poly(A) tail during cytoplasmic polyadenylation, protecting it from deadenylation.165,168
In addition, CPEB1 is bound by the GTP exchange factor xGEF, a member of the family of Rho guanine nucleotide exchange factors closely related to human ARHGEF39 (C9orf100).202,203 Early in oocyte maturation, xGEF recruits a kinase complex consisting of MAP kinase(ERK2) and a complex of CDK1 and its activator Speedy (SPDY, RINGO) to CPEB1.179,180,204 SPDY is required for the induction of cytoplasmic polyadenylation and is itself regulated by translational control.205,126 xGEF and SPDY are therefore likely to be involved in the activation of CPEB1 mediated polyadenylation during early oocyte maturation.
Other mRNA Specific Cytoplasmic Polyadenylation in Vertebrate Oocytes
CPEB-mediated cytoplasmic polyadenylation is by far the best studied case, but there is very good evidence that it is not the only specificity factor that can confer cytoplasmic polyadenylation to mRNAs. Another well-established RNA binding protein that mediates cytoplasmic polyadenylation is Musashi (MSI1, MSI2).206 The MSI-binding element (MBE) (formerly the polyadenylation response element, PRE) was discovered when the role of the CPE was being investigated in a larger section of the Mos 3′ UTR than had been used previously.107,115 This new element conferred early cytoplasmic polyadenylation to the Mos 3′ UTR in oocytes that was maximal prior to meiosis I in the absence of CPEs and CPEB activity. Indeed, the TATA-BP2 mRNA is polyadenylated during oocyte maturation but does not have a recognizable CPE, only an MBE.103 MSI1 was identified as the trans-acting factor and like the MBE, shown to activate translation during oocyte maturation.103,115 A dominant negative MSI1 mutant and knockdown of MSI1 (and the partially compensating MSI2) using antisense oligodeoxynucleotides, prevented the polyadenylation of endogenous Mos mRNA and blocked maturation.103,116 This indicates that in addition to CPEB, MSI also plays an important role in cytoplasmic polyadenylation during oocyte maturation. Moreover, MSI1 is activated by phosphorylation that is dependent on Ringo/Speedy (SPDY) synthesis, and this phosphorylation is co-incident with Mos polyadenylation.116 Nonphosphorylatable MSI1 did not rescue oocyte maturation after removal of endogenous MSI, further strengthening the importance of MSI1 in oocyte maturation. The role of MSI1 in germ cell development and meiotic progression has also been described in Drosophila and mouse.207 Like the CPE, the MBE requires a poly(A) signal for cytoplasmic polyadenylation, but so far no interactions between MSI proteins and the polyadenylation machinery have been described.
The translation control sequence (TCS) is a third cis-element that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. The TCS was discovered when the role of CPEs was being investigated in the WEE1 3′ UTR.58,59 In the absence of CPEs, in both reporter constructs and the endogenous Pcm-1 mRNA, the TCS mediates early cytoplasmic polyadenylation, at least two hours prior to meiosis I. Like the CPE and the MBE, polyadenylation by the TCS requires the poly(A) signal. The TCS also confers translational repression in immature oocytes and translational activation during oocyte maturation. Recently, a trans-acting factor for the TCS has been found. Zygote arrest 2 (ZAR2) binds to the TCS and, like the TCS, represses translation in immature oocytes when tethered to a reporter RNA.63 This repression is relieved during oocyte maturation. ZAR2 and its homolog ZAR1 have been implicated in the early cleavage phase of mouse embryogenesis,208,209 supporting a function for these proteins in the regulation of early developmental processes. The role of ZAR2 in the regulation of polyadenylation has yet to be determined.
The single RRM protein DAZL has been shown to be critical for male and female germ cell differentiation, oocyte maturation and the oocyte-zygotic transition in several vertebrate species.102,210,211 In most cases, DAZL acts by inducing translational activation of its target mRNAs,210 but its binding site, a GUU triplet,125 has not been reported to convey cytoplasmic polyadenylation in Xenopus oocytes.126,127 In zebrafish embryos, DAZL binding regulates the stability of its target mRNAs, presumably to restrict their expression to the future germ cells, as this protein is normally germ cell specific.81,212–214 In one such study, DAZL was reported to overcome miR-430 mediated destabilization by the 3′ UTR of its own mRNA. This was accompanied by cytoplasmic polyadenylation of the injected mRNA.81 In contrast, in mouse oocytes, the translational activation and cytoplasmic polyadenylation of the Dazl mRNA was found to be mediated by CPEB1 and require a poly(A) signal.102 In Xenopus oocytes DAZL mediated translational activation appears not to involve cytoplasmic polyadenylation, but it requires an interaction between DAZL and poly(A) binding proteins.126,127 This interaction is highly conserved throughout the DAZL protein family and across species and cross-rescue experiments indicate that the mechanism of action is also conserved.210,127 However, DAZL can form a complex with the human Pumilio protein PUM2, so some DAZL functions may be mediated by Pumilo.128 More evidence is therefore required before DAZL protein can be truly considered a CyPSF. In addition, one publication indicates that another single RRM protein, RBFOX2 (Rbm9, Fox2), associates with PAPD4 in Xenopus oocytes and therefore also is a candidate CyPSF in this cell type.131
Cytoplasmic Polyadenylation Mediated by Other Elements in Xenopus Embryos
Two more CPEs have been identified by deletion mapping of injected radiolabelled RNAs in Xenopus. Both mediate polyadenylation in early embryogenesis and not during oocyte maturation, which explains why they are less studied: good quality embryos are harder to obtain and inject than oocytes. First discovered, at about the same time as the CPE, was a stretch of 12 U residues that was called the eCPE.106,215,94 Like the CPE, it requires a poly(A) signal to function and the spacing between the eCPE and the poly(A) signal regulates the timing of polyadenylation.79,106,118 The eCPE can bind ELAVL1 (ElrA, HuR), but whether this is the genuine CyPSF is still uncertain.119,216
The second embryonic CPE in Xenopus was found to be a stretch of cytidine residues and called the C-CPE.105,217 Cytoplasmic polyadenylation by the C-CPE also requires a poly(A) signal and the element is bound by PCBP2 (αCP2, hnRNP-E2), which is exclusively cytoplasmic in Xenopus oocytes.218 PCBP2 immunoprecipitates with SYMPK, CPSF2, PAPD4, CPEB1, and PABPL1, but not with PARN, in embryos, and oocytes. This indicates that it is indeed part of a cytoplasmic polyadenylation complex that is pre-assembled in an inactive form in the oocyte. As was predicted by the PCBP2 and CPEB1 association, the C-CPE and the CPE cooperate to increase polyadenylation during early embryogenesis.105
Other CPEs and Factors Found in Invertebrates
PAPD4 was originally discovered as germ-line deficient 2 (GLD-2) in C. elegans91 and a number of PAPD4-interacting RNA binding proteins that activate the translation of target mRNAs in different stages of germ line development have been characterized.82,91,122,219 Although the activity of PAPD4 as a poly(A) polymerase cannot be doubted, it has been much more difficult in this system to prove that cytoplasmic polyadenylation of specific mRNAs actually occurs in the germ cells and to map the sequences required for cytoplasmic polyadenylation on these mRNAs. PAPD4 RNA binding partners in C. elegans germ cell development include the Pumilio homologs FBF-1 and FBF-2 (PUM1, PUM2),122 RNP-8, a single RRM protein with no clear human homologs,82 and GLD-3, a KH domain protein with similarity to the Drosophila deadenylation factor Bicaudal C (BICC1).220–223 In addition to PAPD4, PAPD5 (GLD-4) has been identified as a cytoplasmic poly(A) polymerase in C. elegans and has been shown to promote germ line development with a complex of BICC1 and GLS-1, a protein with no clear human homologs.123,224 BICC1 is also implicated in male fertility in mice, indicating a conserved function in the germline.225
Drosophila Toll mRNA is regulated by cytoplasmic polyadenylation during early embryogenesis. Experiments using an in vitro polyadenylation system were used to map a 180 nucleotide region of the 3′ UTR that is required for this function.226 Competition with this RNA fragment indicated that the polyadenylation factors binding this region are distinct from those found in CPE containing mRNAs. Remarkably, the canonical nuclear poly(A) signal appears not to be required for the polyadenylation of Toll mRNA, indicating it employs a fundamentally different mechanism of cytoplasmic polyadenylation.55 The transacting factors mediating Toll mRNA cytoplasmic polyadenylation have not yet been characterized. Also in Drosophila, interactions of PAPD4 and CPEB (Orb) with fragile X mental retardation protein (FMRP) have been reported in neurons and oocytes, but its function appears to be opposite to that of CPEB.136,137
CYTOPLASMIC POLYADENYLATION OF mRNAS IN SOMATIC CELLS
Elongation of the poly(A) tail has long been known to also take place in the cytoplasm of mammalian tissue culture cells, accounting for approximately 10% of all polyadenylation.227 However, cytoplasmic polyadenylation of specific mRNAs is much harder to study in somatic cells, as the synthesis of new mRNA also can contribute to changes in poly(A) tail size on specific mRNAs. Elongation of the poly(A) tail in a transcriptionally active cell can be due to reduced deadenylation of the poly(A) tail of newly made mRNAs as well as to cytoplasmic polyadenylation. Moreover, if only a relatively small portion of mRNAs in a cell undergoes poly(A) tail changes, for instance those localized under a particular set of synapses, the detection problem is further magnified. Determination of mRNA poly(A) tail sizes in most systems is dependent on techniques that are not very good at detecting rare mRNAs, such as variations on Northern blotting, or on PCR based methods that need to be very carefully controlled to avoid artifacts for a discussion see Ref 50. As a consequence, no new CPEs or specificity factors have so far been firmly identified in somatic cells. Some putative CyPSFs, such as DAZL and ZAR2, are very germ cell specific and unlikely to be of importance in somatic cells, with the possible exception of some tumor cells.228–230 However, a large body of work indicates that the CPEs and factors that function in germ cells and early embryos also are important in other cell types. It is not always clear, however, if cytoplasmic polyadenylation does indeed play a role in their function. Discussion of all cases where CyPSFs or elements have been implicated in mRNA regulation is outside the scope of this review. We have therefore selected some of the examples of regulation by CPEB, Pumilio, Musashi, PCBP and Bicaudal protein families, as these are confirmed or very likely CyPSFs in germ cells and early embryos.
CPEB Family Proteins and PAPD4 in the Nervous System
In neuronal cells, the local regulation of translation in the dendrites is thought to play a role in the remodeling of synapses required for learning.83 CPEB1 protein and polyadenylation factors are present in the cell bodies and dendrites of many neurons and binds to mRNAs that are known to be partially dendritically localized. The CPEs in the 3′ UTRs of mRNAs such as calmodulin kinase II (CAMK2A) were found to be required for dendritic mRNA localization, as well as for translational stimulation and elongation of the poly(A) tail after synaptic stimulation.231–238 The activation of CPEB1 mediated cytoplasmic polyadenylation and translation in dendrites has been attributed to phosphorylation on the same site as in oocyte maturation, and the kinase is thought to be either AURKA or CAMK2A.232,239–241 As CAMK2A mRNA is also targeted by CPEB1, this is potentially another positive feedback loop, which has been proposed to contribute to the generation of a bistable switch.242 Another proposed mechanism of regulation of CPEB proteins in neurons is a prion-like conformation change in CPEB variants in Aplysia and Drosophila, although so far polyadenylation appears not to be involved.243–246
The polyadenylation inhibitor cordycepin has been used to show that polyadenylation is required for normal synaptic function.113,233 However, this type of experiment is no longer valid, because cordycepin is now known to inhibit protein synthesis through effects on the mTOR pathway in some cell types, as well as to have gene-specific effects on de novo mRNA synthesis.247,248 The most compelling early evidence that the poly(A) tail elongation observed in neuronal cells does not represent newly synthesized mRNAs comes from observation of poly(A) tail elongation in vitro, in synapse containing vesicles (synaptoneurosomes).232 Moreover, a recent study shows that a rapid increase in poly(A) (detected by in situ hybridization) does indeed occur in dendrites in response to NMDA receptor stimulation. The timing of this effect coincides with CPEB1 phosphorylation and PARN dissociation and it is abolished by knockdown of PAPD4 (Gld-2).241 Because of the speed of the response and the requirement for PAPD4, it is unlikely that this increase in poly(A) is due to the transport of newly made mRNA into the dendrites. This therefore probably represents a true case of cytoplasmic polyadenylation.
PAPD4 (Gld2) mutants in Drosophila have defects in long-term memory, which indicates that this enzyme is indeed important in synaptic plasticity.136 In addition, Orb2, the somatic CPEB in Drosophila, is required for asymmetric cell division during neuronal development, for normal locomotion and for memory formation.249,250 This suggests a major role for cytoplasmic polyadenylation in the Drosophila nervous system. CPEB1 has also been implicated in axon branching in hippocampal neurons, in cell migration of astrocytes and in motor skill development in rodents using expression of a CPEB1 dominant negative mutant.251–253 The Pumilio proteins, which can cooperate with CPEB1 in oocytes, have also been implicated in neuronal development and synaptic plasticity.254–260 However, CPEB1 knockout mice have no major nervous system defects, although subtle effects on synaptic transmission and memory have been reported.261,262 This may be because other members of the CPEB family also are expressed in mammalian neurons.113,263 In particular, CPEB3 mediated translational control has been linked to changes in synapse function in rodents264,265 and a polymorphism in the CPEB3 gene has been linked to memory in humans as well.266 An increase in poly(A) tail of a CPEB3 targeted mRNA has indeed been reported.264 In contrast to the regulation observed in oocytes, however, activation of translation is thought to be mediated by mono-ubiquitination or calpain mediated degradation.264,265 In addition, the activation of CPEB3 bound mRNAs does not require a poly(A) signal or CPSF.113 Similarly to the situation for CPEB1 in oocytes, CPEB3 is an efficient translational repressor264,265 and CPEB3 (and CPEB4) can associate with the TOB2 protein, which mediates deadenylation through recruitment of the CNOT deadenylation complex.267 The data so far are therefore inconclusive in determining if CPEB3 works as a deadenylation factor that is inactivated or as a CyPSF that is activated during synaptic stimulation in neurons.
CPEB Family Proteins in the Regulation of Cell Proliferation
CPEB proteins have also been implicated in the regulation of mitosis, senescence and tumorigenesis, this has been recently reviewed in depth,151 with only one additional publication to note.171 In most cases where cytoplasmic polyadenylation has been implicated, clear effects of CPEB proteins on translational efficiency were detected. However, actual cytoplasmic polyadenylation in somatic cells other than neurons has not been definitively demonstrated. One approach has been to inject RNA into Xenopus oocytes to show that the 3′ UTR can mediate cytoplasmic polyadenylation.170,231 Unfortunately, this is no definitive guarantee that translational or poly(A) tail changes are caused by cytoplasmic polyadenylation in the tissue of interest. Another study shows a suggestive correlation between phosphorylation of CPEB1 and poly(A) tail elongation.268 Only two articles come close to demonstrating that CPEB mediated cytoplasmic polyadenylation genuinely plays a role, one in mitosis and one in senescence,145,112 these are discussed below.
In early embryos, cell division is atypical, with an absence of the G1 and G2 phases, so the finding that CPEB1 activates translation by cytoplasmic polyadenylation during mitosis in the embryonic cell cycle85,269,270 did not have immediate implications for the normal somatic cell cycle. However, most mRNAs regulated by cytoplasmic polyadenylation in oocytes encode proteins that are also involved in normal mitosis.151 In addition, mRNA transcription is known to be reduced in G2 and blocked in M phase in HeLa cells,271 similar to the situation in oocytes. In the first systematic screen for differentially polyadenylated mRNAs in mammalian cells, Novoa et al.112 compared the poly(A) tail sizes of mRNAs in HeLa cells in the S and G2/M phases. Total poly(A) RNA was isolated using oligo(dT) chromatography and RNA with short poly(A) tails was isolated by low stringency elution of poly(U) chromatography. Hundreds of mRNAs displayed cell cycle dependent changes in the ratios between the total and short poly(A) tail fractions, indicating that poly(A) tail regulation is widespread during the cell cycle. Knockdown of CPEB1 and CPEB4 caused cell cycle defects and affected the polyadenylation of some, but not all of these mRNAs during G2/M. These data indicate that both CPE dependent and other types of cytoplasmic polyadenylation are occurring during the G2/M phase of the cell cycle in HeLa cells.
In primary fibroblasts, knockdown or knockout of CPEB1 prevents senescence, a process that halts cell proliferation in normal cells after a finite number of cell divisions.272,273 The translational activation of the tumor suppressor TP53 (p53), a factor required for senescence, is dependent on CPEB1 and TP53 mRNA has a short poly(A) tail in human fibroblasts with reduced CPEB1.272 Surprisingly, the knockdown of the PAPD4 (Gld2) poly(A) polymerase increased TP53 translation, an effect that was attributed to the role of PAPD4 in the maturation of a microRNA that targets CPEB1 mRNA in these cells, leading to increased CPEB1 levels.145 Knockdown of the PAPD5 (Gld4) poly(A) polymerase did reduce TP53 protein expression and TP53 mRNA polyadenylation, and PAPD5 was found associated with CPEB1 and the TP53 mRNA.272 These data indicate that TP53 mRNA is regulated by polyadenylation mediated by CPEB1 and PAPD5 during the senescence of human fibroblasts. However, since both CPEB proteins and PAPD5 can also be found in the nucleus in somatic cells274–277 and transcription is ongoing in these experiments, an effect on nuclear polyadenylation cannot be excluded.
Other Roles of CyPSFs in Somatic Cells
An exciting recent study demonstrated widespread regulation of poly(A) tail size during the circadian rhythm in the liver, some of which appears to be linked to transcriptional up regulation and some of which are not linked to transcriptional increases.278 The poly(A) tail sizes of some of the mRNAs in the last class are regulated by CPEB1 and CPEB2. Less well characterized and new polyadenylation regulatory elements are likely to be involved in the regulation of the other mRNAs identified in this study.
Musashi (MSI1 and MSI2) was initially characterized as a factor that promoted the self-renewal of neural stem cells by translationally repressing numb, an inhibitor of Notch signaling.279 More recently, MSI has been implicated as a marker for a variety of stem cells including mammary cells,280 normal and malignant gasterointestinal cells,281,282 endometrial stem cells,283 and in normal and malignant hematopoesis.284 The mechanism of translational repression by mammalian MSI in somatic cells is proposed to be the disruption of the closed loop translation initiation complex due to an interaction of MSI1 with PABP.285
The Pumilio family (PUM1 and PUM2) proteins are highly conserved in eukaryotes and well characterized as deadenylation factors and translational repressors in many cell types.286 In addition to their functions in neurons (see above), Pumilio proteins have been implicated in the regulation of MAPK signaling and cell proliferation.287–289 There is so far no evidence that these factors can mediate cytoplasmic polyadenylation in somatic cells, whether on their own or in conjunction with CPEB proteins.
The Bicaudal C homolog BICC1 is linked to polycystic kidney disease in zebrafish, Xenopus, mouse and human,290–292 this is caused by defects in epithelial morphogenesis.292–295 CPEB1 deficiency also causes epithelial defects in the breast tissue of mice.296 These observations complement the long established localization of the polyadenylation factor SYMPK at tight junctions,297,298 and suggest a potential function for local translation mediated by cytoplasmic polyadenylation in the morphogenesis of epithelia. BICC1 appears to be a KH domain RNA binding protein, but a consensus binding element in RNA has so far not been identified.222,299 It has been reported to both to enhance and counteract microRNA mediated repression.300,301 No data are available on the effects of BICC1 on mRNA polyadenylation in somatic cells.
Both PCBP2 (αCP2) and ELAVL 1 (HuR) are best known as proteins that mediate mRNA stabilization in somatic cells,302,303 but there is so far no evidence that this role involves cytoplasmic polyadenylation. PCBP2 and its closely related family member, PCBP1 (hnRNP-E1), have been shown to mediate translational repression in haematopoietic cells.304,305 Strikingly, the C-CPE that can promote cytoplasmic polyadenylation in Xenopus embryos105 and mRNA stabilization in the cytoplasm of somatic cells also mediates gene-specific increased splicing, cleavage and polyadenylation of pre-mRNA in the nucleus.306 In addition, PCBP2 stimulated polyadenylation and was found to associate with three CPSF subunits and SYMPK in nuclear extracts. This observation opens the possibility that other cytoplasmic polyadenylation sequences and factors also can influence nuclear polyadenylation, and conversely, that proteins that bind upstream polyadenylation elements in the nucleus may also have a function in cytoplasmic polyadenylation. In this context it is remarkable that a subunit of the USE binding factor CFIm (NUDT21) has been found together with PCBP1 in a vinculin bound cytoplasmic RNP complex during cell adhesion.307 Moreover, the NUDT21 recognition site is included in the Pumilio binding element, opening the possibility of competitive or subsequent binding.120,308 Interestingly, gene specific increases in splicing and nuclear 3′ mRNA processing have been shown to play a major role in the regulation of cyclin expression during meiosis in yeast,309 and regulation of nuclear polyadenylation by USEs may therefore be the ancestral mechanism that evolved into cytoplasmic polyadenylation in animal germ cells.
CONCLUSIONS
Cytoplasmic polyadenylation was first characterized in Xenopus oocytes, and all well characterized examples of regulatory sequences are still from this system, with the best established specificity factors being CPEB1, CPEB4, MSI1, and PCBP2. However, a role for cytoplasmic polyadenylation is also emerging in the C. elegans germ line, as well as in neuronal plasticity and mitosis in somatic cells. While conclusive evidence of cytoplasmic polyadenylation still is dependent on showing that poly(A) tail elongation is independent of transcription, good circumstantial cases can be made where mRNA activation can be linked to the physical and functional association of an RNA binding protein in complex with one or more poly(A) polymerases, such as has been shown for BicaudalC and Pumilio in C. elegans. However, due caution must be exercised when assigning cytoplasmic mRNA associated functions to poly(A) polymerases, as most are present in the nucleus as well and these enzymes are also involved in the processing and degradation of noncoding RNAs.274,310–312 Increasingly, complexes of multiple RNA binding proteins are implicated in the mRNA specificity as well as the timing of cytoplasmic polyadenylation. The best characterized is the CPEB-Pumilio interaction, but now there are also the CPEB1-PCBP2 and Pumilio-DAZL interactions and many mRNAs that are regulated by cytoplasmic polyadenylation in oocytes have binding sites for multiple factors. This indicates that cooperation and/or antagonism of different poly(A) regulatory factors is common. Cytoplasmic polyadenylation now appears to be a widespread phenomenon, and is increasingly attracting the interest of researchers working in systems other than Xenopus and C. elegans oogenesis and early development. Therefore, we can expect new specificity factors to emerge in the near future.
Acknowledgments
Nicola Gray (Edinburgh) and Claudia Bagni (Leuven) are thanked for prompt answers to specific queries. Any remaining errors are entirely our own.
References
- 1.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagaike T, Manley JL. Transcriptional activators enhance polyadenylation of mRNA precursors. RNA Biol. 2011;8:964–967. doi: 10.4161/rna.8.6.17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinson HG. An active role for splicing in 3′-end formation. Wiley Interdiscip Rev RNA. 2011;2:459–470. doi: 10.1002/wrna.68. [DOI] [PubMed] [Google Scholar]
- 4.Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol. 2011;12:283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tian B, Graber JH. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA. 2011;3:385–396. doi: 10.1002/wrna.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Q, Doublie S. Structural biology of poly(A) site definition. Wiley Interdiscip Rev RNA. 2011;2:732–747. doi: 10.1002/wrna.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2009;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan S, Choi EA, Shi Y. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip Rev RNA. 2011;2:321–335. doi: 10.1002/wrna.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufmann I, Martin G, Friedlein A, Langen H, Keller W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 2004;23:616–626. doi: 10.1038/sj.emboj.7600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Preker PJ, Lingner J, Minvielle-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81:379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 12.Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolev NG, Yario TA, Benson E, Steitz JA. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep. 2008;9:1013–1018. doi: 10.1038/embor.2008.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Q, Gilmartin GM, Doublie S. The structure of human cleavage factor Im hints at functions beyond UGUA-specific RNA binding: A role in alternative polyadenylation and a potential link to 5′ capping and splicing. RNA Biol. 2011;8:748–753. doi: 10.4161/rna.8.5.16040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutz CS, Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip Rev RNA. 2011;2:22–31. doi: 10.1002/wrna.47. [DOI] [PubMed] [Google Scholar]
- 16.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn U, Gundel M, Knoth A, Kerwitz Y, Rudel S, Wahle E. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J Biol Chem. 2009;284:22803–22814. doi: 10.1074/jbc.M109.018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerwitz Y, Kuhn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, Schwarz E, Wahle E. Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J. 2003;22:3705–3714. doi: 10.1093/emboj/cdg347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bardwell VJ, Zarkower D, Edmonds M, Wickens M. The enzyme that adds poly(A) to mRNAs is a classical poly(A) polymerase. Mol Cell Biol. 1990;10:846–849. doi: 10.1128/mcb.10.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topalian SL, Kaneko S, Gonzales MI, Bond GL, Ward Y, Manley JL. Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol Cell Biol. 2001;21:5614–5623. doi: 10.1128/MCB.21.16.5614-5623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, III, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzales ML, Mellman DL, Anderson RA. CKIα is associated with and phosphorylates star-PAP and is also required for expression of select star-PAP target messenger RNAs. J Biol Chem. 2008;283:12665–12673. doi: 10.1074/jbc.M800656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laishram RS, Barlow CA, Anderson RA. CKI isoforms {α} and {varepsilon} regulate Star-PAP target messages by controlling Star-PAP poly(A) polymerase activity and phosphoinositide stimulation. Nucleic Acids Res. 2011;39:7961–7973. doi: 10.1093/nar/gkr549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laishram RS, Anderson RA. The poly A polymerase Star-PAP controls 3′-end cleavage by promoting CPSF interaction and specificity toward the pre-mRNA. EMBO J. 2010;29:4132–4145. doi: 10.1038/emboj.2010.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, Laishram RS, Ji Z, Barlow CA, Tian B, Anderson RA. Star-PAP control of BIK expression and apoptosis is regulated by nuclear PIPKIα and PKCdelta signaling. Mol Cell. 2012;45:25–37. doi: 10.1016/j.molcel.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellman DL, Anderson RA. A novel gene expression pathway regulated by nuclear phosphoinositides. Adv Enzyme Regul. 2009;49:11–28. doi: 10.1016/j.advenzreg.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- 29.Soucek S, Corbett AH, Fasken MB. The long and the short of it: The role of the zinc finger polyadenosine RNA binding protein, Nab2, in control of poly(A) tail length. Biochim Biophys Acta. 1819;2012:546–554. doi: 10.1016/j.bbagrm.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sagawa F, Ibrahim H, Morrison AL, Wilusz CJ, Wilusz J. Nucleophosmin deposition during mRNA 3′ end processing influences poly(A) tail length. EMBO J. 2011;30:3994–4005. doi: 10.1038/emboj.2011.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuehn U, Guendel M, Knoth A, Kerwitz Y, Ruedel S, Wahle E. Poly(A) tail length is controlled by the nuclear poly(A) binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J Biol Chem. 2009;284:22803–22814. doi: 10.1074/jbc.M109.018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson SA, Kim H, Erickson B, Bentley DL. The export factor Yra1 modulates mRNA 3′ end processing. Nat Struct Mol Biol. 2011;18:1164–1171. doi: 10.1038/nsmb.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qu X, Lykke-Andersen S, Nasser T, Saguez C, Bertrand E, Jensen TH, Moore C. Assembly of an export-competent mRNP is needed for efficient release of the 3′ end processing complex after polyadenylation. Mol Cell Biol. 2009;29:5327–5338. doi: 10.1128/MCB.00468-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brook M, Gray NK. The role of mammalian poly(A)-binding proteins in co-ordinating mRNA turnover. Biochem Soc Trans. 2012;40:856–864. doi: 10.1042/BST20120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgess HM, Gray NK. mRNA-specific regulation of translation by poly(A)-binding proteins. Biochem Soc Trans. 2010;38:1517–1522. doi: 10.1042/BST0381517. [DOI] [PubMed] [Google Scholar]
- 36.Burgess HM, Gray NK. An integrated model for the nucleo-cytoplasmic transport of cytoplasmic poly(A)-binding proteins. Commun Integr Biol. 2012;5:243–247. doi: 10.4161/cib.19347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Afonina E, Stauber R, Pavlakis GN. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem. 1998;273:13015–13021. doi: 10.1074/jbc.273.21.13015. [DOI] [PubMed] [Google Scholar]
- 38.Hosoda N, Lejeune F, Maquat LE. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol Cell Biol. 2006;26:3085–3097. doi: 10.1128/MCB.26.8.3085-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Craig AWB, Haghighat A, Yu AT, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 40.Derry MC, Yanagiya A, Martineau Y, Sonenberg N. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb Symp Quant Biol. 2006;71:537–543. doi: 10.1101/sqb.2006.71.061. [DOI] [PubMed] [Google Scholar]
- 41.Martineau Y, Derry MC, Wang X, Yanagiya A, Berlanga JJ, Shyu AB, Imataka H, Gehring K, Sonenberg N. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol Cell Biol. 2008;28:6658–6667. doi: 10.1128/MCB.00738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng S, Gallie DR. Wheat eukaryotic initiation factor 4B organizes assembly of RNA and eIFiso4G, eIF4A, and poly(A)-binding protein. J Biol Chem. 2006;281:24351–24364. doi: 10.1074/jbc.M605404200. [DOI] [PubMed] [Google Scholar]
- 43.Cheng S, Gallie DR. eIF4G, eIFiso4G, and eIF4B bind the poly(A)-binding protein through overlapping sites within the RNA recognition motif domains. J Biol Chem. 2007;282:25247–25258. doi: 10.1074/jbc.M702193200. [DOI] [PubMed] [Google Scholar]
- 44.Bushell M, Wood W, Carpenter G, Pain VM, Morley SJ, Clemens MJ. Disruption of the interaction of mammalian protein synthesis initiation factor 4B with the poly(A) binding protein by caspase- and viral protease-mediated cleavages. J Biol Chem. 2001;276:23922–23928. doi: 10.1074/jbc.M100384200. [DOI] [PubMed] [Google Scholar]
- 45.Wu X, Brewer G. The regulation of mRNA stability in mammalian cells: 2.0. Gene. 2012;500:10–21. doi: 10.1016/j.gene.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schoenberg DR, Maquat LE. Regulation of cytoplasmic mRNA decay. Nat Rev Genet. 2012;13:246–259. doi: 10.1038/nrg3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balagopal V, Fluch L, Nissan T. Ways and means of eukaryotic mRNA decay. Biochim Biophys Acta. 1819;2012:593–603. doi: 10.1016/j.bbagrm.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 48.Masuda A, Andersen HS, Doktor TK, Okamoto T, Ito M, Andresen BS, Ohno K. CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay. Sci Rep. 2012;2:209. doi: 10.1038/srep00209. (Epub ahead of print, January 4, 2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dasgupta T, Ladd AN. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip Rev RNA. 2012;3:104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meijer HA, Bushell M, Hill K, Gant TW, Willis AE, Jones P, De Moor CH. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res. 2007;35:e132. doi: 10.1093/nar/gkm830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, Ascano M, Jr, Tuschl T, Ohler U, Keene JD. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weill L, Belloc E, Bava FA, Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol. 2012;19:577–585. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- 55.Villalba A, Coll O, Gebauer F. Cytoplasmic polyadenylation and translational control. Curr Opin Genet Dev. 2011;21:452–457. doi: 10.1016/j.gde.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 56.De Moor CH, Richter JD. Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J. 1999;18:2294–2303. doi: 10.1093/emboj/18.8.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huarte J, Stutz A, O'Connell ML, Gubler P, Belin D, Darrow AL, Strickland S, Vassalli JD. Transient translational silencing by reversible mRNA deadenylation. Cell. 1992;69:1021–1030. doi: 10.1016/0092-8674(92)90620-r. [DOI] [PubMed] [Google Scholar]
- 58.Charlesworth A, Welk J, MacNicol AM. The temporal control of Wee1 mRNA translation during Xenopus oocyte maturation is regulated by cytoplasmic polyadenylation elements within the 3′-untranslated region. Dev Biol. 2000;227:706–719. doi: 10.1006/dbio.2000.9922. [DOI] [PubMed] [Google Scholar]
- 59.Wang YY, Charlesworth A, Byrd SM, Gregerson R, MacNicol MC, MacNicol AM. A novel mRNA 3′ untranslated region translational control sequence regulates Xenopus Wee1 mRNA translation. Dev Biol. 2008;317:454–466. doi: 10.1016/j.ydbio.2008.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Belloc E, Mendez R. A deadenylation negative feedback mechanism governs meiotic metaphase arrest. Nature. 2008;452:1017–1021. doi: 10.1038/nature06809. [DOI] [PubMed] [Google Scholar]
- 61.Nakahata S, Katsu Y, Mita K, Inoue K, Nagahama Y, Yamashita M. Biochemical identification of Xenopus Pumilio as a sequence-specific Cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog (Xcat-2) and a cytoplasmic polyadenylation element-binding Protein (CPEB) J Biol Chem. 2001;276:20945–20953. doi: 10.1074/jbc.M010528200. [DOI] [PubMed] [Google Scholar]
- 62.Meijer HA, Radford HE, Wilson LS, Lissenden S, De Moor CH. Translational control of maskin mRNA by its 3′ untranslated region. Biol Cell. 2007;99:239–250. doi: 10.1042/BC20060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Charlesworth A, Yamamoto TM, Cook JM, Silva KD, Kotter CV, Carter GS, Holt JW, Lavender HF, MacNicol AM, Ying WY, et al. Xenopus laevis zygote arrest 2 (zar2) encodes a zinc finger RNA-binding protein that binds to the translational control sequence in the maternal Wee1 mRNA and regulates translation. Dev Biol. 2012;369:177–190. doi: 10.1016/j.ydbio.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao Q, Padmanabhan K, Richter JD. Pumilio 2 controls translation by competing with eIF4E for 7-methyl guanosine cap recognition. RNA. 2010;16:221–227. doi: 10.1261/rna.1884610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teplova M, Song J, Gaw HY, Teplov A, Patel DJ. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure. 2010;18:1364–1377. doi: 10.1016/j.str.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edwards J, Malaurie E, Kondrashov A, Long J, De Moor CH, Searle MS, Emsley J. Sequence determinants for the tandem recognition of UGU and CUG rich RNA elements by the two N terminal RRMs of CELF1. Nucleic Acids Res. 2011;39:8638–8650. doi: 10.1093/nar/gkr510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rattenbacher B, Beisang D, Wiesner DL, Jeschke JC, von HM, St Louis-Vlasova IA, Bohjanen PR. Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol Cell Biol. 2010;30:3970–3980. doi: 10.1128/MCB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moretti F, Kaiser C, Zdanowicz-Specht A, Hentze MW. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat Struct Mol Biol. 2012;19:603–608. doi: 10.1038/nsmb.2309. [DOI] [PubMed] [Google Scholar]
- 70.Radford HE, Meijer HA, De Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta. 2008;1779:217–229. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen PJ, Huang YS. CPEB2-eEF2 interaction impedes HIF-1α RNA translation. EMBO J. 2011;31:959–971. doi: 10.1038/emboj.2011.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J Biol Chem. 2010;285:28506–28513. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bethune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in Zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 78.McGrew LL, Dworkin-Rastl E, Dworkin MB, Richter JD. Poly (A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989;3:803–815. doi: 10.1101/gad.3.6.803. [DOI] [PubMed] [Google Scholar]
- 79.Simon R, Richter JD. Further analysis of cytoplasmic polyadenylation in Xenopus embryos and identification of embryonic cytoplasmic polyadenylation element-binding proteins. Mol Cell Biol. 1994;14:7867–7875. doi: 10.1128/mcb.14.12.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barkoff A, Ballantyne S, Wickens M. Meiotic maturation in Xenopus requires polyadenylation of multiple mRNAs. EMBO J. 1998;17:3168–3175. doi: 10.1093/emboj/17.11.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One. 2009;4:e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim KW, Wilson TL, Kimble J. GLD-2/RNP-8 cytoplasmic poly(A) polymerase is a broad-spectrum regulator of the oogenesis program. Proc Natl Acad Sci USA. 2010;107:17445–17450. doi: 10.1073/pnas.1012611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Darnell JC, Richter JD. Cytoplasmic RNA-binding proteins and the control of complex brain function. Cold Spring Harb Perspect Biol. 2012 doi: 10.1101/cshperspect.a012344. 4:a012344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eliscovich C, Peset I, Vernos I, Mendez R. Spindle-localized CPE-mediated translation controls meiotic chromosome segregation. Nat Cell Biol. 2008;10:858–865. doi: 10.1038/ncb1746. [DOI] [PubMed] [Google Scholar]
- 85.Groisman I, Huang YS, Mendez R, Cao Q, Theurkauf W, Richter JD. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–447. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- 86.Sallés FJ, Lieberfarb ME, Wreden C, Gergen JP, Strickland S. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science. 1994;266:1996–1999. doi: 10.1126/science.7801127. [DOI] [PubMed] [Google Scholar]
- 87.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wilt FH. Polyadenylation of maternal RNA of sea urchin eggs after fertilization. Proc Natl Acad Sci USA. 1973;70:2345–2349. doi: 10.1073/pnas.70.8.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fox CA, Sheets MD, Wickens MP. Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev. 1989;3:2151–2162. doi: 10.1101/gad.3.12b.2151. [DOI] [PubMed] [Google Scholar]
- 90.Standart N, Dale M. Regulated polyadenylation of clam maternal mRNAs in vitro. Dev Genet. 1993;14:492–499. doi: 10.1002/dvg.1020140610. [DOI] [PubMed] [Google Scholar]
- 91.Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- 92.Vassalli J-D, Huarte J, Belin D, Gubler P, Vassalli A, O'Connell ML, Parton LA, Rickles RJ, Strickland S. Regulated polyadenylation controls mRNA translation during meiotic maturation of mouse oocytes. Genes Dev. 1989;3:2163–2171. doi: 10.1101/gad.3.12b.2163. [DOI] [PubMed] [Google Scholar]
- 93.Suzuki H, Tsukahara T, Inoue K. Localization of c-mos mRNA around the animal pole in the zebrafish oocyte with Zor-1/Zorba. Biosci Trends. 2009;3:96–104. [PubMed] [Google Scholar]
- 94.Paris J, Osborne HB, Couturier A, Le Guellec R, Philippe M. Changes in the polyadenylation of specific stable RNA during the early development of Xenopus laevis. Gene. 1988;72:169–176. doi: 10.1016/0378-1119(88)90139-4. [DOI] [PubMed] [Google Scholar]
- 95.Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 96.Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 97.Ballantyne S, Daniel DL, Jr, Wickens M. A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol Biol Cell. 1997;8:1633–1648. doi: 10.1091/mbc.8.8.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barkoff AF, Dickson KS, Gray NK, Wickens M. Translational control of cyclin B1 mRNA during meiotic maturation: coordinated repression and cytoplasmic polyadenylation. Dev Biol. 2000;220:97–109. doi: 10.1006/dbio.2000.9613. [DOI] [PubMed] [Google Scholar]
- 99.Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 100.Igea A, Mendez R. Meiosis requires a translational positive loop where CPEB1 ensues its replacement by CPEB4. EMBO J. 2010;29:2182–2193. doi: 10.1038/emboj.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mendez R, Barnard D, Richter JD. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J. 2002;21:1833–1844. doi: 10.1093/emboj/21.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011;25:755–766. doi: 10.1101/gad.2028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25:2792–2801. doi: 10.1038/sj.emboj.7601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McGrew LL, Richter JD. Translational control by cytoplasmic polyadenylation during Xenopus oocyte maturation: Characterization of cis and trans elements and regulation by cyclin/MPF. EMBO J. 1990;9:3743–3751. doi: 10.1002/j.1460-2075.1990.tb07587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Vishnu MR, Sumaroka M, Klein PS, Liebhaber SA. The poly(rC)-binding protein αCP2 is a noncanonical factor in X. laevis cytoplasmic polyadenylation. RNA. 2011;17:944–956. doi: 10.1261/rna.2587411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Simon R, Tassan J-P, Richter JD. Translational control by poly(A) elongation during Xenopus development: differential repression and enhancement by a novel cytoplasmic polyadenylation element. Genes Dev. 1992;6:2580–2591. doi: 10.1101/gad.6.12b.2580. [DOI] [PubMed] [Google Scholar]
- 107.Paris J, Richter JD. Maturation-specific polyadenylation and translational control: diversity of cytoplasmic polyadenylation elements, influence of poly(A) tail size, and formation of stable polyadenylation complexes. Mol Cell Biol. 1990;10:5634–5645. doi: 10.1128/mcb.10.11.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- 109.Tay J, Hodgman R, Richter JD. The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev Biol. 2000;221:1–9. doi: 10.1006/dbio.2000.9669. [DOI] [PubMed] [Google Scholar]
- 110.Oruganty-Das A, Ng T, Udagawa T, Goh EL, Richter JD. Translational control of mitochondrial energy production mediates neuron morphogenesis. Cell Metab. 2012;16:789–800. doi: 10.1016/j.cmet.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- 112.Novoa I, Gallego J, Ferreira PG, Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol. 2010 doi: 10.1038/ncb2046. [DOI] [PubMed] [Google Scholar]
- 113.Huang YS, Kan MC, Lin CL, Richter JD. CPEB3 and CPEB4 in neurons: analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. EMBO J. 2006;25:4865–4876. doi: 10.1038/sj.emboj.7601322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Charlesworth A, Ridge JA, King LA, MacNicol MC, MacNicol AM. A novel regulatory element determines the timing of Mos mRNA translation during Xenopus oocyte maturation. EMBO J. 2002;21:2798–2806. doi: 10.1093/emboj/21.11.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Arumugam K, Wang Y, Hardy LL, MacNicol MC, MacNicol AM. Enforcing temporal control of maternal mRNA translation during oocyte cell-cycle progression. EMBO J. 2009;29:387–397. doi: 10.1038/emboj.2009.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Charlesworth A, Cox LL, MacNicol AM. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J Biol Chem. 2004;279:17650–17659. doi: 10.1074/jbc.M313837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Simon R, Wu L, Richter JD. Cytoplasmic polyadenylation of activin receptor mRNA and the control of pattern formation in Xenopus development. Dev Biol. 1996;179:239–250. doi: 10.1006/dbio.1996.0254. [DOI] [PubMed] [Google Scholar]
- 119.Wu L, Good PJ, Richter JD. The 36-kilodalton embryonic-type cytoplasmic polyadenylation element- binding protein in Xenopus laevis is ElrA, a member of the ELAV family of RNA-binding proteins. Mol Cell Biol. 1997;17:6402–6409. doi: 10.1128/mcb.17.11.6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Campbell ZT, Bhimsaria D, Valley CT, Rodriguez-Martinez JA, Menichelli E, Williamson JR, Ansari AZ, Wickens M. Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity. Cell Rep. 2012;1:570–581. doi: 10.1016/j.celrep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Campbell ZT, Menichelli E, Friend K, Wu J, Kimble J, Williamson JR, Wickens M. Identification of a conserved interface between PUF and CPEB proteins. J Biol Chem. 2012;287:18854–18862. doi: 10.1074/jbc.M112.352815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Suh N, Crittenden SL, Goldstrohm A, Hook B, Thompson B, Wickens M, Kimble J. FBF and its dual control of gld-1 expression in the Caenorhabditis elegans germline. Genetics. 2009;181:1249–1260. doi: 10.1534/genetics.108.099440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Schmid M, Kuchler B, Eckmann CR. Two conserved regulatory cytoplasmic poly(A) polymerases, GLD-4 and GLD-2, regulate meiotic progression in C. elegans. Genes Dev. 2009;23:824–836. doi: 10.1101/gad.494009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Suh N, Jedamzik B, Eckmann CR, Wickens M, Kimble J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc Natl Acad Sci USA. 2006;103:15108–15112. doi: 10.1073/pnas.0607050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jenkins HT, Malkova B, Edwards TA. Kinked β-strands mediate high-affinity recognition of mRNA targets by the germ-cell regulator DAZL. Proc Natl Acad Sci USA. 2011;108:18266–18271. doi: 10.1073/pnas.1105211108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Padmanabhan K, Richter JD. Regulated Pumilio-2 binding controls RINGO/Spy mRNA translation and CPEB activation. Genes Dev. 2006;20:199–209. doi: 10.1101/gad.1383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Collier B, Gorgoni B, Loveridge C, Cooke HJ, Gray NK. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moore FL, Jaruzelska J, Fox MS, Urano J, Firpo MT, Turek PJ, Dorfman DM, Pera RA. Human Pumilio-2 is expressed in embryonic stem cells and germ cells and interacts with DAZ (Deleted in AZoospermia) and DAZ-like proteins. Proc Natl Acad Sci USA. 2003;100:538–543. doi: 10.1073/pnas.0234478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nakahata S, Kawamoto S. Tissue-dependent isoforms of mammalian Fox-1 homologs are associated with tissue-specific splicing activities. Nucleic Acids Res. 2005;33:2078–2089. doi: 10.1093/nar/gki338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Papin C, Rouget C, Mandart E. Xenopus Rbm9 is a novel interactor of XGld2 in the cytoplasmic polyadenylation complex. FEBS J. 2008;275:490–503. doi: 10.1111/j.1742-4658.2007.06216.x. [DOI] [PubMed] [Google Scholar]
- 132.Didiot MC, Tian Z, Schaeffer C, Subramanian M, Mandel JL, Moine H. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008;36:4902–4912. doi: 10.1093/nar/gkn472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zalfa F, Adinolfi S, Napoli I, Kuhn-Holsken E, Urlaub H, Achsel T, Pastore A, Bagni C. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J Biol Chem. 2005;280:33403–33410. doi: 10.1074/jbc.M504286200. [DOI] [PubMed] [Google Scholar]
- 134.Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ascano M, Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kwak JE, Drier E, Barbee SA, Ramaswami M, Yin JC, Wickens M. GLD2 poly(A) polymerase is required for long-term memory. Proc Natl Acad Sci USA. 2008;105:14644–14649. doi: 10.1073/pnas.0803185105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Costa A, Wang Y, Dockendorff TC, Erdjument-Bromage H, Tempst P, Schedl P, Jongens TA. The Drosophila fragile X protein functions as a negative regulator in the orb autoregulatory pathway. Dev Cell. 2005;8:331–342. doi: 10.1016/j.devcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 138.Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci USA. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Benoit P, Papin C, Kwak JE, Wickens M, Simonelig M. PAP- and GLD-2-type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development. 2008;135:1969–1979. doi: 10.1242/dev.021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gebauer F, Richter JD. Cloning and characterization of a Xenopus poly(A) polymerase. Mol Cell Biol. 1995;15:1422–1430. doi: 10.1128/mcb.15.3.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bilger A, Fox CA, Wahle E, Wickens M. Nuclear polyadenylation factors recognize cytoplasmic polyadenylation elements. Genes Dev. 1994;8:1106–1116. doi: 10.1101/gad.8.9.1106. [DOI] [PubMed] [Google Scholar]
- 142.Zhao WQ, Manley JL. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol Cell Biol. 1996;16:2378–2386. doi: 10.1128/mcb.16.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nakanishi T, Kubota H, Ishibashi N, Kumagai S, Watanabe H, Yamashita M, Kashiwabara S, Miyado K, Baba T. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev Biol. 2006;289:115–126. doi: 10.1016/j.ydbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 144.Nakanishi T, Kumagai S, Kimura M, Watanabe H, Sakurai T, Kimura M, Kashiwabara S, Baba T. Disruption of mouse poly(A) polymerase mGLD-2 does not alter polyadenylation status in oocytes and somatic cells. Biochem Biophys Res Commun. 2007;364:14–19. doi: 10.1016/j.bbrc.2007.09.096. [DOI] [PubMed] [Google Scholar]
- 145.Burns DM, D'Ambrogio A, Nottrott S, Richter JD. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011;473:105–108. doi: 10.1038/nature09908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Choi HS, Lee SH, Kim H, Lee Y. Germ cell-specific gene 1 targets testis-specific poly(A) polymerase to the endoplasmic reticulum through protein-protein interactions. FEBS Lett. 2008;582:1203–1209. doi: 10.1016/j.febslet.2008.01.065. [DOI] [PubMed] [Google Scholar]
- 147.Kashiwabara S, Zhuang T, Yamagata K, Noguchi J, Fukamizu A, Baba T. Identification of a novel isoform of poly(A) polymerase, TPAP, specifically present in the cytoplasm of spermatogenic cells. Dev Biol. 2000;228:106–115. doi: 10.1006/dbio.2000.9894. [DOI] [PubMed] [Google Scholar]
- 148.Kashiwabara S, Noguchi J, Zhuang T, Ohmura K, Honda A, Sugiura S, Miyamoto K, Takahashi S, Inoue K, Ogura A, et al. Regulation of spermatogenesis by testis-specific, cytoplasmic poly(A) polymerase TPAP. Science. 2002;298:1999–2002. doi: 10.1126/science.1074632. [DOI] [PubMed] [Google Scholar]
- 149.Kashiwabara SI, Nakanishi T, Kimura M, Baba T. Non-canonical poly(A) polymerase in mammalian gametogenesis. Biochim Biophys Acta. 2008;1779:230–238. doi: 10.1016/j.bbagrm.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 150.Zhuang T, Kashiwabara S, Noguchi J, Baba T. Transgenic expression of testis-specific poly(A) polymerase TPAP in wild-type and TPAP-deficient mice. J Reprod Dev. 2004;50:207–213. doi: 10.1262/jrd.50.207. [DOI] [PubMed] [Google Scholar]
- 151.Fernandez-Miranda G, Mendez R. The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res Rev. 2012;11:460–472. doi: 10.1016/j.arr.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 152.Wang XP, Cooper NG. Comparative in silico analyses of cpeb1-4 with functional predictions. Bioinform Biol Insights. 2010;4:61–83. doi: 10.4137/bbi.s5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hake LE, Mendez R, Richter JD. Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol Cell Biol. 1998;18:685–693. doi: 10.1128/mcb.18.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Stebbins-Boaz B, Hake LE, Richter JD. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J. 1996;15:2582–2592. [PMC free article] [PubMed] [Google Scholar]
- 155.Groisman I, Huang YS, Mendez R, Cao Q, Richter JD. Translational control of embryonic cell division by CPEB and maskin. Cold Spring Harb Symp Quant Biol. 2001;66:345–351. doi: 10.1101/sqb.2001.66.345. [DOI] [PubMed] [Google Scholar]
- 156.Tay J, Richter JD. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell. 2001;1:201–213. doi: 10.1016/s1534-5807(01)00025-9. [DOI] [PubMed] [Google Scholar]
- 157.Castagnetti S, Ephrussi A. Orb and a long poly(A) tail are required for efficient oskar translation at the posterior pole of the Drosophila oocyte. Development. 2003;130:835–843. doi: 10.1242/dev.00309. [DOI] [PubMed] [Google Scholar]
- 158.Jin SW, Arno N, Cohen A, Shah A, Xu Q, Chen N, Ellis RE. In Caenorhabditis elegans, the RNA-binding domains of the cytoplasmic polyadenylation element binding protein FOG-1 are needed to regulate germ cell fates. Genetics. 2001;159:1617–1630. doi: 10.1093/genetics/159.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Luitjens C, Gallegos M, Kraemer B, Kimble J, Wickens M. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 2000;14:2596–2609. doi: 10.1101/gad.831700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chang JS, Tan L, Schedl P. The Drosophila CPEB homolog, orb, is required for oskar protein expression in oocytes. Dev Biol. 1999;215:91–106. doi: 10.1006/dbio.1999.9444. [DOI] [PubMed] [Google Scholar]
- 161.Juge F, Zaessinger S, Temme C, Wahle E, Simonelig M. Control of poly(A) polymerase level is essential to cytoplasmic polyadenylation and early development in Drosophila. EMBO J. 2002;21:6603–6613. doi: 10.1093/emboj/cdf633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dickson KS, Bilger A, Ballantyne S, Wickens MP. The cleavage and polyadenylation specificity factor in Xenopus laevis oocytes is a cytoplasmic factor involved in regulated polyadenylation. Mol Cell Biol. 1999;19:5707–5717. doi: 10.1128/mcb.19.8.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 164.van EttenJ, Schagat TL, Hrit J, Weidmann C, Brumbaugh J, Coon JJ, Goldstrohm AC. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem. 2012;287:36370–36383. doi: 10.1074/jbc.M112.373522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Friend K, Brook M, Bezirci FB, Sheets MD, Gray NK, Seli E. Embryonic poly(A)-binding protein (ePAB) phosphorylation is required for Xenopus oocyte maturation. Biochem J. 2012;445:93–100. doi: 10.1042/BJ20120304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell. 2000;6:1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- 167.Barnard DC, Cao Q, Richter JD. Differential phosphorylation controls Maskin association with eukaryotic translation initiation factor 4E and localization on the mitotic apparatus. Mol Cell Biol. 2005;25:7605–7615. doi: 10.1128/MCB.25.17.7605-7615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kim JH, Richter JD. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev. 2007;21:2571–2579. doi: 10.1101/gad.1593007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cao Q, Richter JD. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 2002;21:3852–3862. doi: 10.1093/emboj/cdf353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hägele S, Kuhn U, Boning M, Katschinski DM. Cytoplasmic polyadenylation-element-binding protein (CPEB)1 and 2 bind to the HIF-1α mRNA 3′-UTR and modulate HIF-1α protein expression. Biochem J. 2009;417:235–246. doi: 10.1042/BJ20081353. [DOI] [PubMed] [Google Scholar]
- 171.Nairismagi ML, Vislovukh A, Meng Q, Kratassiouk G, Beldiman C, Petretich M, Groisman R, Fuchtbauer EM, Harel-Bellan A, Groisman I. Translational control of TWIST1 expression in MCF-10A cell lines recapitulating breast cancer progression. Oncogene. 2012;13:4960–4966. doi: 10.1038/onc.2011.650. [DOI] [PubMed] [Google Scholar]
- 172.Dickson KS, Thompson SR, Gray NK, Wickens M. Poly(A) polymerase and the regulation of cytoplasmic polyadenylation. J Biol Chem. 2001;276:41810–41816. doi: 10.1074/jbc.M103030200. [DOI] [PubMed] [Google Scholar]
- 173.Nakahata S, Kotani T, Mita K, Kawasaki T, Katsu Y, Nagahama Y, Yamashita M. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech Dev. 2003;120:865–880. doi: 10.1016/s0925-4773(03)00160-6. [DOI] [PubMed] [Google Scholar]
- 174.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development. 1997;124:3015–3023. doi: 10.1242/dev.124.15.3015. [DOI] [PubMed] [Google Scholar]
- 176.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 177.Hodgman R, Tay J, Mendez R, Richter JD. CPEB phosphorylation and cytoplasmic polyadenylation are catalyzed by the kinase IAK1/Eg2 in maturing mouse oocytes. Development. 2001;128:2815–2822. doi: 10.1242/dev.128.14.2815. [DOI] [PubMed] [Google Scholar]
- 178.Tay J, Hodgman R, Sarkissian M, Richter JD. Regulated CPEB phosphorylation during meiotic progression suggests a mechanism for temporal control of maternal mRNA translation. Genes Dev. 2003;17:1457–1462. doi: 10.1101/gad.1071403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Keady BT, Kuo P, Martinez SE, Yuan L, Hake LE. MAPK interacts with XGef and is required for CPEB activation during meiosis in Xenopus oocytes. J Cell Sci. 2007;120:1093–1103. doi: 10.1242/jcs.03416. [DOI] [PubMed] [Google Scholar]
- 180.Kuo P, Runge E, Lu X, Hake LE. XGef influences XRINGO/CDK1 signaling and CPEB activation during Xenopus oocyte maturation. Differentiation. 2010;81:133–140. doi: 10.1016/j.diff.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 181.Frank-Vaillant M, Haccard O, Thibier C, Ozon R, Arlot-Bonnemains Y, Prigent C, Jessus C. Progesterone regulates the accumulation and the activation of Eg2 kinase in Xenopus oocytes. J Cell Sci. 2000;113:1127–1138. doi: 10.1242/jcs.113.7.1127. [DOI] [PubMed] [Google Scholar]
- 182.Ma C, Cummings C, Liu XJ. Biphasic activation of Aurora-A kinase during the meiosis I- meiosis II transition in Xenopus oocytes. Mol Cell Biol. 2003;23:1703–1716. doi: 10.1128/MCB.23.5.1703-1716.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Maton G, Thibier C, Castro A, Lorca T, Prigent C, Jessus C. Cdc2-cyclin B triggers H3 kinase activation of Aurora-A in Xenopus oocytes. J Biol Chem. 2003;278:21439–21449. doi: 10.1074/jbc.M300811200. [DOI] [PubMed] [Google Scholar]
- 184.Wong LC, Costa A, McLeod I, Sarkeshik A, Yates J, III, Kyin S, Perlman D, Schedl P. The functioning of the Drosophila CPEB protein Orb is regulated by phosphorylation and requires casein kinase 2 activity. PLoS One. 2011;6:e24355. doi: 10.1371/journal.pone.0024355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Vardy L, Pesin JA, Orr-Weaver TL. Regulation of Cyclin A protein in meiosis and early embryogenesis. Proc Natl Acad Sci USA. 2009;106:1838–1843. doi: 10.1073/pnas.0813237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Bouvet P, Omilli F, Arlot-Bonnemains Y, Legagneux V, Roghi C, Bassez T, Osborne HB. The deadenylation conferred by the 3′ untranslated region of a developmentally controlled mRNA in Xenopus embryos is switched to polyadenylation by deletion of a short sequence element. Mol Cell Biol. 1994;14:1893–1900. doi: 10.1128/mcb.14.3.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rouhana L, Wickens M. Autoregulation of GLD-2 cytoplasmic poly(A) polymerase. RNA. 2007;13:188–199. doi: 10.1261/rna.333507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tan L, Chang JS, Costa A, Schedl P. An autoregulatory feedback loop directs the localized expression of the Drosophila CPEB protein Orb in the developing oocyte. Development. 2001;128:1159–1169. doi: 10.1242/dev.128.7.1159. [DOI] [PubMed] [Google Scholar]
- 189.Wong LC, Schedl P. Cup blocks the precocious activation of the orb autoregulatory loop. PLoS One. 2011;6:e28261. doi: 10.1371/journal.pone.0028261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Reverte CG, Ahearn MD, Hake LE. CPEB degradation during Xenopus oocyte maturation requires a PEST domain and the 26S proteasome. Dev Biol. 2001;231:447–458. doi: 10.1006/dbio.2001.0153. [DOI] [PubMed] [Google Scholar]
- 191.Setoyama D, Yamashita M, Sagata N. Mechanism of degradation of CPEB during Xenopus oocyte maturation. Proc Natl Acad Sci USA. 2007;104:18001–18006. doi: 10.1073/pnas.0706952104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lin CL, Huang YT, Richter JD. Transient CPEB dimerization and translational control. RNA. 2012;18:1050–1061. doi: 10.1261/rna.031682.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sagata N, Oskarsson M, Copeland T, Brumbaugh J, Vande Woude GF. Function of c-mos proto-oncogene product in meiotic maturation in Xenopus oocytes. Nature. 1988;335:519–525. doi: 10.1038/335519a0. [DOI] [PubMed] [Google Scholar]
- 194.Sheets MD, Wu M, Wickens M. Polyadenylation of c-mos mRNA as a control point in Xenopus meiotic maturation. Nature. 1995;374:511–516. doi: 10.1038/374511a0. [DOI] [PubMed] [Google Scholar]
- 195.De Moor CH, Richter JD. The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol Cell Biol. 1997;17:6419–6426. doi: 10.1128/mcb.17.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Belloc E, Pique M, Mendez R. Sequential waves of polyadenylation and deadenylation define a translation circuit that drives meiotic progression. Biochem Soc Trans. 2008;36:665–670. doi: 10.1042/BST0360665. [DOI] [PubMed] [Google Scholar]
- 197.Racki WJ, Richter JD. CPEB controls oocyte growth and follicle development in the mouse. Development. 2006;133:4527–4537. doi: 10.1242/dev.02651. [DOI] [PubMed] [Google Scholar]
- 198.Gebauer F, Xu W, Cooper GM, Richter JD. Translational control by cytoplasmic polyadenylation of c-mos mRNA is necessary for oocyte maturation in the mouse. EMBO J. 1994;13:5712–5720. doi: 10.1002/j.1460-2075.1994.tb06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Seli E, Lalioti MD, Flaherty SM, Sakkas D, Terzi N, Steitz JA. An embryonic poly(A)-binding protein (ePAB) is expressed in mouse oocytes and early preimplantation embryos. Proc Natl Acad Sci USA. 2005;102:367–372. doi: 10.1073/pnas.0408378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Guzeloglu-Kayisli O, Pauli S, Demir H, Lalioti MD, Sakkas D, Seli E. Identification and characterization of human embryonic poly(A) binding protein (EPAB) Mol Hum Reprod. 2008;14:581–588. doi: 10.1093/molehr/gan047. [DOI] [PubMed] [Google Scholar]
- 201.Guzeloglu-Kayisli O, Lalioti MD, Aydiner F, Sasson I, Ilbay O, Sakkas D, Lowther KM, Mehlmann LM, Seli E. Embryonic poly(A)-binding protein (EPAB) is required for oocyte maturation and female fertility in mice. Biochem J. 2012;446:47–58. doi: 10.1042/BJ20120467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wang H, Li Y, Wang Y, Han ZG, Cai B. C9orf100, a new member of the Dbl-family guanine nucleotide exchange factors, promotes cell proliferation and migration in hepatocellular carcinoma. Mol Med Report. 2012;5:1169–1174. doi: 10.3892/mmr.2012.783. [DOI] [PubMed] [Google Scholar]
- 203.Reverte CG, Yuan L, Keady BT, Lacza C, Attfield KR, Mahon GM, Freeman B, Whitehead IP, Hake LE. XGef is a CPEB-interacting protein involved in Xenopus oocyte maturation. Dev Biol. 2003;255:383–398. doi: 10.1016/s0012-1606(02)00089-1. [DOI] [PubMed] [Google Scholar]
- 204.Martinez SE, Yuan L, Lacza C, Ransom H, Mahon GM, Whitehead IP, Hake LE. XGef mediates early CPEB phosphorylation during Xenopus oocyte meiotic maturation. Mol Biol Cell. 2005;16:1152–1164. doi: 10.1091/mbc.E04-07-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ferby I, Blazquez M, Palmer A, Eritja R, Nebreda AR. A novel p34(cdc2)-binding and activating protein that is necessary and sufficient to trigger G(2)/M progression in Xenopus oocytes. Genes Dev. 1999;13:2177–2189. doi: 10.1101/gad.13.16.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, Ueda S, Uchiyama Y, Noda T, Okano H. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci USA. 2002;99:15194–15199. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Gunter KM, McLaughlin EA. Translational control in germ cell development: a role for the RNA-binding proteins Musashi-1 and Musashi-2. IUBMB Life. 2011;63:678–685. doi: 10.1002/iub.499. [DOI] [PubMed] [Google Scholar]
- 208.Wu X, Viveiros MM, Eppig JJ, Bai Y, Fitzpatrick SL, Matzuk MM. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet. 2003;33:187–191. doi: 10.1038/ng1079. [DOI] [PubMed] [Google Scholar]
- 209.Hu J, Wang F, Zhu X, Yuan Y, Ding M, Gao S. Mouse ZAR1-like (XM_359149) colocalizes with mRNA processing components and its dominant-negative mutant caused two-cell-stage embryonic arrest. Dev Dyn. 2010;239:407–424. doi: 10.1002/dvdy.22170. [DOI] [PubMed] [Google Scholar]
- 210.Brook M, Smith JW, Gray NK. The DAZL and PABP families: RNA-binding proteins with interrelated roles in translational control in oocytes. Reproduction. 2009;137:595–617. doi: 10.1530/REP-08-0524. [DOI] [PubMed] [Google Scholar]
- 211.Reynolds N, Collier B, Bingham V, Gray NK, Cooke HJ. Translation of the synaptonemal complex component Sycp3 is enhanced in vivo by the germ cell specific regulator Dazl. RNA. 2007;13:974–981. doi: 10.1261/rna.465507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kosaka K, Kawakami K, Sakamoto H, Inoue K. Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis. Mech Dev. 2007;124:279–289. doi: 10.1016/j.mod.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 213.Wiszniak SE, Dredge BK, Jensen KB. HuB (elavl2) mRNA is restricted to the germ cells by post-transcriptional mechanisms including stabilisation of the message by DAZL. PLoS One. 2011;6:e20773. doi: 10.1371/journal.pone.0020773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Zeng M, Lu Y, Liao X, Li D, Sun H, Liang S, Zhang S, Ma Y, Yang Z. DAZL binds to 3′UTR of Tex19.1 mRNAs and regulates Tex19.1 expression. Mol Biol Rep. 2009;36:2399–2403. doi: 10.1007/s11033-009-9470-1. [DOI] [PubMed] [Google Scholar]
- 215.Osborne HB, Duval C, Ghoda L, Omilli F, Bassez T, Coffino P. Expression and post-transcriptional regulation of ornithine decarboxylase during early Xenopus development. Eur J Biochem. 1991;202:575–581. doi: 10.1111/j.1432-1033.1991.tb16410.x. [DOI] [PubMed] [Google Scholar]
- 216.Slevin MK, Gourronc F, Hartley RS. ElrA binding to the 3′UTR of cyclin E1 mRNA requires polyadenylation elements. Nucleic Acids Res. 2007;35:2167–2176. doi: 10.1093/nar/gkm084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Paillard L, Maniey D, Lachaume P, Legagneux V, Osborne HB. Identification of a C-rich element as a novel cytoplasmic polyadenylation element in Xenopus embryos. Mech Dev. 2000;93:117–125. doi: 10.1016/s0925-4773(00)00279-3. [DOI] [PubMed] [Google Scholar]
- 218.Gravina P, Campioni N, Loreni F, Pierandrei-Amaldi P, Cardinali B. Complementary DNA analysis, expression and subcellular localization of hnRNP E2 gene in Xenopus laevis. Gene. 2002;290:193–201. doi: 10.1016/s0378-1119(02)00561-9. [DOI] [PubMed] [Google Scholar]
- 219.Kim KW, Nykamp K, Suh N, Bachorik JL, Wang L, Kimble J. Antagonism between GLD-2 binding partners controls gamete sex. Dev Cell. 2009;16:723–733. doi: 10.1016/j.devcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Eckmann CR, Crittenden SL, Suh N, Kimble J. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics. 2004;168:147–160. doi: 10.1534/genetics.104.029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Eckmann CR, Kraemer B, Wickens M, Kimble J. GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev Cell. 2002;3:697–710. doi: 10.1016/s1534-5807(02)00322-2. [DOI] [PubMed] [Google Scholar]
- 222.Nakel K, Hartung SA, Bonneau F, Eckmann CR, Conti E. Four KH domains of the C. elegans bicaudal-C ortholog GLD-3 form a globular structural platform. RNA. 2010;16:2058–2067. doi: 10.1261/rna.2315010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko P. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev Cell. 2007;13:691–704. doi: 10.1016/j.devcel.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 224.Rybarska A, Harterink M, Jedamzik B, Kupinski AP, Schmid M, Eckmann CR. GLS-1, a novel P granule component, modulates a network of conserved RNA regulators to influence germ cell fate decisions. PLoS Genet. 2009;5:e1000494. doi: 10.1371/journal.pgen.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Weiss J, Hurley LA, Harris RM, Finlayson C, Tong M, Fisher LA, Moran JL, Beier DR, Mason C, Jameson JL. ENU mutagenesis in mice identifies candidate genes for hypogonadism. Mamm Genome. 2012;23:346–355. doi: 10.1007/s00335-011-9388-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Coll O, Villalba A, Bussotti G, Notredame C, Gebauer F. A novel, noncanonical mechanism of cytoplasmic polyadenylation operates in Drosophila embryogenesis. Genes Dev. 2010;24:129–134. doi: 10.1101/gad.568610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Brawerman G, Diez J. Metabolism of the polyadenylate sequence of nuclear RNA and messenger RNA in mammalian cells. Cell. 1975;5:271–280. doi: 10.1016/0092-8674(75)90102-6. [DOI] [PubMed] [Google Scholar]
- 228.Watanabe T, Yachi K, Ohta T, Fukushima T, Yoshino A, Katayama Y, Shinojima Y, Terui T, Nagase H. Non-promoter hypermethylation of zygote arrest 1 (ZAR1) in human brain tumors. Brain Tumor Pathol. 2011;28:199–202. doi: 10.1007/s10014-011-0019-3. [DOI] [PubMed] [Google Scholar]
- 229.Shinojima Y, Terui T, Hara H, Kimura M, Igarashi J, Wang X, Kawashima H, Kobayashi Y, Muroi S, Hayakawa S, et al. Identification and analysis of an early diagnostic marker for malignant melanoma: ZAR1 intra-genic differential methylation. J Dermatol Sci. 2010;59:98–106. doi: 10.1016/j.jdermsci.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ezeh UI, Turek PJ, Reijo RA, Clark AT. Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer. 2005;104:2255–2265. doi: 10.1002/cncr.21432. [DOI] [PubMed] [Google Scholar]
- 231.Du L, Richter JD. Activity-dependent polyadenylation in neurons. RNA. 2005;11:1340–1347. doi: 10.1261/rna.2870505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Huang YS, Jung MY, Sarkissian M, Richter JD. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and α CaMKII mRNA polyadenylation at synapses. EMBO J. 2002;21:2139–2148. doi: 10.1093/emboj/21.9.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, Richter JD, Fallon JR. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J Neurosci. 2001;21:9541–9548. doi: 10.1523/JNEUROSCI.21-24-09541.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron. 1998;21:1129–1139. doi: 10.1016/s0896-6273(00)80630-3. [DOI] [PubMed] [Google Scholar]
- 235.Zearfoss NR, Alarcon JM, Trifilieff P, Kandel E, Richter JD. A molecular circuit composed of CPEB-1 and c-Jun controls growth hormone-mediated synaptic plasticity in the mouse hippocampus. J Neurosci. 2008;28:8502–8509. doi: 10.1523/JNEUROSCI.1756-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003;17:638–653. doi: 10.1101/gad.1053003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Bestman JE, Cline HT. The relationship between dendritic branch dynamics and CPEB-labeled RNP granules captured in vivo. Front Neural Circuits. 2009;3:10. doi: 10.3389/neuro.04.010.2009. (Epub ahead of print, September 1, 2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Bestman JE, Cline HT. The RNA binding protein CPEB regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proc Natl Acad Sci USA. 2008;105:20494–20499. doi: 10.1073/pnas.0806296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Atkins CM, Davare MA, Oh MC, Derkach V, Soderling TR. Bidirectional regulation of cytoplasmic polyadenylation element-binding protein phosphorylation by Ca2+/calmodulin-dependent protein kinase II and protein phosphatase 1 during hippocampal long-term potentiation. J Neurosci. 2005;25:5604–5610. doi: 10.1523/JNEUROSCI.5051-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Atkins CM, Nozaki N, Shigeri Y, Soderling TR. Cytoplasmic polyadenylation element binding protein-dependent protein synthesis is regulated by calcium/calmodulin-dependent protein kinase II. J Neurosci. 2004;24:5193–5201. doi: 10.1523/JNEUROSCI.0854-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell. 2012;47:253–266. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Aslam N, Shouval HZ. Regulation of cytoplasmic polyadenylation can generate a bistable switch. BMC Syst Biol. 2012;6:12. doi: 10.1186/1752-0509-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- 244.Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. doi: 10.1016/s0092-8674(03)01020-1. [DOI] [PubMed] [Google Scholar]
- 245.Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 246.Kruttner S, Stepien B, Noordermeer JN, Mommaas MA, Mechtler K, Dickson BJ, Keleman K. Drosophila CPEB Orb2A mediates memory independent of Its RNA-binding domain. Neuron. 2012;76:383–395. doi: 10.1016/j.neuron.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kondrashov A, Meijer HA, Barthet-Barateig A, Parker HN, Khurshid A, Tessier S, Sicard M, Knox AJ, Pang L, De Moor CH. Inhibition of polyadenylation reduces inflammatory gene induction. RNA. 2012;18:2236–2250. doi: 10.1261/rna.032391.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wong YY, Moon A, Duffin R, Barthet-Barateig A, Meijer HA, Clemens MJ, De Moor CH. Cordycepin inhibits protein synthesis and cell adhesion through effects on signal transduction. J Biol Chem. 2010;285:2610–2621. doi: 10.1074/jbc.M109.071159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hafer N, Xu S, Bhat KM, Schedl P. The Drosophila CPEB protein Orb2 has a novel expression pattern and is important for asymmetric cell division and nervous system function. Genetics. 2011;189:907–921. doi: 10.1534/genetics.110.123646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Keleman K, Kruttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci. 2007;10:1587–1593. doi: 10.1038/nn1996. [DOI] [PubMed] [Google Scholar]
- 251.Jones KJ, Korb E, Kundel MA, Kochanek AR, Kabraji S, McEvoy M, Shin CY, Wells DG. CPEB1 regulates β-catenin mRNA translation and cell migration in astrocytes. Glia. 2008;56:1401–1413. doi: 10.1002/glia.20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Kundel M, Jones KJ, Shin CY, Wells DG. Cytoplasmic polyadenylation element-binding protein regulates neurotrophin-3-dependent β-catenin mRNA translation in developing hippocampal neurons. J Neurosci. 2009;29:13630–13639. doi: 10.1523/JNEUROSCI.2910-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.McEvoy M, Cao G, Llopis PM, Kundel M, Jones K, Hofler C, Shin C, Wells DG. Cytoplasmic polyadenylation element binding protein 1-mediated mRNA translation in Purkinje neurons is required for cerebellar long-term depression and motor coordination. J Neurosci. 2007;27:6400–6411. doi: 10.1523/JNEUROSCI.5211-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Vessey JP, Schoderboeck L, Gingl E, Luzi E, Riefler J, Di LF, Karra D, Thomas S, Kiebler MA, Macchi P. Mammalian Pumilio 2 regulates dendrite morphogenesis and synaptic function. Proc Natl Acad Sci USA. 2010;107:3222–3227. doi: 10.1073/pnas.0907128107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Kuo MW, Wang SH, Chang JC, Chang CH, Huang LJ, Lin HH, Yu AL, Li WH, Yu J. A novel puf-A gene predicted from evolutionary analysis is involved in the development of eyes and primordial germ-cells. PLoS One. 2009;4:e4980. doi: 10.1371/journal.pone.0004980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Menon KP, Andrews S, Murthy M, Gavis ER, Zinn K. The translational repressors Nanos and Pumilio have divergent effects on presynaptic terminal growth and postsynaptic glutamate receptor subunit composition. J Neurosci. 2009;29:5558–5572. doi: 10.1523/JNEUROSCI.0520-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Chen G, Li W, Zhang QS, Regulski M, Sinha N, Barditch J, Tully T, Krainer AR, Zhang MQ, Dubnau J. Identification of synaptic targets of Drosophila pumilio. PLoS Comput Biol. 2008;4:e1000026. doi: 10.1371/journal.pcbi.1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Baines RA. Neuronal homeostasis through translational control. Mol Neurobiol. 2005;32:113–121. doi: 10.1385/MN:32:2:113. [DOI] [PubMed] [Google Scholar]
- 259.Zhong J, Zhang T, Bloch LM. Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neurosci. 2006;7:17. doi: 10.1186/1471-2202-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol. 2004;14:314–321. doi: 10.1016/j.cub.2004.01.052. [DOI] [PubMed] [Google Scholar]
- 261.Alarcon JM, Hodgman R, Theis M, Huang YS, Kandel ER, Richter JD. Selective modulation of some forms of schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn Mem. 2004;11:318–327. doi: 10.1101/lm.72704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Berger-Sweeney J, Zearfoss NR, Richter JD. Reduced extinction of hippocampal-dependent memories in CPEB knockout mice. Learn Mem. 2006;13:4–7. doi: 10.1101/lm.73706. [DOI] [PubMed] [Google Scholar]
- 263.Theis M, Si K, Kandel ER. Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc Natl Acad Sci USA. 2003;100:9602–9607. doi: 10.1073/pnas.1133424100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Pavlopoulos E, Trifilieff P, Chevaleyre V, Fioriti L, Zairis S, Pagano A, Malleret G, Kandel ER. Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell. 2011;147:1369–1383. doi: 10.1016/j.cell.2011.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wang CF, Huang YS. Calpain 2 activated through N-Methyl-D-aspartic acid receptor signaling cleaves CPEB3 and abrogates CPEB3-repressed translation in neurons. Mol Cell Biol. 2012;32:3321–3332. doi: 10.1128/MCB.00296-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Vogler C, Spalek K, Aerni A, Demougin P, Muller A, Huynh KD, Papassotiropoulos A, de Quervain DJ. CPEB3 is associated with human episodic memory. Front Behav Neurosci. 2009;3:4. doi: 10.3389/neuro.08.004.2009. (Epub ahead of print, May 4, 2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hosoda N, Funakoshi Y, Hirasawa M, Yamagishi R, Asano Y, Miyagawa R, Ogami K, Tsujimoto M, Hoshino S. Anti-proliferative protein Tob negatively regulates CPEB3 target by recruiting Caf1 deadenylase. EMBO J. 2011;30:1311–1323. doi: 10.1038/emboj.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sasayama T, Marumoto T, Kunitoku N, Zhang D, Tamaki N, Kohmura E, Saya H, Hirota T. Over-expression of Aurora-A targets cytoplasmic polyadenylation element binding protein and promotes mRNA polyadenylation of Cdk1 and cyclin B1. Genes Cells. 2005;10:627–638. doi: 10.1111/j.1365-2443.2005.00870.x. [DOI] [PubMed] [Google Scholar]
- 269.Cao Q, Kim JH, Richter JD. CDK1 and calcineurin regulate Maskin association with eIF4E and translational control of cell cycle progression. Nat Struct Mol Biol. 2006;13:1128–1134. doi: 10.1038/nsmb1169. [DOI] [PubMed] [Google Scholar]
- 270.Groisman I, Jung MY, Sarkissian M, Cao Q, Richter JD. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/s0092-8674(02)00733-x. [DOI] [PubMed] [Google Scholar]
- 271.Pederson T, Robbins E. RNA synthesis in HeLa cells. Pattern in hypertonic medium and its similarity to synthesis during G2-prophase. J Cell Biol. 1970;47:734–744. doi: 10.1083/jcb.47.3.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Burns DM, Richter JD. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 2008;22:3449–3460. doi: 10.1101/gad.1697808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Groisman I, Ivshina M, Marin V, Kennedy NJ, Davis RJ, Richter JD. Control of cellular senescence by CPEB. Genes Dev. 2006;20:2701–2712. doi: 10.1101/gad.1438906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Berndt H, Harnisch C, Rammelt C, Stohr N, Zirkel A, Dohm JC, Himmelbauer H, Tavanez JP, Huttelmaier S, Wahle E. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012;18:958–972. doi: 10.1261/rna.032292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Lin CL, Evans V, Shen S, Xing Y, Richter JD. The nuclear experience of CPEB: implications for RNA processing and translational control. RNA. 2010;16:338–348. doi: 10.1261/rna.1779810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Kan MC, Oruganty-Das A, Cooper-Morgan A, Jin G, Swanger S, Bassell G, Florman H, Van LK, Richter JD. CPEB4 is a cell survival protein retained in the nucleus upon ischemia or endoplasmic reticulum calcium depletion. Mol Cell Biol. 2010;30:5658–5671. doi: 10.1128/MCB.00716-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ernoult-Lange M, Wilczynska A, Harper M, Aigueperse C, Dautry F, Kress M, Weil D. Nucleocytoplasmic traffic of CPEB1 and accumulation in Crm1 nucleolar bodies. Mol Biol Cell. 2009;20:176–187. doi: 10.1091/mbc.E08-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 2012;26:2724–2736. doi: 10.1101/gad.208306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 280.Clarke RB. Isolation and characterization of human mammary stem cells. Cell Prolif. 2005;38:375–386. doi: 10.1111/j.1365-2184.2005.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Todaro M, Francipane MG, Medema JP, Stassi G. Colon cancer stem cells: promise of targeted therapy. Gastroenterology. 2010;138:2151–2162. doi: 10.1053/j.gastro.2009.12.063. [DOI] [PubMed] [Google Scholar]
- 282.Yen TH, Wright NA. The gastrointestinal tract stem cell niche. Stem Cell Rev. 2006;2:203–212. doi: 10.1007/s12015-006-0048-1. [DOI] [PubMed] [Google Scholar]
- 283.Cervello I, Mirantes C, Santamaria X, Dolcet X, Matias-Guiu X, Simon C. Stem cells in human endometrium and endometrial carcinoma. Int J Gynecol Pathol. 2011;30:317–327. doi: 10.1097/PGP.0b013e3182102754. [DOI] [PubMed] [Google Scholar]
- 284.de Andres-Aguayo L, Varas F, Graf T. Musashi 2 in hematopoiesis. Curr Opin Hematol. 2012;19:268–272. doi: 10.1097/MOH.0b013e328353c778. [DOI] [PubMed] [Google Scholar]
- 285.Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, Okano H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J Cell Biol. 2008;181:639–653. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 287.Kim SY, Kim JY, Malik S, Son W, Kwon KS, Kim C. Negative regulation of EGFR/MAPK pathway by Pumilio in Drosophila melanogaster. PLoS One. 2012;7:e34016. doi: 10.1371/journal.pone.0034016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Bermudez O, Jouandin P, Rottier J, Bourcier C, Pages G, Gimond C. Post-transcriptional regulation of the DUSP6/MKP-3 phosphatase by MEK/ERK signalling and hypoxia. J Cell Physiol. 2010;226:276–284. doi: 10.1002/jcp.22339. [DOI] [PubMed] [Google Scholar]
- 289.Lee MH, Hook B, Pan G, Kershner AM, Merritt C, Seydoux G, Thomson JA, Wickens M, Kimble J. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007;3:e233. doi: 10.1371/journal.pgen.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Bouvrette DJ, Sittaramane V, Heidel JR, Chandrasekhar A, Bryda EC. Knockdown of bicaudal C in zebrafish (Danio rerio) causes cystic kidneys: a nonmammalian model of polycystic kidney disease. Comp Med. 2010;60:96–106. [PMC free article] [PubMed] [Google Scholar]
- 291.Cogswell C, Price SJ, Hou X, Guay-Woodford LM, Flaherty L, Bryda EC. Positional cloning of jcpk/bpk locus of the mouse. Mamm Genome. 2003;14:242–249. doi: 10.1007/s00335-002-2241-0. [DOI] [PubMed] [Google Scholar]
- 292.Kraus MR, Clauin S, Pfister Y, Di MM, Ulinski T, Constam D, Bellanne-Chantelot C, Grapin-Botton A. Two mutations in human BICC1 resulting in Wnt pathway hyperactivity associated with cystic renal dysplasia. Hum Mutat. 2012;33:86–90. doi: 10.1002/humu.21610. [DOI] [PubMed] [Google Scholar]
- 293.Maisonneuve C, Guilleret I, Vick P, Weber T, Andre P, Beyer T, Blum M, Constam DB. Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development. 2009;136:3019–3030. doi: 10.1242/dev.038174. [DOI] [PubMed] [Google Scholar]
- 294.Ryan S, Verghese S, Cianciola NL, Cotton CU, Carlin CR. Autosomal recessive polycystic kidney disease epithelial cell model reveals multiple basolateral epidermal growth factor receptor sorting pathways. Mol Biol Cell. 2010;21:2732–2745. doi: 10.1091/mbc.E09-12-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Fu Y, Kim I, Lian P, Li A, Zhou L, Li C, Liang D, Coffey RJ, Ma J, Zhao P, et al. Loss of Bicc1 impairs tubulomorphogenesis of cultured IMCD cells by disrupting E-cadherin-based cell-cell adhesion. Eur J Cell Biol. 2010;89:428–436. doi: 10.1016/j.ejcb.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Nagaoka K, Udagawa T, Richter JD. CPEB-mediated ZO-1 mRNA localization is required for epithelial tight-junction assembly and cell polarity. Nat Commun. 2012;3:675. doi: 10.1038/ncomms1678. doi: 10.1038/ncomms1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Kiessling F, Kartenbeck J, Haller C. Cell-cell contacts in the human cell line ECV304 exhibit both endothelial and epithelial characteristics. Cell Tissue Res. 1999;297:131–140. doi: 10.1007/s004410051340. [DOI] [PubMed] [Google Scholar]
- 298.Paffenholz R, Kuhn C, Grund C, Stehr S, Franke WW. The arm-repeat protein NPRAP (neurojungin) is a constituent of the plaques of the outer limiting zone in the retina, defining a novel type of adhering junction. Exp Cell Res. 1999;250:452–464. doi: 10.1006/excr.1999.4534. [DOI] [PubMed] [Google Scholar]
- 299.Bouvrette DJ, Price SJ, Bryda EC. K homology domains of the mouse polycystic kidney disease-related protein, Bicaudal-C (Bicc1), mediate RNA binding in vitro. Nephron Exp Nephrol. 2008;108:e27–e34. doi: 10.1159/000112913. [DOI] [PubMed] [Google Scholar]
- 300.Piazzon N, Maisonneuve C, Guilleret I, Rotman S, Constam DB. Bicc1 links the regulation of cAMP signaling in polycystic kidneys to microRNA-induced gene silencing. J Mol Cell Biol. 2012;4:398–408. doi: 10.1093/jmcb/mjs027. [DOI] [PubMed] [Google Scholar]
- 301.Tran U, Zakin L, Schweickert A, Agrawal R, Doger R, Blum M, De Robertis EM, Wessely O. The RNA-binding protein bicaudal C regulates polycystin 2 in the kidney by antagonizing miR-17 activity. Development. 2010;137:1107–1116. doi: 10.1242/dev.046045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.von Roretz C, Di MS, Mazroui R, Gallouzi IE. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdiscip Rev RNA. 2011;2:336–347. doi: 10.1002/wrna.55. [DOI] [PubMed] [Google Scholar]
- 303.Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Ostareck-Lederer A, Ostareck DH. Control of mRNA translation and stability in haematopoietic cells: The function of hnRNPs K and E1/E2. Biol Cell. 2004;96:407–411. doi: 10.1016/j.biolcel.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 305.Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, Liu S, Schwind S, Santhanam R, Hickey CJ, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Ji X, Kong J, Carstens RP, Liebhaber SA. The 3′ untranslated region complex involved in stabilization of human α-globin mRNA assembles in the nucleus and serves an independent role as a splice enhancer. Mol Cell Biol. 2007;27:3290–3302. doi: 10.1128/MCB.02289-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.de Hoog CL, Foster LJ, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–662. doi: 10.1016/s0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- 308.Yang Q, Gilmartin GM, Doublie S. Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc Natl Acad Sci USA. 2010;107:10062–10067. doi: 10.1073/pnas.1000848107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.McPheeters DS, Cremona N, Sunder S, Chen HM, Averbeck N, Leatherwood J, Wise JA. A complex gene regulatory mechanism that operates at the nexus of multiple RNA processing decisions. Nat Struct Mol Biol. 2009;16:255–264. doi: 10.1038/nsmb.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Houseley J, Tollervey D. The nuclear RNA surveillance machinery: the link between ncRNAs and genome structure in budding yeast? Biochim Biophys Acta. 2008;1779:239–246. doi: 10.1016/j.bbagrm.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 311.Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, Suzuki T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010;11:106–111. doi: 10.1038/embor.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]