Abstract
Combinatorial approaches that integrate conventional pathology with genomic profiling and functional genomics have begun to enhance our understanding of the genetic basis of breast cancer. These methods have identified key genotypic-phenotypic correlations in different breast cancer subtypes that have led to the discovery of genetic dependencies that drive their behavior. Moreover, this knowledge has been applied to define novel tailored therapies for these groups of cancer patients. With the current emphasis on characterizing the mutational repertoire of breast cancers by next generation sequencing, the question remains as to what constitutes a driver event. By focusing efforts on homogenous subgroups of breast cancer and integrating orthogonal data-types combined with functional approaches, we can begin to unravel the heterogeneity and identify aberrations that can be therapeutically targeted.
Introduction
Traditionally, breast cancers have been characterized into biologically and clinically meaningful subgroups according to histological grade and type (i.e. growth pattern)(1), with the majority of breast cancers being classified by histological exclusion i.e. invasive carcinomas of no special type (IC-NST). On the other hand the remaining tumors can be histologically classified according to their distinctive growth patterns and are termed ‘special’ histological types. Over the last decade seminal class discovery expression profiling studies have identified a number of molecular subtypes of breast cancer defined at the transcriptomic level that are characterized by distinct histological features, clinical behaviors and responses to therapy(2). Indeed, different breast cancer subtypes (both histological and molecular) harbor distinct patterns of genetic aberrations and are driven by alterations in distinct molecular pathways and networks(3, 4). It is now widely accepted that breast cancer heterogeneity may be underpinned by myriad mechanisms of genetic aberration (e.g. gene amplifications, in-frame fusion genes or mutations and homozygous deletions, disrupting fusions or deleterious mutations causing gene activation or inactivation respectively), and that phenotypic subgroups harbor distinct patterns of genomic aberrations(3, 5). Moreover, targeting these genomic alterations has proven an effective way of developing tailored therapies for subgroups of breast cancers(5-9).
The use of high-throughput technologies has enabled the investigation of biological phenomena and allowed its correlation to specific disease behavior. Research has focused on integrative approaches combining high-throughput genomic data through the use of microarray-based comparative genomic hybridization (aCGH), gene expression profiling and more recently the use of next generation sequencing, to define the genetic underpinning of different subtypes of cancer with the ultimate goal of identifying novel therapeutic targets. However the main challenges for the translation of the genetic alterations identified by massively parallel sequencing into benefit for cancer patients lie in the identification of biologically relevant aberrations among the deluge of sequencing data being produced, which can be used as therapeutic targets or predictive biomarkers.
Exploring genotypic-phenotypic correlations
There is evidence to suggest that at least some subtypes of breast cancer are underpinned by distinct arrays of genomic alterations. In fact some special histological types of breast cancer harbor specific pathognomonic alterations such as the ETV6–NTRK3 oncogenic fusion gene in secretory carcinomas, the MYB-NFIB fusion gene in adenoid cystic carcinomas, and inactivation of E-cadherin through mutation and gene methylation in lobular carcinomas of the breast (for a review on special histological types of breast cancer see(10)). Perhaps the best example in breast cancer is the characterization of ERBB2 (HER2) as the driver of the 17q12 amplification, which has spurred the hunt for additional amplified driver events. We, and others have explored the genotypic–phenotypic correlations of different molecular subgroups of breast cancers through the use of high-throughput genomic analyses using aCGH(3, 5). Through aCGH profiling of a series of 95 high-grade breast cancers, we have shown that distinct patterns of copy number alterations are found in different molecular subtypes(5). These analyses highlighted the genotypic-phenotypic association between specific amplifications and subtypes of breast cancer(3, 4).
Integrating data-types to identify therapeutic targets
By using a combination of aCGH and gene expression profiling, we have shown that canonical pathways involved in estrogen receptor (ER) signaling, proliferation and DNA repair are enriched for genes whose expression is driven by copy number in basal-like, HER2 and luminal tumors(3), suggesting that the diversity of breast cancer and the molecular subtypes may stem, to some degree, from the different patterns of genetic aberrations found in these cancers. Moreover, biological phenomena characteristic of each subtype (e.g. proliferation, HER2 and ER signaling) may be driven by specific patterns of copy number aberrations. This approach has also led to the identification of genes that are consistently overexpressed when amplified, which are considered potential ‘amplicon drivers’. However not all genes within an amplicon are overexpressed, and an amplicon may harbor more than one driver(6). The expression of some driver genes is also more pervasive i.e. are overexpressed by other mechanisms in addition to amplification. That said, such approaches have been successful in identifying novel targets for subgroups of breast cancer, by exploiting the concepts of oncogene addiction. For instance, FGFR1, one of the genes mapping to the 8p11-p12 amplicon, is amplified in 10-15% of breast cancers and is associated with ER-positive disease and poor survival(11). FGFR1 is consistently overexpressed in tumors harboring FGFR1 amplification both of which have been shown to constitute a mechanism of resistance to endocrine therapy(9). A phase II clinical trial is currently testing the efficacy of small molecule FGFR inhibitors for these patients. By performing genome wide correlations between amplifications in different subgroups of breast cancer, we have identified a number of subgroup specific amplifications. This approach, coupled with integrating these data with matched gene expression data led to the identification of PPM1D as a putative amplicon driver(5). RNA interference-induced silencing and chemical inhibition of PPM1D in a panel of phenotypically matched PPM1D amplified and non-amplified cells showed that PPM1D expression and phosphatase activity is selectively required for the survival of cells harboring PPM1D gene amplification(5, 12). These data suggest that PPM1D may prove a viable therapeutic target for the subset of HER2-positive breast cancers harboring amplification at 17q23.2. Through a similar approach, we identified 38 genes that were significantly overexpressed when amplified in a series of 56 triple negative breast cancers, including FGFR2 amplifications in approximately 4%. Our work demonstrated that cancer cells harboring FGFR2 amplification are exquisitely sensitive to inhibition of FGFR2 in vitro and in vivo through the use of RNA interference and treatment with FGFR small molecule inhibitors [8], suggesting that FGFR inhibitors may constitute a tailored therapy approach for a subgroup of triple negative tumors. More recently, we have shown that 5% of ER-negative high-grade breast cancers that harbor amplification of CCNE1 within the 19q12 amplicon are dependent on CCNE1 and CDK2 kinase activity for their survival. Cancer cells with CCNE1 gene amplification are sensitive to CDK2 inhibitors, providing a rationale for the testing of these chemical inhibitors in a subgroup of patients with ER-negative grade III breast cancers in the context of clinical trials(6).
As well as using genetic and transcriptomic data to identify potential therapeutic targets in a candidate driven approach, integrating functional profiling data offers an unbiased way of identifying genetic dependencies. This approach has been used to identify additional amplicon drivers in HER2 amplified tumors by systematically assessing cell viability in a panel of HER2 amplified cell lines after silencing of all genes that were significantly overexpressed when amplified identified in a cohort of primary HER2 amplified breast cancers. This approach identified the transcription factor TFAP2C as a novel genetic dependency in 5% of HER2 amplified breast cancer cells(13). Whilst such screening approaches as these can identify novel amplicon drivers, many of the targets identified (e.g. transcription factors) are not directly targetable. By exploiting the concept of synthetic lethality(7), (where loss of either gene is compatible with cell survival, however loss or inhibition of both genes results in cell death), the alterations in the cells’ physiology that arise as a consequence of aberrant activation of oncogenes or tumor suppressor gene loss, rather than oncogene/tumor suppressor proteins themselves, are targeted to achieve tumor selectivity. This concept has been successfully applied to identify novel therapeutic targets including PARP inhibitors in BRCA1/2 mutant patients, further corroborated by the identification of a resistance mechanism to PARP inhibitors(14). High-throughput RNA interference screening of the kinome (i.e. pharmacologically tractable genes), in a panel of commonly used breast cancer cell line models, identified a series of novel genetic dependencies, in basal, luminal and HER2 subgroups(15). This approach also led to the identification of genetic dependencies of cells with specific mutations, e.g. PTEN-null breast tumor cells were found to be dependent on signaling through mitotic checkpoint kinases. Integration of viability data with transcript and protein profiling also identified a correlation between sensitivity to ADCK2 silencing and high ADCK2 mRNA and protein levels in ER-positive cells(15). Such unbiased approaches provide a framework upon which additional dependencies and candidate therapeutic targets may be identified.
The next generation
The advent of next generation sequencing has increased our understanding of the complexity of cancer genomes tremendously and has identified a number of subtype-specific mutations associated with different cancer types. Massively parallel sequencing studies in breast cancer have identified a plethora of novel mutations, including MAP3K1 mutations in ER-positive cancers(16), and PARK2 mutations in triple negative disease(17). In addition RNA-sequencing studies have enabled the identification of novel recurrent targetable expressed fusion genes, involving the MAST kinase and NOTCH gene family members(18). Large-scale sequencing efforts currently being undertaken with consortia such as The International Cancer Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA), are leading to an unprecedented amount of data. However, the main challenges that lie ahead are for the translation of the genetic alterations identified by next-generation sequencing into benefit for cancer patients. These mainly depend on i) the identification of ‘driver’ mutations and ii) the targeting of ‘driver’ genetic aberrations. While the identification of ‘driver’ genetic aberrations so far has been largely based on statistical algorithms(19), the targeting of the ‘driver’ aberrations has proven difficult. However, the majority of the novel mutations observed in the common types of breast cancer are at relatively low frequency, and the main challenge lies in the distinction of what constitutes a ‘driver’ mutation event versus a ‘passenger’ event (i.e., has no biological significance on the cell harboring its mutation at a given point in time)(7).
Traditionally, the identification of driver events stems from the fact that they are recurrent at a significant frequency above the background mutation rate within the tumor cohort studied. We can integrate different sorts of genetic alterations to aid the identification of recurrent activation or tumor suppressive events, such as mutation and homozygous deletions, or gross DNA rearrangements of a tumor suppressor gene, or amplification and activating mutations of an oncogene (Fig. 1). We have used this approach to identify novel candidate cancer genes in BRCA1 mutant tumors, by integrating a list of mutations identified from whole genome sequencing, with published aCGH data for the presence of homozygous deletions(20). This can also be taken further to look for functional recurrences in the form of genetic alterations in members of the same gene family or members of the same signaling network or pathway. For example mutations in chromatin remodeling genes appear to be a common alteration in many types of solid tumors(19, 21), and identifying ways of targeting these tumors with chromatin remodeling defects is a key challenge that needs to be explored in future studies. There are a number of computational tools that exist to predict the functional effect of a mutation of interest on a protein and to identify pathways that are deregulated in cancer and therefore are likely to contain significant driver genes. Algorithms that identify key transcriptional regulators of oncogenic programs can be used to prioritize mutations for follow-up studies (for a review of these see(22)). Algorithms that predict the pathogenicity of somatic mutations based on the selection pressure and type of mutation have also been developed(19, 23). However, novel predicted ‘drivers’, still need to be functionally investigated in appropriate model systems before they can be definitively defined as driver events. The recent functional validation of HER2 mutations in breast carcinomas without HER2 amplification has highlighted the importance of this step(24). Through the use of functional genomic screens we can begin to identify driver events in a high-throughput manner. This can be achieved a number of ways, from cross-species comparative approaches identifying driver genes as those that are conserved in human and mouse tumors, high-throughput insertional mutagenesis screens through to whole genome shRNA screens(22). Perhaps a more intuitive approach, which aids in the identification of both tumor suppressor genes and oncogenic drivers lies in the generation of cancer genome focused screens by generation of overexpression libraries of mutant open –reading frames (ORF’s) and short hairpin RNA’s (shRNA’s) that target the same set of genes identified by sequencing primary tumor samples. These libraries can then be screened for their ability to transform pre-malignant cells. In addition, wild-type ORF libraries generated from primary tumors without prior knowledge of the mutations may also provide an effective approach for gain of function screens.
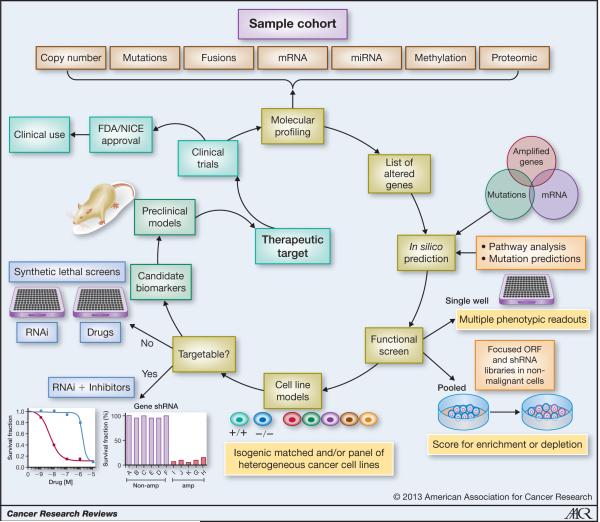
FIGURE 1. Schematic of data integration to identify therapeutic targets.
Molecular profiling of a cohort of primary breast cancers allows the identification of genomic alterations. Integration of this data is useful to identify potential oncogenic and tumor suppressive events in silico; e.g. genes that are overexpressed when amplified, in-frame fusion genes where the 3′ partner is overexpressed, mutations that are expressed at the mRNA level; genes that are under-expressed when deleted or methylated, disrupting fusions/structural rearrangements or mutations resulting in under-expression respectively. Additional in silico analyses can be performed to identify potential candidate driver genes by using prediction algorithms that ascribe biological meaning to genomic data. For example searching for significantly altered pathways that are more likely to contain driver genes, prediction of key transcriptional regulators of oncogenic programs and prediction of which missense mutations are likely to have a biological effect on the protein. Construction of cancer-focused screens can be a useful tool to investigate which candidate driver genes confer tumor specific dependencies. Oncogenic drivers and tumor suppressors are simultaneously assessed by constructing parallel libraries of cDNA ORF’s and shRNA’s, which are expressed in pre-malignant cells, and subsequently assayed for tumorigenicity either in vitro through the use of 3D models or in vivo. In addition, other measures of phenotypic alterations can also be assayed in tandem e.g. invasion, migration and resistance to anoikis. Both pooled screening using next generation sequencing for deconvolution and identification of biologically active ORF’s and shRNA’s, or single well screening can be used. To take forward hits from these screens, the use of appropriate cancer cell line models constitutes a more translationally relevant platform for drug discovery and development. Either a panel of phenotypically matched breast cancer cell lines (ER, PR, HER2, TP53) with and without the aberration of interest are used, or an isogenic cell pair to investigate the selectivity of the genomic alteration of interest. Genes that are directly targetable with validated inhibitors (e.g. kinases) can then be taken forward for further evaluation in the cell line panel e.g. for oncogenic events assessment with RNAi and available inhibitors, can be used to assess tumor dependency (cells with the aberration will be sensitive to gene inhibition, whereas those without will not). Aberrations that are not directly targetable with available inhibitors can be assessed through synthetic lethal screens using siRNA druggable libraries and drug screens. Candidate dependencies can be subsequently validated in pre-clinical models before evaluation in clinical trials. By identifying the genetic alteration and then identifying ways of targeting it, allows the genomic biomarker to be established a priori, cutting down the time to identify biomarkers of sensitivity during the drug development process. As is sometimes the case, promising preclinical data do not readily transfer to positive outcomes from early clinical trials. While inter- and intra-tumor genetic heterogeneity almost certainly play a role in resistance to targeted therapies, other molecular mechanisms can be teased out using the same approach described above. We can change the cohort of samples interrogated with molecular profiling to identify biomarkers for resistance or sensitivity to the targeted agent in question (i.e. before or after treatment, responders or non-responders). Identification of targets that are selective to inhibitors already in clinical trials will enhance the time to routine clinical use.
As many mutations and fusion genes identified may not be directly targetable, synthetic lethality approaches constitute an alternative for the identification of novel targets. These can be achieved through screening of isogenic cell line models with and without the genomic alteration of interest and/or a panel of heterogeneous cell lines with and without the alteration, with si/shRNA screens of druggable genes and high-throughput small molecule drug screens. Through the use of drug screens using small molecule inhibitors that are already FDA approved, the time needed from target identification to phase II clinical trials is much shorter. In fact there are a plethora of small molecule inhibitors available that have no useful predictive biomarker. Identifying these biomarkers through these integrated approaches would ultimately lead to patient benefit more quickly. Concerted efforts within the scientific community are being aimed at addressing these issues, and interrogation of systematic pharmacogenomic screening data for an aberration of interest is becoming a reality(25, 26). In parallel, the growing field of metabolomics is yielding interesting possibilities for classifying tumors based on their metabolic signatures, and in identifying pathways related to drug resistance or toxicity through metabolomic profiling. Furthermore, metabolic dependencies resultant from specific genomic alterations, offer novel therapeutic opportunities. Readers are directed to excellent reviews on the subject(27). In addition, other factors such as the importance of epigenetic mechanisms (including methylation and acetylation)(28) and non-coding RNAs upon gene regulation(29), and the role of the tumor micro-environment need also to be considered(30, 31).
However, not all recurrent mutations and fusion genes are represented by the available breast cancer cell line models, and pathognomonic events underlying some types of breast cancer can only be studied in the context of forced expression models, making the use of synthetic lethal approaches limited. Such models may not recapitulate the network state space of primary tumors harboring the genetic aberration of interest. These caveats must be born in mind when interpreting pre-clinical functional validation data. That said, there are a number of common genetic aberrations which are not directly targetable (e.g. TP53 and KRAS mutations, and PTEN loss of function), where adequate models are in abundance. This provides an opportunity to leverage the power of synthetic lethal screens in multiple isogenic models, thereby providing some control for the context-dependent nature of many genetic dependencies. Furthermore, by subjecting samples with and without the mutation of interest to deep sequencing one might identify a pattern of co-mutation (e.g. are there a set of genes frequently mutated in TP53-mutant triple negative breast cancers but not in TP53 wild type cancers), which could be modeled in vitro through synthetic lethal screens to interrogate potential cooperative interactions. Systems biology approaches would likely prove invaluable in these strategies.
Finally, next generation sequencing studies have highlighted the scope(32) and important role of intra-tumor genetic heterogeneity in cancer evolution and emergence of drug resistance(33). High depth multi-region sequencing and single cell sequencing can be used to characterise the repertoire somatic variants or patterns of copy number changes in non-modal clones within a tumor. Of course, not all these mutations will be biologically relevant. Integration of these data with pathway analysis tools and on-line resources such as the Connectivity Map(34) which identifies connections between drugs, disease and genes, aids prioritization of mutations and subsequent compound library screening, using chemical libraries of drugs currently in clinical trials. This approach would identify which mutations confer resistance to which drug; the ideal scenario would then be to analyze pre- and post-treatment samples from neoadjuvant trials to confirm the role of these non-modal clones in the evolution of drug resistance. Focused high depth sequencing could be effectively employed as a screening strategy to exclude patients from treatment with agents they are likely to develop resistance to, or early relapse after.
Summary
Integration of multiple data-types is becoming increasingly useful for the identification of therapeutic targets, within different subtypes of breast cancer. With the advent of next generation sequencing technologies and the vast amounts of data being generated, it is possible to identify recurrent mutational patterns within breast cancer. However, given the relatively low frequency of novel mutations and fusion genes in breast cancer and to fully understand the biology and therapeutic responses of some patients, the clonal genotypes of the individual tumors will need to be determined. It is evident that these large-scale sequencing projects need to be integrated with functional screens to achieve the goal of developing novel therapeutic strategies. For functional screening to be useful in identifying key driver events, researchers need to account for the fact that many gene alterations will be context dependent; either through epistatic interactions, or dependence on a particular developmental stage of the tumor. It will be necessary to develop more complex models to assess interactions in a more network-driven approach. The goal of individualized patient management will be a step closer with the inception of clinical trials designed to perform genome-wide or targeted sequencing of cancers to identify targetable aberrations and to determine the mechanisms of resistance to specific therapeutic agents.
Acknowledgments
Grant Support This work is supported in part by Breakthrough Breast Cancer. Rachael Natrajan is supported by a Breast Cancer Campaign Career Development Fellowship. Paul Wilkerson is in receipt of a Wellcome Trust Clinical Research Fellowship grant.
Footnotes
Disclosure of Potential Conflicts of Interest None
References
- 1.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 2002;41:154–61. [PubMed] [Google Scholar]
- 2.Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010;220:263–80. doi: 10.1002/path.2648. [DOI] [PubMed] [Google Scholar]
- 3.Natrajan R, Weigelt B, Mackay A, Geyer FC, Grigoriadis A, Tan DS, et al. An integrative genomic and transcriptomic analysis reveals molecular pathways and networks regulated by copy number aberrations in basal-like, HER2 and luminal cancers. Breast Cancer Res Treat. 2010;121:575–89. doi: 10.1007/s10549-009-0501-3. [DOI] [PubMed] [Google Scholar]
- 4.Nikolsky Y, Sviridov E, Yao J, Dosymbekov D, Ustyansky V, Kaznacheev V, et al. Genome-wide functional synergy between amplified and mutated genes in human breast cancer. Cancer Res. 2008;68:9532–40. doi: 10.1158/0008-5472.CAN-08-3082. [DOI] [PubMed] [Google Scholar]
- 5.Natrajan R, Lambros MB, Rodriguez-Pinilla SM, Moreno-Bueno G, Tan DS, Marchio C, et al. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res. 2009;15:2711–22. doi: 10.1158/1078-0432.CCR-08-1878. [DOI] [PubMed] [Google Scholar]
- 6.Natrajan R, Mackay A, Wilkerson PM, Lambros MB, Wetterskog D, Arnedos M, et al. Functional characterization of the 19q12 amplicon in grade III breast cancers. Breast Cancer Research. 2012;14:R53. doi: 10.1186/bcr3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashworth A, Lord CJ, Reis-Filho JS. Genetic interactions in cancer progression and treatment. Cell. 2011;145:30–8. doi: 10.1016/j.cell.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 8.Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–23. doi: 10.1038/onc.2009.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res. 2010;70:2085–94. doi: 10.1158/0008-5472.CAN-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weigelt B, Geyer FC, Reis-Filho JS. Histological types of breast cancer: how special are they? Mol Oncol. 2010;4:192–208. doi: 10.1016/j.molonc.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuny M, Kramar A, Courjal F, Johannsdottir V, Iacopetta B, Fontaine H, et al. Relating genotype and phenotype in breast cancer: an analysis of the prognostic significance of amplification at eight different genes or loci and of p53 mutations. Cancer Res. 2000;60:1077–83. [PubMed] [Google Scholar]
- 12.Rayter S, Elliott R, Travers J, Rowlands MG, Richardson TB, Boxall K, et al. A chemical inhibitor of PPM1D that selectively kills cells overexpressing PPM1D. Oncogene. 2008;27:1036–44. doi: 10.1038/sj.onc.1210729. [DOI] [PubMed] [Google Scholar]
- 13.Shiu KK, Wetterskog D, Mackay A, Natrajan R, Lambros M, Sims D, et al. Integrative molecular and functional profiling of ERBB2-amplified breast cancers identifies new genetic dependencies. Oncogene. 2013 doi: 10.1038/onc.2012.625. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–5. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 15.Brough R, Frankum JR, Sims D, Mackay A, Mendes-Pereira AM, Bajrami I, et al. Functional Viability Profiles of Breast Cancer. Cancer Discov. 2011;1:260–73. doi: 10.1158/2159-8290.CD-11-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–60. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–9. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17:1646–51. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–4. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Natrajan R, Mackay A, Lambros MB, Weigelt B, Wilkerson PM, Manie E, et al. A whole-genome massively parallel sequencing analysis of BRCA1 mutant oestrogen receptor-negative and -positive breast cancers. J Pathol. 2012;227:29–41. doi: 10.1002/path.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu JN, Roberts CW. ARID1A Mutations in Cancer: Another Epigenetic Tumor Suppressor? Cancer Discov. 2012;3(1):35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eifert C, Powers RS. From cancer genomes to oncogenic drivers, tumour dependencies and therapeutic targets. Nat Rev Cancer. 2012;12(8):572–8. doi: 10.1038/nrc3299. [DOI] [PubMed] [Google Scholar]
- 23.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bose R, Kavuri SM, Searleman AC, Shen W, Shen D, Koboldt DC, et al. Activating HER2 Mutations in HER2 Gene Amplification Negative Breast Cancer. Cancer Discov. 2013 Feb;3(2):224–37. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Claudino WM, Goncalves PH, di Leo A, Philip PA, Sarkar FH. Metabolomics in cancer: a bench-to-bedside intersection. Critical Rev Oncol Hematol. 2012;84:1–7. doi: 10.1016/j.critrevonc.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Huang Y, Nayak S, Jankowitz R, Davidson NE, Oesterreich S. Epigenetics in breast cancer: what’s new? Breast Cancer Res. 2011;13:225. doi: 10.1186/bcr2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Quesne J, Caldas C. Micro-RNAs and breast cancer. Mol Oncol. 2010;4:230–41. doi: 10.1016/j.molonc.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Artacho-Cordon A, Artacho-Cordon F, Rios-Arrabal S, Calvente I, Nunez MI. Tumor microenvironment and breast cancer progression: a complex scenario. Cancer Biol Ther. 2012;13:14–24. doi: 10.4161/cbt.13.1.18869. [DOI] [PubMed] [Google Scholar]
- 31.Kenny PA, Lee GY, Bissell MJ. Targeting the tumor microenvironment. Front Biosci. 2007;12:3468–74. doi: 10.2741/2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Eng J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–10. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–35. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]