Abstract
Background
The mechanism of CD4+ T-cell decline in HIV-1 infection is unclear, but the association with plasma viral RNA load suggests viral replication is involved. Indeed, viremic controller patients with low viral RNA loads typically maintain high CD4+ T-cell counts. Within a local cohort of 86 viremic controllers, we identify a subgroup (18 “discord controllers”) with low CD4+ T-cell counts that present clinical uncertainty. The underlying mechanism accounting for CD4+ T-cell decline in the face of low or undetectable plasma (RNA) viral load remains unresolved. The objective of this study was to investigate the viral and host immune system dynamics in discord controllers by measuring cellular HIV-1 DNA load, T-cell populations, and T-cell activation markers.
Methods
We compared discord controllers (viral RNA load <2000 copies/mL, <450 CD4+ T-cells/mm3) with typical controllers (viral RNA load <2000 copies/mL, >450 CD4+ T-cells/mm3) and progressors (viral RNA load >10,000 copies/mL, <450 CD4+ T-cells/mm3). We quantified CD4+/CD8+ naive/central memory/effector memory subsets (CD45RA/RO ± CD62L), activation levels (CD38+HLA-DR+), and HIV-1 DNA load.
Results
Discord controllers resembled progressors showing high viral DNA load, depletion of naive CD4+ T-cells, and higher activation in all CD4+ T-cell subsets, compared with typical controllers. They were similar to typical controllers with lower CD8+ T-cell activation compared with progressors.
Conclusions
Our data are consistent with a relationship between CD4+ T-cell activation and disease progression. HIV-1 DNA load may be a better marker of viral replication and disease progression than viral RNA load. Lower level CD8+ T-cell activation correlates with low viral RNA load but not with disease progression or viral DNA load.
Keywords: HIV-1, viremic controller, HIV-1 DNA, plasma RNA load, activation
INTRODUCTION
After HIV-1 seroconversion, CD4+ T-cells, the main target cell population, decline over the asymptomatic phase leading to progressive immunodeficiency. Although it is known that HIV-1 is cytopathic for CD4+ T-cells, the mechanisms leading to CD4+ T-cell depletion are not fully understood. It is suggested that the proportion of infected CD4+ T-cells is too small to fully account for the extent of CD4+ T-cell decline,1 and an increased death rate is observed in nontarget cells, such as CD8+ T-cells.2 In keeping with this, Rodriguez et al3 showed that in untreated individuals, plasma HIV-1 RNA load only minimally predicts CD4+ T-cell decline, with current evidence suggesting that immune activation is the major predictor of disease progression.4–11 Studies also suggest that naive T-cell compartments are preferentially depleted in progressive disease10,12,13; it is not known whether this is due to impaired production, sequestration to another site or phenotype, or destruction of naive T-cells. In addition, HIV-1 DNA load has been shown to predict disease progression.14–16
Some patients, viremic controllers, maintain low plasma viral RNA loads without antiviral medications. The vast majority of these patients are infected with replication-competent virus, and yet typically maintain high CD4+ T-cell counts.17 However, cases of progressive CD4+ T-cell depletion despite continued control of viral RNA load have been described.18–20 We term these patients “discord controllers” to reflect the discrepancy in viral RNA load and CD4+ T-cell counts. This patient subset presents uncertainty in clinical management because parameters that are normally associated with progression (viral RNA load and CD4+ T-cell count) are uncoupled. The mechanism leading to this discrepancy is unclear, although aberrant immune activation causing T-cell decline in the face of low plasma viral RNA load is one potential explanation.
We hypothesized that those changes typically associated with disease progression, depleted naive CD4+ and CD8+ T-cells, high level CD4+ and CD8+ T-cell activation, and high HIV-1 DNA loads, would be more marked in discord controllers as compared with typical controllers (low viral RNA load, high CD4+ T-cells). Furthermore, we hypothesized that analysis of discord controllers with comparison to typical controllers would allow immunologic changes associated with disease progression and those due to high plasma viral RNA load to be distinguished; this is not possible with analysis of patients with the typical pattern of progression (high HIV-1 RNA load, low CD4+ T-cells). Our aims based on these hypotheses were to analyze these factors typically associated with disease progression in discord controllers as compared with typical controllers and patients with a typical progression pattern.
METHODS
Study Groups
Patients were recruited, with informed written consent, from outpatient clinics at Barts and The London and Homerton University Hospital Trusts. All patients were HIV-1 seropositive and HIV-2 seronegative. Serology was not available for 1 patient, but sequence analysis as part of antiretroviral resistance testing demonstrated HIV-1 infection. Viremic controllers were defined as having plasma HIV-1 RNA loads below 2000 copies per milliliter [≥3 measurements over 12 months without antiretroviral therapy (ART)], definition in line with that used by the International HIV Controller Consortium. The controller cohort was divided into 2 groups based on the geometric mean of the last 3 CD4+ T-cell counts: typical controllers >450 cells/μL (mm3), discord controllers <450 cells/mm3. The reason 450 cells/mm3 was used as the cutoff was based on the fact many laboratories define a normal CD4+ T-cell count to be above this value.21 In addition, the “When to start consortium”22 have found decreased mortality when ART is started in the range 351–450 cells/mm3 instead of waiting until they are below 350 cells/mm3, further supporting the suggestion that this range is abnormally low. The geometric mean of the 3 most recent % CD4+ T-cells for each controller cohort was calculated to determine whether the lower absolute CD4+ T-cell count in the discord controllers was reflected in a lower % CD4+ T-cells. Epidemiological data were collected including age, sex, time since first positive test, ethnicity, country of birth, and blood-borne virus risk behavior.
Controls were (1) Viremic noncontrollers, for viral DNA load studies: ART-naive HIV-1–infected persons, infected >12 months with a viral RNA load >10,000 copies per milliliter; for T-cell studies controls comprised (2) HIV-1 progressors, definition as for viremic noncontrollers but with CD4+ T-cell counts <450 cells/mm3, and (3) uninfected subjects.
Plasma HIV-1 RNA loads and CD4+ T-cell counts were stable for most controller patients. Two typical controllers, however, with low viral RNA load for 3.7 and 3.5 years subsequently showed a significant rise in viral RNA load while maintaining good CD4+ T-cell counts (geometric means of 732 and 708 cells/mm3, respectively). Another patient with low viral RNA loads and good CD4+ T-cell counts for 2.3 years subsequently experienced a fall in CD4+ T-cell count, but without an viral RNA load measurement, prompting initiation of ART. Thirteen patients (3 discord controllers and 10 typical controllers) were treated with ART during pregnancy but controlled plasma viral RNA load for at least 12 months both before and after ART. Controllers mentioned in this paragraph are included in the analysis of patient characteristics and clade analysis but were not used for viral DNA load and T-cell analysis. In addition, T-cell work was carried out only on those discord controllers with CD4+ T-cell geometric mean of ≤410 cells/mm3, and those typical controllers with a mean of ≥500 cells/mm3, to avoid those patients too near the cutoff.
CD4+ T-Cell Counts and Viral RNA Loads
Routine laboratory CD4+ T-cell counts were performed using BD FACS Sample Prep Assistant with Trucount beads, acquiring samples on a BD FACSCanto II flow cytometer. FACSCanto clinical software was used for data analysis. Plasma viral RNA load was measured using Roche Ampliprep/COBAS Taqman HIV-1 Test v1.0 (Roche Molecular Systems, Inc, Pleasanton, CA; detection limit of 40 copies/mL). At least 1 sample from each patient was also tested using a different assay to confirm the low viral RNA load. Individual patient viral RNA loads and CD4+ T-cell counts are reported as the geometric mean of last 3 measurements or last 3 before ART. If a patient had achieved viremic control (<2000 copies/mL, >12 months), but subsequently viral RNA load rose above 2000 copies per milliliter, the last date of viremic control was determined to be the last date an viral RNA load measurement would still contribute to a geometric mean of <2000 copies per milliliter.
Rate of change of CD4+ T-cell count was calculated using the geometric mean of the 3 most recent values (x) and the mean of the oldest recorded values (y), and the time between them (z): [(y – x)/z]. Patients were only included in this calculation if the values spanned at least 3 years.
Clade Analysis
Some patients’ clades were obtained during routine genotypic antiretroviral resistance testing (RNA sequence from protease and AA 1–335 of reverse transcriptase). This assay has demonstrated efficacy in picking up a wide variety of subtypes including recombinant subtypes.23 For patients on whom genotypic antiretroviral resistance testing was unavailable, previously described env24 and gag25 polymerase chain reactions (PCRs) were used in parallel to accurately determine recombinant subtypes. Amplified products were sequenced using the second round forward and reverse primers from each PCR on an ABI 3100 Genetic Analyzer (Applied Biosystems Inc., Foster City, CA). Sequences were analyzed using SeqScape V2.1.1 software and subtyped using the REGA online typing tool (Stanford University, 2006).
HIV-1 DNA Load Determination
DNA was extracted from cryopreserved peripheral blood mononuclear cells using Qiagen EZ1 DNA Blood Kit on the Qiagen BioRobot EZ1. A fixed concentration of phocine herpes virus was added to each sample before extraction as an internal amplification control. A quantitative HIV-1 PCR, amplifying the long-terminal repeat (LTR) region, was designed based on previously described reverse transcriptase–polymerase chain reaction.26,27 A final 25 μL PCR mixture contained 5 μL of peripheral blood mononuclear cells DNA, 2× PCR QuantiTect Multiplex RT-PCR No Rox Mix (Qiagen), 0.2 μM of each primer (F1 5′-AGCCTCAATAAAGCTTGCCTTGA-3′; R1 5′-GGCGCCACTGCTAGAGATTTT-3′), and 0.2 μM probe (AAGTAGTGTGTGCCCGTCTGT, fluorescent label). Thermocycling conditions (ABI 7500) were 95°C for 15 minutes, then 45 cycles of 95°C for 15 seconds, 60°C for 1 minute, and 72°C for 5 minutes. Mean values were calculated from duplicate PCRs. Quantitative PCR for β-globin (adapted from Lo et al28) was run in parallel. An external plasmid dilution series was run to construct a standard curve allowing viral DNA loads to be expressed as per cell equivalent.
T-cell Immunophenotyping Using Flow Cytometry
Seven-color flow cytometry was used to quantify CD4+ and CD8+ T-cell populations and their naive (CD45RA+CD62L+), central memory (CD45RO+CD62L+), and effector memory (CD45RO+CD62L−) subsets in EDTA-anticoagulated blood. Activation levels were determined by measuring the percentage of T-cells coexpressing CD38 and HLA-DR. The following monoclonal antibodies and fluorochromes were used: CD3-Pacific Blue, CD4-allophycocyanin (APC)-H7, CD45-RA-phycoerythrin(PE)-Cy7, CD45-RO-PE-Cy7, CD62L-PE, HLA-DR-peridinin chlorophyll protein (PerCP), CD38-PerCP-Cy5.5 (BD Biosciences, San Jose, CA).
Saturating amounts of monoclonal antibodies/isotype controls were incubated with 100 μL of whole blood (within 4 hours of collection) for 15 minutes at room temperature. Optilyse C (Immunotech, Marseilles, France) was added for 15 minutes at room temperature. Blood aliquots with single-color staining were processed with each sample for compensation. After 2 washes with phosphate-buffered saline containing 2% fetal calf serum, 0.02% NaN3, and 1 mM EDTA, stained cells were resuspended and fixed in 300 μL phosphate-buffered saline 4% paraformaldehyde. A constant volume of Flow-count Fluorospheres (Beckman Coulter, Brea, CA) was added to enable absolute quantification of cells. The fixed stained cells were acquired within 12 hours of fixing on a BD LSR II cell analyzer (BD Biosciences) with a minimum of 50,000 (usually 100,000) events acquired per sample.
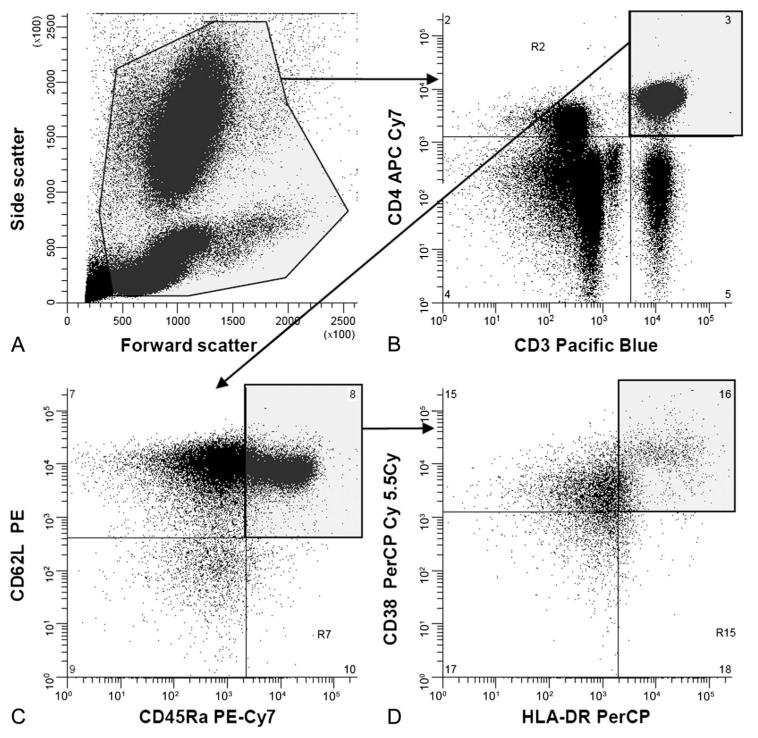
Winlist 6.0 software (Verity Software House, Topsham, ME) was used for analyses and color compensation (gating strategy shown in Fig. 1). CD4+ T-cells were identified within a viable cell gate, set on the basis of light scatter, as cells that were CD3+CD4+. CD8+ T-cells were identified as mononuclear cells that were CD3+CD4−. Two-dimensional dot plots, in which quadrant gates were set on the isotype controls, were used to define naive, central memory, and effector memory populations in both CD4+ and CD8+ T-cell populations. In turn, each of these populations were viewed on further 2-dimensional plots to determine activation level (percentage of cells coexpressing CD38 and HLA-DR). The number of flow-count spheres acquired allowed a precise determination of sample volume acquired, allowing expression of number of cells per volume of blood.
FIGURE 1.
Gating strategy for flow cytometric analysis of T-cell subsets. A, Forward and side scatter to define viable cell gate. B, Double staining of viable cells with anti-CD3 and anti-CD4. CD4+ T-cells are in the top right quadrant; CD4− T-cells (predominately CD8+ T-cells) are in the bottom right quadrant. C, Double staining of CD4+ T-cells with anti-CD45RA and anti-CD62L, naive CD62L+CD45RA+ T-cells, defined with reference to staining with isotype-matched control antibodies, are in the top right quadrant. D, Double staining of naive CD4+ T-cells with anti-CD38 and anti-HLA-DR, activated CD38+HLA-DR+ T-cells, defined with reference to staining with isotype-matched control antibodies, are in the upper right quadrant.
Statistical Analysis
Due to the rarity of these patients even in a large total HIV-1–infected cohort, power calculations were not under-taken to determine sample sizes; instead, all patients fulfilling the controller inclusion criteria and available to consent to the study were included. Therefore, it is necessary to interpret non-statistically different results with caution.
Analysis was performed using Prism (version 4.0; Graphpad Software, San Diego, CA) and results considered significant if P < 0.05. Viral RNA and DNA loads were log10 transformed before statistical analysis. To determine differences in HIV-1 RNA/DNA loads, age, time since first positive test and T-cell flow data, a 2-tailed Mann–Whitney U test was applied. Fisher’s exact test (2-tailed) was used when comparing sex distribution, ethnicity, country of birth, risk behavior, and clade in each cohort. A correction for multiple comparisons was employed using the false discovery rate calculation.
RESULTS
Controller Phenotype
A cohort of 82 HIV-1 viremic controllers was established: 64 typical controllers and 18 discord controllers (Table 1). Approximately 3000 patients attend the clinics, thus viremic controllers represent 2.7% (discord controllers 0.6%, typical controllers 2.1%) of all infected patients. Comparing the 2 controller cohorts, plasma viral RNA loads were indistinguishable (P = 0.71). Nine typical controllers and 1 discord controller were elite controllers (viral RNA load below 50 copies/mL, occasional nonconsecutive blips). There was no difference in median age (P = 0.44), sex distribution (P = 0.79), ethnicity, region of birth, or risk behavior. No significant difference was found comparing time since first positive test (best available surrogate for time since seroconversion) between the 2 controller cohorts (P = 0.25), but a larger cohort may reveal a difference. The %CD4+ T-cells in the discord controllers [median 22.3, interquartile ratio (IQR) 17.6–26.0] was significantly lower than that seen in the typical controllers (median 33.7, IQR 23.8–40.0, P < 0.0001). The rate of change of CD4+ T-cell count was not significantly different comparing the 2 cohorts (discord controllers, median −2.9, IQR −26.4 to −1.0; typical controllers, median 2.1, IQR −16.5 to −34.6, P = 0.2174); however, again, a larger study could reveal a difference here.
TABLE 1.
Patient Characteristics and Clade of Infecting Virus in Controller Cohorts
| Typical Controllers (CD4+ Count >450 cells/mm3), n = 64 |
Discord Controllers (CD4+ Count <450 cells/mm3), n = 18* |
|
|---|---|---|
| Age, yrs | 37.5 (32.0–43.0) | 38.5 (32.5–49.0) |
| Female sex, n (%) | 35 (54.7) | 11 (61.1) |
| CD4+ T–cell count, cells/mm3† | 699.3 (550.4–843.1) | 347.2 (298.0–406.6) |
| Plasma viral RNA load, log10 copies/mL† | 408.5 (160.7–1000.0) | 428.6 (100.3–1043.0) |
| Place of birth, n (%): | ||
| Europe | 25 (39.1) | 7 (38.9) |
| Africa (Sub-Sahara) | 28 (43.7) | 10 (55.6) |
| Caribbean | 6 (9.4) | 0 (0) |
| South Asia | 3 (4.7) | 0 (0) |
| Other | 2 (3.1) (Australia, Ecuador) | 1 (5.5) (New Zealand) |
| Ethnicity, n (%) | ||
| White British | 12 (18.8) | 3 (16.7) |
| White other | 8 (12.5) | 2 (11.1) |
| Black African (Sub Saharan) | 28 (43.7) | 11 (61.1) |
| Black Caribbean | 11 (17.2) | 2 (11.1) |
| South Asian | 3 (4.7) | 0 (0) |
| Other | 2 (3.1) (Mixed white and black African; South American) |
0 (0) |
| Time since first positive test, yrs | 4.5 (2.5–6.5) | 4.6 (3.3–9.3) |
| Risk behavior | ||
| Heterosexual | 42 (65.6) | 11 (61.1) |
| Specified abroad | 26 (40.6) | 5 (27.8) |
| Homosexual | 20 (31.3) | 6 (33.3) |
| Blood Products | 1 (16) | 1 (5.6) |
| IVDU | 1 (16) | 0 (0) |
| Clade, n‡ (%) | n = 41 | n = 15 |
| B | 14 (34.1) | 6 (40.0) |
| CRF02_AG | 8 (19.5) | 3 (20.0)§ |
| C | 7 (17.1) | 6 (40.0) |
| A | 4 (9.7) | 0 (0) |
| F | 2 (4.9) | 0 (0) |
| CRF14_BG | 2 (4.9) | 0 (0) |
| Other | D; A/C; A/D; B/D | 0 (0) |
Data presented in this table, when not stated to be n (%) are median (IQR).
Three patients in this cohort started ART due to their low CD4+ T–cell count (viral RNA load/CD4+ values before starting ART were used).
Median (IQR) of geometric mean calculated for each subject using 3 most recent CD4+ T–cell count and plasma viral RNA load values.
Data obtained using genotypic antiretroviral resistance testing for 28 typical controllers and 10 discord controllers, and using nested PCR for 13 typical controllers and 5 discord controllers; a product was not amplified for 2 samples tested using the nested PCRs (plasma viral RNA load <50 and 111 copies/mL).
For 1 sample, sequence analysis of nested PCR products indicated either CRF02_AG or CRF09_CPX. Statistical analysis employed a Mann–Whitney U test for plasma viral RNA load, age, time since first positive test, and a Fisher’s exact test for sex distribution, ethnicity, country of birth, and clade in each cohort. IVDU indicates intravenous drug use.
Clade Distribution in Controller Cohorts
The clade distribution in the East London HIV-1–infected population is diverse,23 with around 64% of isolates being non-clade B (D. A. Clark, PhD, and D.F. Bibby, PhD, unpublished data). We investigated whether there was a skew regarding infecting viral clade in discord controllers. It was not possible to obtain a product for 2 samples tested (plasma viral RNA load <50 and 111 copies/mL). As shown in Table 1, there was a non-significant trend towards an increased frequency of clade C in the discord controllers (40.0%) compared with typical controllers (17.1%) and also compared with the total tested HIV-1 population (25.1%) (D. A. Clark, PhD, and D.F. Bibby, PhD, unpublished data over 5 years at Department of Virology, Barts and the London NHS Trust).
Depleted Naive CD4+ T-Cell Compartment in Discord Controllers
HIV-1 infection, as it progresses, leads to decline of CD4+ naive T-cells.10,12 We measured the number and percentage of these naive cells (CD45RA+CD62L+), central memory T-cells (CD45RO+CD62L+), and effector memory T-cells (CD45RO+CD62L−) in blood from participants in each cohort.
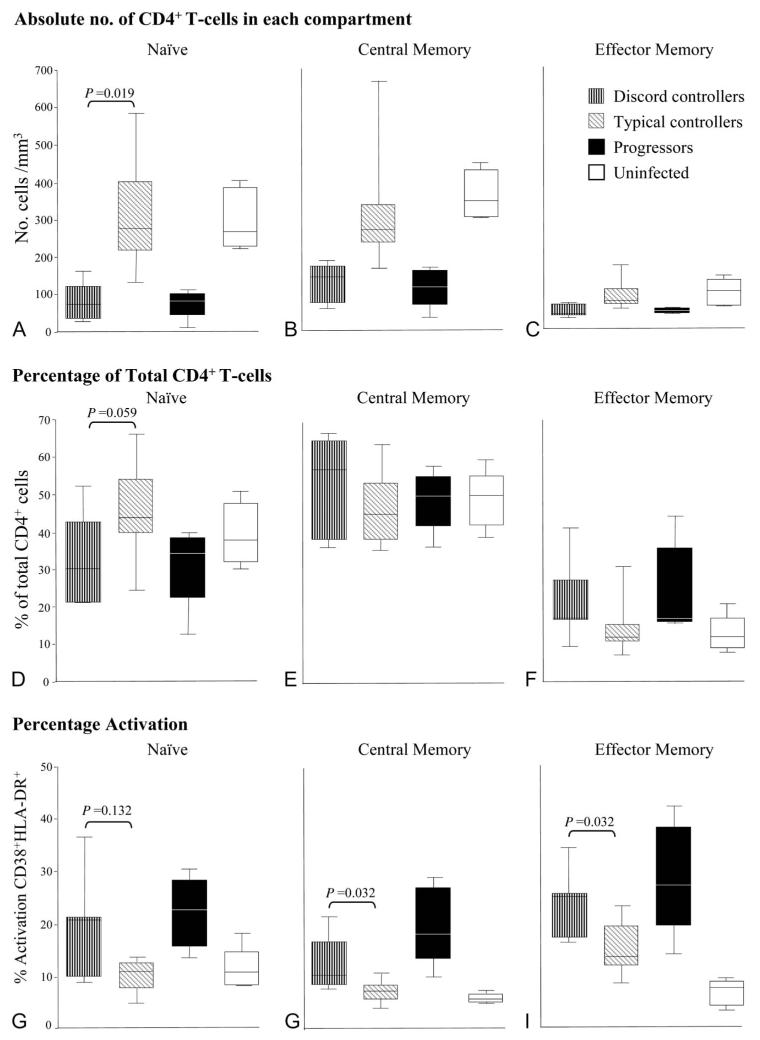
As expected, lower CD4+ T-cell numbers were seen in each subset in progressors and discord controllers compared with typical controllers and uninfected patients (Figs. 2A–C). A more marked depletion of the naive subset was seen in both discord controllers and progressors compared with the other patient groups (percentage of total CD4+ T-cells, Fig. 2D). The higher percentage of effector memory cells in progressors and discord controllers compared with typical controllers (Figs. 2C, F) represents a reciprocal change due to marked loss of naive cells. The percentage of central memory T-cells was similar in all 3 patient groups (Fig. 2E).
FIGURE 2.
Representation of CD4+ T-cell subsets by patient group. Patients analyzed included 7 patients with discord controller phenotype [median (IQR) for viral RNA load 675 (128–1320) copies/mL, CD4+ T-cells/mm3 337 (267–400)]; 12 patients showing the typical controller phenotype [viral RNA load 268 (116–926) copies/mL, CD4+ T-cells/mm3 815 (691–971)], 5 progressors [viral RNA load 60,683 (28,603–155,518) copies/mL, CD4+ T-cells/mm3 309 (296–339)], and 5 uninfected subjects. Progressors: age 35 (30–48) years, female 40%, CD4+ T-cell count 309 (287–346) cells/mm3, HIV-1 RNA load 60,682 (23,947–232,854) copies per milliliter, all reported as median (range). Absolute numbers of cells in CD4+ T-cell subsets are shown in (A) naive (CD45RA+CD62L+), (B) central memory (CD45RO+CD62L+), and (C) effector memory (CD45RO+CD62L−). Percentage of cells in CD4+ T-cell subsets compared with total CD4+ T-cell pool are shown for (D) naive, (E) central memory, and (F) effector memory. Percentage activation of cells in CD4+ T-cell subsets (as percentage of total cells in that subset) by patient group is shown in (G–I). Each central bar represents the median value, each box represents the IQR, and the whiskers represent the minimum and maximum values. Statistical analysis employed a Mann–Whitney U test.
Increased CD4+ T-Cell Activation in Discord Controllers
High-level CD4+ T-cell activation is associated with disease progression.5,8 We, therefore, measured T-cell activation level (percentage of cells coexpressing CD38+ and HLA-DR+) in blood. Progressors and discord controllers had comparably increased levels of activation in naive, central memory, and effector memory CD4+ T-cell subsets (Figs. 2G–I) compared with typical controllers and uninfected subjects. Typical controllers demonstrated a trend toward increased activated effector memory CD4+ T-cells as previously reported.29
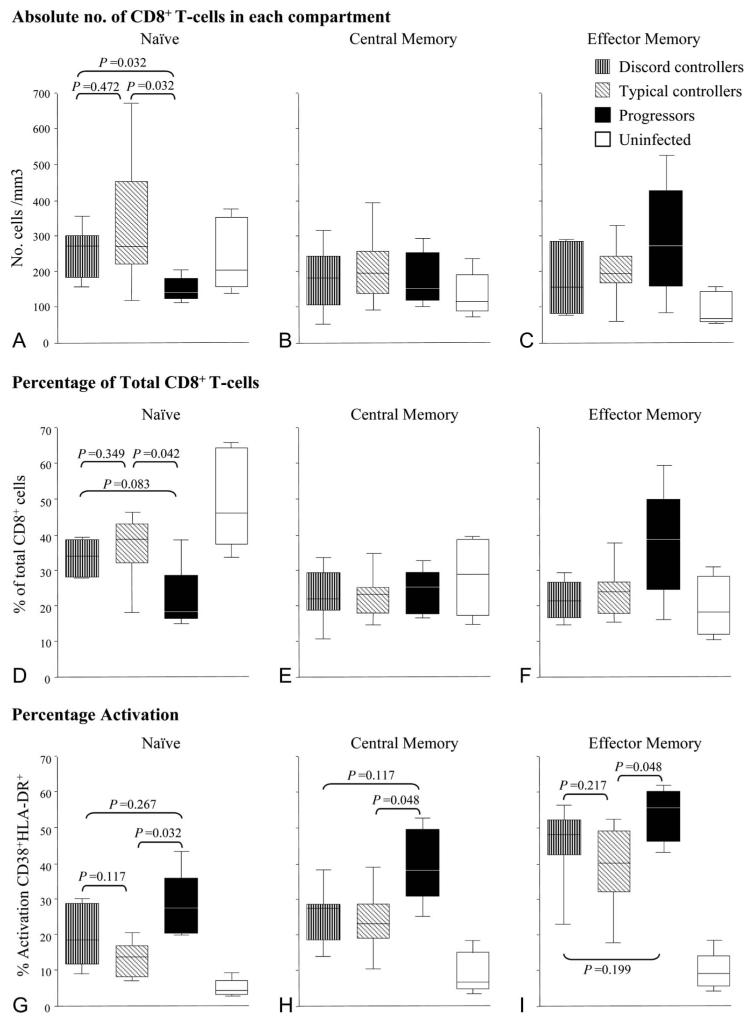
Naive CD8+ T-Cell Levels Are Not Depleted Compared With Typical Controllers
So far, our results suggest that the CD4+ T-cell compartment is similar in discord controllers and progressors. We next characterized the CD8+ T-cell population. Controller groups had similar numbers and percentages of CD8+ naive T-cells compared with uninfected controls, whereas the progressors had a marked depletion of this cell type (Figs. 3A, D). Progressors had an expanded number of effector memory CD8+ T-cells compared with all other groups (Fig. 3F).
FIGURE 3.
Representation of CD8+ T-cell subsets by patient group. Patients analyzed as described in Figure 2: discord controllers, typical controllers, progressors, and uninfected subjects. Absolute numbers of cells in CD8+ T-cell subsets are shown in (A) naive (CD45RA+CD62L+), (B) central memory (CD45RO+CD62L+), and (C) effector memory (CD45RO+CD62L−). Percentage of cells in CD8+ T-cell subsets compared with total CD8+ T-cell pool are shown for (D) naive, (E) central memory, and (F) effector memory. Percentage activation of cells in CD8+ T-cell subsets (as percentage of total cells in that subset) by patient group is shown in (G–I). Each central bar represents the median value, each box represents the IQR, and the whiskers represent the minimum and maximum values. Statistical analysis employed a Mann–Whitney U test.
CD8+ T-Cell Activation Is Not Higher in Discord Controllers Compared With Typical Controllers
CD8+ T-cell activation (CD38+ HLA-DR+) was increased in all T-cell subsets of all HIV-positive patients, even in typical controllers, compared with uninfected controls as has been demonstrated by Lopez et al.30 This is in keeping with the proposition that CD8+ T-cell subset alterations are a highly sensitive marker of HIV-1 infection. This increased activation was more marked in progressors in all subsets, especially in the memory compartments (Figs. 3G–I); in keeping with a previous report showing increased memory CD8+ T-cell activation in noncontrollers compared with controllers.31 In comparison, activation was equivalently low in both controller cohorts (Figs. 3G–I). Thus, despite the fact that discord controllers have low CD4+ T-cell counts (like progressors), their CD8+ T-cell activation pattern more closely resembles that of typical controllers.
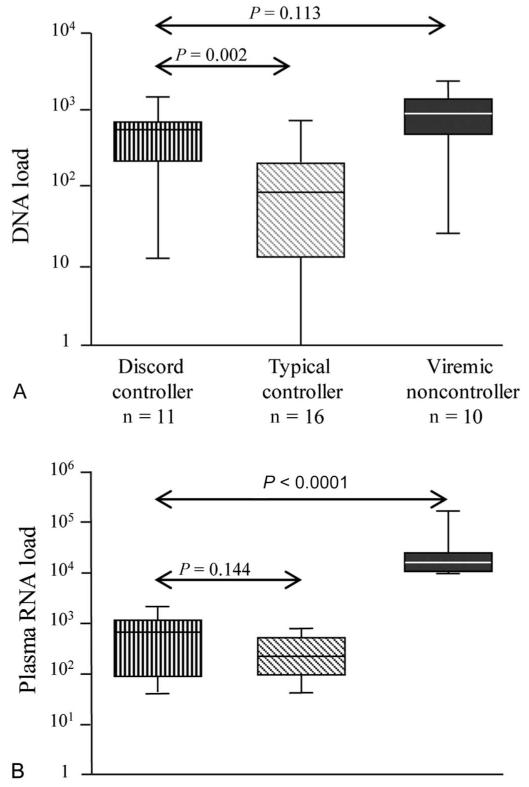
Higher HIV-1 DNA Load in the Discord Controller Cohort Compared With Typical Controllers
Viremic controllers typically have low HIV-1 DNA loads in addition to low viral RNA loads.32 Because discord controllers have low CD4+ T-cell counts, we hypothesized that high levels of viral replication were nevertheless ongoing and that this may be reflected in higher viral DNA loads. Plasma viral RNA load and cellular viral DNA load were determined for controllers and a group of noncontrollers (Fig. 4). We found discord controllers to have significantly higher viral DNA loads (median 601, range 13–1529 copies per 106 CD4+ T-cells) than typical controllers (median 87, range 0–755 copies per 106 CD4+ T-cells) and that viral DNA loads were as high as those seen in noncontrollers (high viral RNA loads) (median 852, range 27–2188 copies per 106 CD4+ T-cells), suggesting significant ongoing viral replication.
FIGURE 4.
HIV-1 DNA load compared with HIV-1 RNA load in patient groups. A, viral DNA load (DNA copies per 106 CD4+ T-cells; log10 scale) as compared with B, Plasma viral RNA load (copies/mL; log10 scale) by patient group. Each central bar represents the median value, each box represents the IQR, and the whiskers represent the minimum and maximum values. Viremic noncontrollers: age 42 (28–66) years, female 10%, CD4+ T-cell count 399 (258–716) cells/mm3, HIV-1 RNA load 15,809 (10,041–169,049) copies per milliliter, all reported as median (range). Median (range) of CD4+ T-cell count values (cells/mm3) within 12 months before the sample tested for plasma HIV-1 DNA load were 339 (251–424) for discord controllers; 775 (499–1326) for typical controllers; 399 (258–716) for viremic noncontrollers. Statistical analysis employed a Mann–Whitney U test.
ART in Discord Controllers Leads to Modest Recovery of CD4+ T-Cell Counts
Typically, after initiation of ART in a patient, plasma viral RNA load becomes undetectable and CD4+ T-cell count increases.33 During this study, 5 patients in the discord controller group started ART. This resulted in an increase from 308 to 353 CD4+ cells/mm3 over 12 months of ART, 269 to 319 CD4+ cells/mm3 over 19 months, 249 to 414 CD4+ cells/ mm3 over 19 months, 269 to 460 CD4+ cells/mm3 over 23 months, and 284 to 441 CD4+ cells/mm3 over 22 months of ART. An early rise was seen for only 1 patient where CD4+ T-cell count rose from 249 to 411 cells/mm3 after just 1 month of ART; CD4+ T-cell recovery occurred over many months for the other patients, and indeed counts continued to increase past 12 months of ART for 2 of the patients.
It was observed that although absolute CD4+ T-cell counts rose early for only 1 patient, %CD4+ T-cells did rise early for 3 patients due to the fact that absolute CD8+ T-cell count dropped markedly after 1 month of ART (760–536, 1429–1005, and 892–421 cells/mm3). Other studies have described a decrease in either memory CD8+ T-cells,34 activated CD8+ T-cells,35 or HIV-1-specific CD8+ T-cells36 within the first few weeks of ART that may account for the decrease we see in total CD8+ T-cells.
In summary, discord controllers share many characteristics with progressors, namely high HIV-1 DNA loads, a depleted naive CD4+ T-cell population with increased CD4+ T-cell activation levels; in contrast, the CD8+ T-cell compartment in discord controllers is more similar to that seen in typical controllers.
DISCUSSION
In HIV-1 infection, low viral RNA loads usually predict good CD4+ T-cell counts.37 We describe a cohort of HIV-1–infected patients, who, despite maintaining low viral RNA loads (similar to typical controllers), have low CD4+ T-cell counts (similar to typical progressors) indicating disease progression. We show that these patients, termed discord controllers, in addition to having low CD4+ T-cell counts have high viral DNA loads, preferentially depleted naive CD4+ T-cells and increased CD4+ T-cell activation, similar to typical progressors. This finding supports Hunt et al18 who show that lower CD4+ T-cell counts correlate with higher CD4+ T-cell activation even among controllers. Of note, the increased CD4+ T-cell activation was similar in discord controllers and progressors, despite progressors having much higher plasma viral RNA loads of the latter groups, suggesting that factors independent of viral RNA load are driving CD4+ T-cell activation.
With regard to the CD8+ T-cell compartment, discord controllers were more similar to typical controllers. Both controller cohorts have higher numbers of naive CD8+ T-cells and lower levels of CD8+ T-cell activation compared with progressors. These observations suggest that a more preserved CD8+ T-cell compartment is associated with control of plasma viremia but not necessarily with lack of disease progression as is shown in discord controllers. Some studies suggest that CD8+ T-cell activation is a predictor of disease progression, independent of CD4+ T-cell count and HIV-1 RNA.4,6,38 However, we do not find this to be the case in our controller population. Indeed, our observations are more in keeping with alternate studies, which show that higher plasma viral RNA load positively correlates with increased CD8+ T-cell activation,5,30 and that this correlation holds regardless of CD4+ T-cell counts.30 Functional analysis of the CD8+ T-cells present in these patient cohorts may highlight differences in cellular response to HIV-1, which confer protection against disease progression.
There was no association between the discord phenotype and age or time since first positive test (best available surrogate for time since seroconversion). Although time since diagnosis is not a perfect surrogate for time since seroconversion, this data suggests that the discord controller phenotype is not associated with a longer duration of infection compared with typical controllers. However, this study was not powered or designed to determine this fully. The fact that there is a trend toward lower %CD4+ T-cells in the discord controller cohort argues against suggestions that lower absolute CD4+ T-cell counts in the discord controller group are merely a feature of lower total lymphocyte counts associated with ethnic variation. The finding that the rate of change of CD4+ T-cell count was indistinguishable comparing the 2 controller cohorts might also support this premise. There was a trend toward an over-representation of clade C in the discord cohort; however, larger studies are required to confirm this association and determine whether this clade predisposes to the discord controller phenotype.
Initiation of ART in 5 patients achieved a modest gain in CD4+ T-cell numbers similar to those described recently by Okulicz et al39 further supporting the notion that low CD4+ T-cell counts in the discord controller population are not merely due to a lower “normal” CD4+ T-cell count, for example, due to ethnic variation. An increase over the first 10 weeks of ART has been suggested to be due to “redistribution” of CD4+ T-cells trapped in lymphoid.40 The fact that this type of early rise occurred for only 1 of the 5 patients suggests that redistribution was not the predominant mechanism leading to CD4+ T-cell recovery in these patients.
Unintegrated HIV-1 DNA is less stable than integrated “proviral” DNA, and as such has been proposed, together with total HIV-1 DNA, to be a marker for ongoing replication in vivo.41 This notion is further supported by observations that DNA reservoirs are maintained by viral replication that may be suppressed by ART42,43 and that total HIV-1 DNA correlates inversely with CD4+ T-cell counts and predicts disease progression.15,16 Compared with typical controllers, we found significantly higher viral DNA loads in the discord controllers, which was equivalent to that seen in progressors suggesting high levels of ongoing replication in vivo. It is unclear why this high cellular viral DNA load does not result in a higher-level plasma viral RNA load in the discord controllers. Given the crucial role of the CD8+ compartment in control of plasma viral RNA load,44–46 the low plasma viral RNA load in the discord controllers may be contingent on a CD8+ T-cell compartment that is maintained in a subset distribution and activation pattern similar to typical controllers. Another possibility is that viral replication is strictly compartmentalized; the gastrointestinal tract (lamina propria and organized lymphoid tissue) is a potential candidate because it has been shown to be an important site of HIV-1 replication and of CD4+ T-cell sequestration and depletion.47 Alternatively, discord controllers may be infected with a virus that predominantly spreads directly from cell to cell. Another scenario is that virus may be released efficiently into the blood but cleared by humoral immune mechanisms. These possible scenarios that may not be mutually exclusive are currently under investigation.
With respect to the mechanism of CD4+ T-cell loss in discord controllers, our observations favor a model where CD4+ T-cell depletion is driven, at least partly, by CD4+ T-cell activation. This activation seems to be associated with high-level viral replication as suggested by high total HIV-1 DNA loads. This is in keeping with a large body of evidence, which suggests that immune activation is associated with more rapid clinical progression and CD4+ T-cell decline.4–11 This may result from clonal exhaustion and drainage of memory T-cell pools. Concomitantly, naive CD4+ T-cell depletion may result from this high immune activation driving cells into another phenotype but may also be due to a deficiency of CD4+ T-cell regeneration or sequestration to another site. Sequential studies in individual patients are required to further elucidate this, given the dynamic nature of HIV-1 infection.
In summary, we have identified a subset of HIV-1–infected patients, discord controllers, who despite maintaining low plasma viral RNA loads experience disease progression. With the exception of CD8+ T-cell subset distribution and activation, we show that discord controllers are similar to progressors, based on the CD4+ T-cell compartment and HIV-1 DNA load, further supporting data that plasma viral RNA load alone does not lead to CD4+ T-cell decline. Rather, we have demonstrated increased CD4+ T-cell activation in discord controllers, which may relate to the higher viral DNA load in this group. This higher viral DNA load suggests comparatively higher levels of virus turnover in the discord controllers, despite controlled plasma viral RNA loads, possibly explaining disease progression. Moreover, the fact that spontaneous control of viral replication does not necessarily confer protection against disease progression highlights the need to intensify the search for new therapies aimed at normalizing perturbations of the T-cell compartment.
ACKNOWLEDGMENTS
We thank Eithne O’Sullivan, Corinna Pade, Carl DeSouza, James Hand, and Meaghan Kall for consenting patients and for sample collection. We also thank Professor Judy Breuer for useful discussions and pointing out the discord controller phenotype.
Supported by MRC Senior Non-Clinical Fellowship awarded to A.M. (G117/547), Wellcome Trust grant (WT075853MA), Barts and the London Charity Grant (MMBG1E7R), BHIVA SpR Research Grant (MMBG1F2R).
Footnotes
Presented at the 15th Conference on Retroviruses and Opportunistic Infections, February 3–6, 2008, Boston, MA (poster 352) and the AIDS Vaccine Conference, October 19–22, 2009, Paris, France (poster).
The authors have no conflicts of interests to disclose.
REFERENCES
- 1.Haase AT. Population biology of HIV-1 infection: viral and CD4+ T cell demographics and dynamics in lymphatic tissues. Annu Rev Immunol. 1999;17:625–656. doi: 10.1146/annurev.immunol.17.1.625. [DOI] [PubMed] [Google Scholar]
- 2.Hellerstein M, Hanley MB, Cesar D, et al. Directly measured kinetics of circulating T lymphocytes in normal and HIV-1-infected humans. Nat Med. 1999;5:83–89. doi: 10.1038/4772. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez B, Sethi AK, Cheruvu VK, et al. Predictive value of plasma HIV RNA level on rate of CD4 T-cell decline in untreated HIV infection. JAMA. 2006;296:1498–1506. doi: 10.1001/jama.296.12.1498. [DOI] [PubMed] [Google Scholar]
- 4.Deeks SG, Kitchen CMR, Liu L, et al. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood. 2004;104:942–947. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- 5.Giorgi JV, Hultin LE, McKeating JA, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis. 1999;179:859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, Cumberland WG, Hultin LE, et al. Elevated CD38 antigen expression on CD8+ T cells is a stronger marker for the risk of chronic HIV disease progression to AIDS and death in the Multicenter AIDS Cohort Study than CD4+ cell count, soluble immune activation markers, or combinations of HLA-DR and CD38 expression. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;16:83–92. doi: 10.1097/00042560-199710010-00003. [DOI] [PubMed] [Google Scholar]
- 7.Silvestri G, Paiardini M, Pandrea I, et al. Understanding the benign nature of SIV infection in natural hosts. J Clin Invest. 2007;117:3148–3154. doi: 10.1172/JCI33034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hazenberg MD, Otto SA, van Benthem BH, et al. Persistent immune activation in HIV-1 infection is associated with progression to AIDS. AIDS. 2003;17:1881–1888. doi: 10.1097/00002030-200309050-00006. [DOI] [PubMed] [Google Scholar]
- 9.Bofill M, Mocroft A, Lipman M, et al. Increased numbers of primed activated CD8+CD38+CD45RO+ T cells predict the decline of CD4+ T cells in HIV-1-infected patients. AIDS. 1996;10:827–834. doi: 10.1097/00002030-199607000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Sousa AE, Carneiro J, Meier-Schellersheim M, et al. CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. J Immunol. 2002;169:3400–3406. doi: 10.4049/jimmunol.169.6.3400. [DOI] [PubMed] [Google Scholar]
- 11.Choudhary SK, Vrisekoop N, Jansen CA, et al. Low immune activation despite high levels of pathogenic human immunodeficiency virus type 1 results in long-term asymptomatic disease. J Virol. 2007;81:8838–8842. doi: 10.1128/JVI.02663-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roederer M, Dubs JG, Anderson MT, et al. CD8 naive T cell counts decrease progressively in HIV-infected adults. J Clin Invest. 1995;95:2061–2066. doi: 10.1172/JCI117892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabin RL, Roederer M, Maldonado Y, et al. Altered representation of naive and memory CD8 T cell subsets in HIV-infected children. J Clin Invest. 1995;95:2054–2060. doi: 10.1172/JCI117891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goujard C, Bonarek M, Meyer L, et al. CD4 cell count and HIV DNA level are independent predictors of disease progression after primary HIV type 1 infection in untreated patients. Clin Infect Dis. 2006;42:709–715. doi: 10.1086/500213. [DOI] [PubMed] [Google Scholar]
- 15.Avettand-Fenoel V, Boufassa F, Galimand J, et al. HIV-1 DNA for the measurement of the HIV reservoir is predictive of disease progression in seroconverters whatever the mode of result expression is. J Clin Virol. 2008;42:399–404. doi: 10.1016/j.jcv.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Rouzioux C, Hubert J-B, Burgard M, et al. Early levels of HIV-1 DNA in peripheral blood mononuclear cells are predictive of disease progression independently of HIV-1 RNA levels and CD4+ T cell counts. J Infect Dis. 2005;192:46–55. doi: 10.1086/430610. [DOI] [PubMed] [Google Scholar]
- 17.Walker BD. Elite control of HIV Infection: implications for vaccines and treatment. Top HIV Med. 2007;15:134–136. [PubMed] [Google Scholar]
- 18.Hunt PW, Brenchley J, Sinclair E, et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis. 2008;197:126–133. doi: 10.1086/524143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereyra F, Addo MM, Kaufmann DE, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis. 2008;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 20.Andrade A, Bailey JR, Xu J, et al. CD4+ T cell depletion in an untreated HIV type 1-infected human leukocyte antigen-B*5801-positive patient with an undetectable viral load. Clin Infect Dis. 2008;46:e78–e82. doi: 10.1086/529387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandalia S, Westrop SJ, Beck EJ, et al. Are long-term non-progressors very slow progressors? Insights from the Chelsea and Westminster HIV cohort, 1988-2010. PLoS One. 2012;7:e29844. doi: 10.1371/journal.pone.0029844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sterne JA, May M, Costagliola D, et al. Timing of initiation of antiretroviral therapy in AIDS-free HIV-1-infected patients: a collaborative analysis of 18 HIV cohort studies. Lancet. 2009;373:1352–1363. doi: 10.1016/S0140-6736(09)60612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dreja H, O’Sullivan E, Pade C, et al. Neutralization activity in a geographically diverse East London cohort of human immunodeficiency virus type 1-infected patients: clade C infection results in a stronger and broader humoral immune response than clade B infection. J Gen Virol. 2010;91:2794–2803. doi: 10.1099/vir.0.024224-0. [DOI] [PubMed] [Google Scholar]
- 24.Yang C, Dash BC, Simon F, et al. Detection of diverse variants of human immunodeficiency virus-1 groups M, N, and O and simian immunodeficiency viruses from chimpanzees by using generic pol and env primer pairs. J Infect Dis. 2000;181:1791–1795. doi: 10.1086/315439. [DOI] [PubMed] [Google Scholar]
- 25.Ndembi N, Takehisa J, Zekeng L, et al. Genetic diversity of HIV type 1 in rural eastern Cameroon. J Acquir Immune Defic Syndr. 2004;37:1641–1650. doi: 10.1097/00126334-200412150-00019. [DOI] [PubMed] [Google Scholar]
- 26.Drosten C, Panning M, Drexler JF, et al. Ultrasensitive monitoring of HIV-1 viral load by a low-cost real-time reverse transcription-PCR assay with internal control for the 5′ long terminal repeat domain. Clin Chem. 2006;52:1258–1266. doi: 10.1373/clinchem.2006.066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo W, Yang H, Rathbun K, et al. Detection of human immunodeficiency virus type 1 DNA in dried blood spots by a duplex real-time PCR assay. J Clin Microbiol. 2005;43:1851–1857. doi: 10.1128/JCM.43.4.1851-1857.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo YM, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potter SJ, Lacabaratz C, Lambotte O, et al. Preserved central memory and activated effector memory CD4+ T-cell subsets in human immunodeficiency virus controllers: an ANRS EP36 study. J Virol. 2007;81:13904–13915. doi: 10.1128/JVI.01401-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez M, Soriano V, Peris-Pertusa A, et al. Elite controllers display higher activation on central memory CD8 T cells than HIV patients successfully on HAART. AIDS Res Hum Retroviruses. 2011;27:157–165. doi: 10.1089/aid.2010.0107. [DOI] [PubMed] [Google Scholar]
- 31.Emu B, Sinclair E, Favre D, et al. Phenotypic, functional, and kinetic parameters associated with apparent T-cell control of human immunodeficiency virus replication in individuals with and without antiretroviral treatment. J Virol. 2005;79:14169–14178. doi: 10.1128/JVI.79.22.14169-14178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambotte O, Boufassa F, Madec Y, et al. HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin Infect Dis. 2005;41:1053–1056. doi: 10.1086/433188. [DOI] [PubMed] [Google Scholar]
- 33.Hammer SM, Squires KE, Hughes MD, et al. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 34.Dyrhol-Riise AM, Voltersvik P, Røsok BI, et al. Normalization of CD4+ cell numbers and reduced levels of memory CD8+ cells in blood and tonsillar tissue after highly active antiretroviral therapy in early HIV type-1 infection. AIDS Res Hum Retroviruses. 2000;16:191–201. doi: 10.1089/088922200309287. [DOI] [PubMed] [Google Scholar]
- 35.Tilling R, Kinloch S, Goh LE, et al. Parallel decline of CD8+/CD38++ T cells and viraemia in response to quadruple highly active antiretroviral therapy in primary HIV infection. AIDS. 2002;16:589–596. doi: 10.1097/00002030-200203080-00010. [DOI] [PubMed] [Google Scholar]
- 36.Casazza JP, Betts MR, Picker LJ, et al. Decay kinetics of human immunodeficiency virus-specific CD8+ T cells in peripheral blood after initiation of highly active antiretroviral therapy. J Virol. 2001;75:6508–6516. doi: 10.1128/JVI.75.14.6508-6516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mellors JW, Rinaldo CR, Gupta P, et al. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 38.Liu Z, Cumberland WG, Hultin LE, et al. CD8+ T-lymphocyte activation in HIV-1 disease reflects an aspect of pathogenesis distinct from viral burden and immunodeficiency. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;18:332–340. doi: 10.1097/00042560-199808010-00004. [DOI] [PubMed] [Google Scholar]
- 39.Okulicz JF, Grandits GA, Weintrob AC, et al. CD4 T cell count reconstitution in HIV controllers after highly active antiretroviral therapy. Clin Infect Dis. 2010;50:1187–1191. doi: 10.1086/651421. [DOI] [PubMed] [Google Scholar]
- 40.Bucy RP, Hockett RD, Derdeyn CA, et al. Initial increase in blood CD4 (+) lymphocytes after HIV antiretroviral therapy reflects redistribution from lymphoid tissues. J Clin Invest. 1999;103:1391–1398. doi: 10.1172/JCI5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cara A, Vargas J, Keller M, et al. Circular viral DNA and anomalous junction sequence in PBMC of HIV-infected individuals with no detectable plasma HIV RNA. Virology. 2002;292:1–5. doi: 10.1006/viro.2001.1243. [DOI] [PubMed] [Google Scholar]
- 42.Ibanez A, Puig T, Elias J, et al. Quantification of integrated and total HIV-1 DNA after long-term highly active antiretroviral therapy in HIV-1-infected patients. AIDS. 1999;13:1045–1049. doi: 10.1097/00002030-199906180-00007. [DOI] [PubMed] [Google Scholar]
- 43.Viard J-P, Burgard M, Hubert J-B, et al. Impact of 5 years of maximally successful highly active antiretroviral therapy on CD4 cell count and HIV-1 DNA level. AIDS. 2004;18:45–49. doi: 10.1097/00002030-200401020-00005. [DOI] [PubMed] [Google Scholar]
- 44.Benito JM, Lopez M, Soriano V. The role of CD8+ T-cell response in HIV infection. AIDS Rev. 2004;6:79–88. [PubMed] [Google Scholar]
- 45.Ogg GS, Jin X, Bonhoeffer S, et al. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 46.Jin X, Bauer DE, Tuttleton SE, et al. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guadalupe M, Reay E, Sankaran S, et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol. 2003;77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]