Abstract
The aim of this study was to explore the role of vitamin-D in rheumatoid arthritis (RA) pathogenesis by investigating enrichment of vitamin-D response elements (VDREs) in confirmed RA susceptibility loci and testing variants associated with vitamin-D levels for association with RA.
Bioinformatically, VDRE genomic positions were overlaid with non HLA confirmed RA susceptibility regions. The number of VDREs at RA loci was compared to a randomly selected set of genomic loci to calculate an average relative risk (RR). SNPs in the DHCR7/NADSYN1 and CYP2R1 loci, previously associated with circulating vitamin-D levels, were tested in UK RA cases (n = 3870) and controls (n = 8430).
Significant enrichment of VDREs was seen at RA loci (p=9.23×10−8) when regions were defined either by gene (RR 5.50) or position (RR 5.86). SNPs in the DHCR7/NADSYN1 locus showed evidence of positive association with RA, rs4944076 (p=0.008, OR 1.14 95% CI 1.03-1.24).
The significant enrichment of VDREs at RA associated loci and the modest association of variants in loci controlling levels of circulating vitamin-D, supports the hypothesis that vitamin-D plays a role in the development of RA.
INTRODUCTION
Vitamin D is a steroid hormone which plays an important role in many processes 1. Vitamin D acts through its nuclear receptor, the vitamin D receptor (VDR), the expression of which on immune cells raises the question of a role for the hormone in the control of the immune system. Numerous lines of evidence support a regulatory role for vitamin D; indeed the active form, calcitriol or 1,25,dihydroxyvitamin D3 (VitD3), has been found to control over 200 genes including those involved in cell differentiation, proliferation, apoptosis and angiogenesis 2-4. The VDR acts by binding to specific sequences in DNA known as vitamin D response elements (VDREs), which result in transcriptional regulation of vitamin D responsive genes.
Rheumatoid arthritis (RA) is a chronic inflammatory autoimmune disease which causes inflammation of synovial joints. RA is thought to arise as a result of an autoimmune reaction due to a breach in self tolerance inducing a type 1 T helper cell (Th1) driven immune response which causes infiltration of immune cells into the joint and secretion of pro-inflammatory cytokines 5.
Vitamin D deficiency is common in RA and the active form of vitamin D can alter several aspects of the immune responses which are pivotal to RA pathogenesis. The cytokine profile of immune cells can be affected, for example production of IL-2 and IFN- γ which are important immune mediators by Th1 lymphocytes is inhibited resulting in a shift towards a regulatory type 2 T helper cell (Th2) phenotype. Inhibition of IFN- γ production also reduces antigen presentation by antigen presenting cells (APCs) and in turn reduces T cell activation, acting as a negative feedback regulator 6. Vitamin D inhibits the differentiation of monocytes to dendritic cells 7, reduces the production of IL-12 by dendritic cells, and stimulates phagacytosis of bacteria by macrophages 8;9. Boonstra et al showed that vitamin D increased the production of IL-4, IL-5, and IL-10 by Th2 cells, IL-4 and IL-10 normally act to inhibit Th1 function (10). Vitamin D has also been shown to inhibit the activation of Th17 cells by inhibiting the expression of IL-6 11;12. In addition vitamin D can affect the invasiveness of cultured RA fibroblast-like synoviocytes (FLS), where higher concentrations of calcitriol were shown to significantly decrease the invasion of RA FLS by 53%13. The expression of matrix metalloproteinases (MMP), such as MMP-1 and MMP-2 can also be affected by vitamin D. MMPs are key molecules involved in the destruction of cartilage and bone in RA. IL-1β is known to increase the expression of MMP-1 by RA FLS, Laragione et al have shown that in both human and rats treatment of activated FLS with calcitriol significantly inhibited IL-1β induced expression of MMP-1 by 73-5%13.
Given its immunoregulatory potential, a role for Vitamin D has been postulated in autoimmune diseases (AID). Circumstantial evidence supports this possibility; for example, circulating vitamin D levels have been reported to correlate with the risk of multiple sclerosis (MS) 14, type 1 diabetes (T1D) 15 and Crohn’s disease (CD) 16. In rheumatoid arthritis (RA), decreased serum levels of precursor vitamin D (25(OH)D) and active VitD3 have been reported in cross-sectional studies 17. However findings may be confounded by the decreased physical activity and reduced sun exposure that RA patients with disability experience, which means that it is not clear whether the associations observed are cause or effect. With regards to disease onset, the evidence surrounding vitamin D intake is conflicting. Merlino et al showed an inverse prospective association between high vitamin D intake and the risk of RA in women from the Iowa health study 18, however a large prospective study of 180,000 women showed no association between vitamin D intake and subsequent development of RA 19.
Low serum levels of vitamin D have also been associated with increased disease severity 20-25; however the regulatory genes associated with disease severity may be different to those associated with susceptibility, therefore there may be a different set of genes regulating disease severity which remain unexplored for VDRE enrichment.
Thus, there is conflicting epidemiological evidence for a role of vitamin D in influencing disease susceptibility to RA and differentiating cause from effect is challenging. However, there is emerging genetic evidence that vitamin D levels may be linked to the onset of AIDs. First, a recent study by Ramagopalan et al determined VDREs throughout the genome using chromatin immunoprecipitation (ChIP) followed by massively parallel sequencing (ChIP-seq)26. The authors identified VDREs in lymphoblastoid cell lines (LCLs) from two individuals of European ancestry before and after calcitriol (VitD3) stimulation. In the basal state, before stimulation, 623 genomic sites were identified, whilst after calcitriol stimulation, the number of VDRE sites increased to 2776. The authors demonstrated significant enrichment of VDREs in known AID susceptibility loci, including CD, systemic lupus erythematosus (SLE), T1D and MS (p value < 0.001 in all diseases). Interestingly, the study also tested 16 RA susceptibility loci and found significant VDRE enrichment (p < 0.001). However only 9 of the 16 loci have been confirmed to be associated with RA, in addition there are now 45 non-HLA loci confirmed to be associated with RA susceptibility 27
The second piece of genetic evidence to support a role for vitamin D in the aetiology of AID comes from a recent study in a T1D cohort, this study confirmed the association of four vitamin D metabolism genes (GC, DHCR7/NADSYN1, CYP2R1, CYP24A1) with vitamin D levels in healthy controls; in addition, two of these genes showed association with T1D susceptibility (DHCR7 p1.2×10−3 and CYP2R1 p=3.0×10−3) 28.
The association of the same variants in the same genes that control circulating levels of vitamin D with an AID provides an unbiased test of the hypothesis that vitamin D levels are important in the aetiology of AIDs (mendelian randomisation) 29. Therefore the aims of the current study were, first, to investigate potential enrichment of VDREs in RA susceptibility loci that have been confirmed to date 27; second, to test variants previously associated with circulating levels of vitamin D and T1D susceptibility for association with RA susceptibility.
RESULTS
Enrichment of VDREs at RA loci
Out of the 46 RA regions defined by gene a total of 39 VDREs were identified in 17 RA regions, showing that 37% of RA regions contain VDREs. The relative risk for RA associated genes harbouring VDRES compared to random genes was 5.5 (95% CI 2.55-26.33).
Analysing the data using a simple 2×2 table also showed a significant increase in VDREs in RA associated loci compared to the rest of the protein coding genes in the genome obtained from ensembl build 65, p=1.20×10−10 (OR 5.72, 95% CI 2.95-10.79).
However this 2×2 table method does not take into account the number of VDREs in each gene region; therefore a trend test was carried out using a 2×16 table (online supplementary Table 2), which also showed a significant enrichment of VDREs (p=9.23×10−8)
When defining regions by gene, the RA defined gene may not be the true causal gene. To overcome this issue we also defined regions by position, extending 50kb up and downstream of the associated SNP site. Defining regions by SNP position identified 25 VDREs in 16 out of 46 regions (35.5% of RA associated SNPs contain VDREs within 50kb) again showing a significant enrichment of VDREs at RA loci (after 100,000 randomisations RR 5.86, 95% CI 2.04-51). However only one RA associated SNP was shown to lie directly within a VDRE; rs947474 is located on chromosome 10 and maps within the PRKCQ locus.
Association of SNPs in vitamin D gene regions with RA
DHCR7/NADSYN1
After QC 360 SNPs in 3870 UK RA cases and 8430 UK controls remained for analysis capturing 59% of variation across the DHCR7/NADSYN1 locus (r2>0.8). Weak association (p=0.04) was seen at rs11600569, which is in complete linkage disequilibrium (LD) with the vitamin D SNP associated with T1D (rs12785878), the odds ratio is the same in both diseases (OR 1.07) (Table 2) 28.
Table 2. Logistic regression results showing top SNP associations in the DHCR7/NADSYN1 and CYP2R1 regions with RA.
| DHCR7/NADSYN1 | |||||||
|---|---|---|---|---|---|---|---|
| SNP | SNP position |
SNP type |
MAF in controls |
MAF in cases |
P value | OR | 95% CI |
| rs4944076 | 70889302 | intronic | 0.09 | 0.10 | 0.008 | 1.135 | 1.034:1.246 |
| rs4944997 | 70884016 | intronic | 0.09 | 0.10 | 0.008 | 1.135 | 1.033:1.246 |
| rs2919722 | 70631179 | intronic | 0.40 | 0.42 | 0.014 | 1.072 | 1.014:1.133 |
| rs1002171 | 70895219 | intronic | 0.05 | 0.05 | 0.014 | 1.166 | 1.031:1.319 |
| rs11600569 | 70851395 | intronic | 0.22 | 0.23 | 0.040 | 1.071 | 1.003:1.143 |
| CYP2R1 | |||||||
| rs117162870 | 14782929 | intronic | 0.04 | 0.05 | 0.04 | 1.15 | 1.009:1.305 |
| rs116856365 | 14726866 | intronic | 0.04 | 0.05 | 0.04 | 1.42 | 1.004:1.300 |
| rs7116978 | 14838347 | intronic | 0.37 | 0.38 | 0.05 | 1.06 | 0.9996:1.118 |
| rs6486205 | 14837832 | intronic | 0.37 | 0.38 | 0.05 | 1.06 | 0.999:1.118 |
MAF: minor allele frequency, OR: odds ratio, CI: confidence interval. rs11600569 shown in bold is perfectly correlated (r2=1) with rs12785878 previously associated with vitamin D levels and T1D 28.
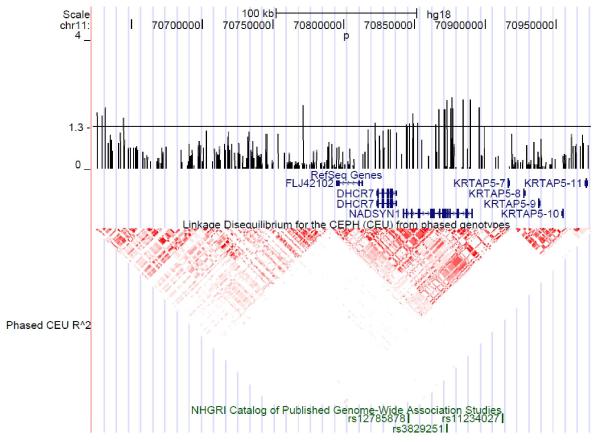
Association was seen between rs4944076, located in an intron of nicotinamide adenine dinucleotide synthase 1 (NADSYN1) and RA (p=0.008, OR 1.14, 95% CI 1.03-1.24) after PCA correction for geographical variation (Figure 1, Table 2, full results supplementary table 3). This SNP was modestly correlated (r2 0.3) with rs12785878 28.
Figure 1.
Fine mapping results showing the −log10 p values of SNPs in the DHCR7/NADSYN1 region associated with RA. The p value threshold is set at 1.3 which is equivalent to a p value of 0.05. Reference sequence genes are shown in blue, HapMap LD is shown in red. SNPs in the region previously associated with circulating levels of vitamin D through GWAS are shown in green.
CYP2R1
After QC 177 SNPs remained for analysis, capturing 47% of the variation across the CYP2R1 region (r2>0.8). Weak association was seen at 4 SNPs (Table 2): two of these SNPs, rs7116978 and rs6486205 (p=0.05 OR 1.06 95% CI 0.99:1.18) are highly correlated with rs10741657 and rs2060793 (r2 0.87), previously associated with vitamin D levels and T1D 28 (Supplementary figure 1).
DISCUSSION
Although RA loci have previously been shown to be enriched for VDREs, this was in a total of 16 non confirmed loci, of which only 9 have since been validated as RA susceptibility loci. Here we perform the first comprehensive analysis of VDREs in all 45 confirmed non-HLA RA susceptibility loci. We have shown that genes or regions surrounding SNPs associated with susceptibility to RA are significantly enriched for VDREs. Indeed the VDR has a binding site at 17 out of 46 confirmed non HLA RA associated genes, and binds within 50kb of 35.5% of RA associated SNPs resulting in a relative risk of 5.86 compared with the rest of the genome. We have also shown that SNPs within the vitamin D associated loci DHCR7/NADSYN1 show modest association with RA susceptibility, suggesting that vitamin D may play a role in the development of RA.
There are some points that should be considered when interpreting these results. Primarily, the RA associated genes as defined by this study may not represent the true causal gene. Indeed when genes are assigned to loci any functionality that is associated is largely speculative, and involves variants being assigned to the nearest gene, or to a gene in the region which is a plausible biological candidate. Therefore we used GRAIL which is the best method by which to assign genes to loci in a non biased fashion. We also used a position based-approach as well as a gene-based approach with the assumption that a causal gene will lie reasonably close to the currently defined associated variant. However, associated variants may not exert their effects on nearby genes, but may be acting on genes some distance away in cis or trans 30.
Although most regions have been fine-mapped using the immunochip (a custom SNP array designed for dense genotyping of 186 loci identified through genome wide association studies)27, in large sample collections, it is possible that the true causal variants are not the currently defined RA associated variants or those in tight linkage disequilibrium with them and, indeed, independent associations may exist in already confirmed susceptibility loci. Therefore more causal variants may exist within VDREs than indicated here and functional studies to define causal SNPs will be necessary to understand the full picture.
Finally, it is possible that some genes and SNPs included in the randomised control groups will ultimately be associated with RA once all contributing genetic factors have been identified. However, that would reduce the likelihood of a difference being observed between currently defined RA loci and random non-RA, associated loci and would be more likely to lead to a type 2 error.
Reassuringly, results from the two methods to define RA regions produced very similar results. In addition defining region by position showed an increased RR compared to defining regions by gene (RR 5.86, RR5.50 respectively) suggesting that as the regions are more specifically centred on the RA associated variant, VDRE enrichment increases.
Several studies have found reduced vitamin D levels in individuals with RA and other AIDs 14;16;17;19 but determining whether their association with disease is cause or effect is challenging. However, several SNPs have been reproducibly associated with vitamin D levels 28;31: if the same SNPs are also associated with disease, it provides strong evidence that the pathway is causal (Mendelian randomisation)29. In the current study, the genetic results are of borderline significance; a SNP perfectly correlated (r2=1) with the T1D-associated variant showed modest association (rs11600569 p=0.04) after correction for geographical variation. Although this association would not remain significant after correction for multiple testing, it is interesting to note that the effect size seen for the association of this SNP with RA is the same as was previously identified in T1D (OR 1.07) 28 increasing the plausibility of this result. In addition we have shown association between other SNPs in the DHCR7/NADSYN1 locus and RA (p=0.008), and although these SNPs have not been previously associated with vitamin D levels, since the association between SNPs in this region and vitamin D levels was identified by genome wide association studies, the causal variant in the region responsible for altering vitamin D levels has not yet been determined and it is possible that a secondary association in the region could be identified.
All variants associated with vitamin D levels (table 2) are intronic variants. The most associated variant rs4944076 lies in an intron of NADSYN1. Bioinformatics analysis using the program ASSIMILATOR 32 which retrieves and queries the experimental data generated by ENCODE which is available on the UCSC web browser, has shown that rs4944076 lies within a region of open chromatin, DNase 1 hypersensitivity and histone modification suggesting possible regulatory potential. In addition one SNP, rs4944997 has been found to be an expression quantitative trait locus, potentially regulating the expression of NADSYN133. However the associations at this locus are modest and will require replication and identification of the functional variant before speculation of the functional effect of the variant can be determined.
The main source of vitamin D comes from endogenous production in the skin after exposure to UVB light from the sun which results in the conversion of 7-dehydrocholesterol (present in the skin) to pre-vitamin D3. This is then metabolised in the liver by CYP2R1 and CYP27A1 enzymes to form 25(OH)D, the major circulating form of vitamin D which is converted to its active form 1,25,(OH)2D3 (calcitriol) in the kidneys by CYP27B1. DHCR7 encodes an enzyme which catalyses the conversion of 7-dehydroxycholesterol o cholesterol, which removes pro-cholesterol from the vitamin D pathway, reducing the availability of 7-dehydrocholesterol for conversion to 25(OH)D. We could speculate that variants resulting in increased DHCR7 activity could therefore lead to increased removal of 7-dehydrocholesterol from the vitamin D pathway causing vitamin D deficiency.
In conclusion analysis of all 45 confirmed RA non-HLA susceptibility loci to date has shown a significant enrichment of VDREs at RA loci; in addition we have shown the modest association of SNPs previously associated with vitamin D levels, with RA. Validation of this finding is required in larger, independent studies. If confirmed, the genetic association along with the significant enrichment of VDREs in RA associated loci would provide supportive evidence for the involvement of vitamin D in RA and may pave the way for future trials of vitamin D therapy to prevent RA.
METHODS
Analysis of enrichment of VDREs at RA loci
The enrichment of VDREs at confirmed RA loci 27 was investigated bioinformatically to compare the genomic location of defined RA loci with the genomic positions of VDREs identified by Ramagopalan et al. A total of 2776 VDREs identified by ChIP-seq in LCLs after calcitriol (VitD3) stimulation were obtained from Ramagopalan et al 26, and were assigned to the nearest gene within 100kb using the Ensembl genome browser (www.ensembl.org) build 65. The 100kb threshold was selected arbitrarily; it is possible that VDREs act on more distant genes however it has been shown that calcitriol responsive genes have a VDRE at a median distance of 66.6Kb from the transcription start site.
RA loci were defined using the ensembl genome sequence in two ways:
First, regions were defined based on the most plausible candidate gene, a query gene list was created using the 45 non HLA loci defined in a recent study by Eyre et al 27 (supplementary table 1). Each associated single nucleotide polymorphism (SNP) from this study was assigned to the most functionally plausible candidate gene using GRAIL (Gene relationships across implicated loci) 34. GRAIL uses the literature to identify relationships between genes and selects the best candidate gene in a region in relation to a particular phenotype. One locus contained multiple candidate genes (IL2-IL21) and therefore was included twice, creating a final list of 46 genes in the query list.
Second, regions were defined using the base pair position of each associated SNP from Eyre et al and defining the RA loci by extending 50kb up and downstream of each of the associated SNP (one locus IKZF3 had two candidate SNPs, see supplementary table 1).
The genomic locations of these defined RA loci obtained from Ensembl were then compared to the genomic positions of the VDREs from Ramagopalan et al 26 using perl scripts, to determine the number of VDREs present in RA loci.
To identify an enrichment of VDREs a comparison set of the same number of random loci was created. Perl scripts were used to select random genes from all protein coding genes in the genome (except those already associated with RA) or base pair positions from the ensembl genome sequence and regions were defined in the same way as described for the RA loci. In total 100,000 randomly selected comparison gene sets were generated and VDRE enrichment was determined by assessing the relative risk. This was calculated by dividing the number of VDREs in the query gene list by the average number of VDREs in 100,000 randomisations. Some random gene sets will undoubtedly contain zero VDREs but the risk ratio cannot be calculated if there are no events in the control group. To overcome this problem, 0.5 was added to the total number of VDREs in all gene sets including the query list.
STATA version 11 was then used to calculate p values. A chi2 test was performed to test the null hypothesis that the probability of a VDRE existing at a given locus did not depend on whether that locus was close to a region associated with RA risk, by creating a 2×2 table cross-classifying genes by association with RA and presence of a VDRE. As this method does not take into account the number of VDREs in each region a trend test was performed using a 2×N table.
Analysis of SNPs associated with circulating levels of Vitamin D
The regions surrounding two genes previously associated with circulating levels of vitamin D and T1D were fine mapped as part of a larger study 27 (DHCR7/NADSYN1 and CYP2R1) 28;31. All known SNPs in the region from HapMap and the 1000 genomes project low coverage whole genome sequencing pilot, were included for fine mapping. Genotyping was carried out using a custom Illumina infinium genotyping chip, in 4752 UK RA cases and 9006 UK RA controls (described previously 27).
As the DHCR7/NADSYN1 locus lies in a region identified by the Wellcome Trust Case Control Consortium (WTCCC) 35 to be a region in which allele frequencies show geographic differentiation in the UK, principle components analysis (PCA) 36 was carried out (described previously 27) to correct for this. The study was approved by the North West Ethics Committee (MREC 99/8/84).
Supplementary Material
Table 1. VDRE enrichment results when regions are defined by gene or SNP position.
| Chi2 | |||||
|---|---|---|---|---|---|
| RR | 95% CI | P value | OR | 95% CI | |
| Defining regions by gene | 5.50 | 2.55-26.33 | 1.21×10−10 | 5.72 | 2.95-10.79 |
|
Defining regions by position
(50kb) |
5.86 | 2.04-51 |
RR: relative risk, CI: confidence interval, OR: odds ratio.
Acknowledgements
We thank Arthritis Research UK for their support (grant ref 17552). This work was supported by the innovative medicines initiative joint undertaking (IMI JU) funded project BeTheCure, (contract number 115142-2).
Footnotes
Conflict of interests: The authors declare that they have no conflicting interests.
Supplementary information is available at http://www.nature.com/gene/index.html
Reference List
- (1).Holick MF. Vitamin D deficiency. N Engl J Med. 2007;357(3):266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- (2).Holick MF. Resurrection of vitamin D deficiency and rickets. J Clin Invest. 2006;116(8):2062–2072. doi: 10.1172/JCI29449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).DeLuca HF. Overview of general physiologic features and functions of vitamin D. Am J Clin Nutr. 2004;80(6 Suppl):1689S–1696S. doi: 10.1093/ajcn/80.6.1689S. [DOI] [PubMed] [Google Scholar]
- (4).Holick MF. High prevalence of vitamin D inadequacy and implications for health. Mayo Clin Proc. 2006;81(3):353–373. doi: 10.4065/81.3.353. [DOI] [PubMed] [Google Scholar]
- (5).McInnes IB, O’Dell JR. State-of-the-art: rheumatoid arthritis. Ann Rheum Dis. 2010;69(11):1898–1906. doi: 10.1136/ard.2010.134684. [DOI] [PubMed] [Google Scholar]
- (6).Cippitelli M, Santoni A. Vitamin D3: a transcriptional modulator of the interferon-gamma gene. Eur J Immunol. 1998;28(10):3017–3030. doi: 10.1002/(SICI)1521-4141(199810)28:10<3017::AID-IMMU3017>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- (7).DeLuca HF, Cantorna MT. Vitamin D: its role and uses in immunology. FASEB J. 2001;15(14):2579–2585. doi: 10.1096/fj.01-0433rev. [DOI] [PubMed] [Google Scholar]
- (8).Griffin MD, Lutz WH, Phan VA, Bachman LA, McKean DJ, Kumar R. Potent inhibition of dendritic cell differentiation and maturation by vitamin D analogs. Biochem Biophys Res Commun. 2000;270(3):701–708. doi: 10.1006/bbrc.2000.2490. [DOI] [PubMed] [Google Scholar]
- (9).Griffin MD, Lutz W, Phan VA, Bachman LA, McKean DJ, Kumar R. Dendritic cell modulation by 1alpha,25 dihydroxyvitamin D3 and its analogs: a vitamin D receptor-dependent pathway that promotes a persistent state of immaturity in vitro and in vivo. Proc Natl Acad Sci U S A. 2001;98(12):6800–6805. doi: 10.1073/pnas.121172198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Boonstra A, Barrat FJ, Crain C, Heath VL, Savelkoul HFJ, O’Garra A. 1{alpha},25-Dihydroxyvitamin D3 Has a Direct Effect on Naive CD4+ T Cells to Enhance the Development of Th2 Cells. J Immunol. 2001;167(9):4974–4980. doi: 10.4049/jimmunol.167.9.4974. [DOI] [PubMed] [Google Scholar]
- (11).Stockinger B. Th17 cells: An orphan with influence. Immunol Cell Biol. 2007;85(2):83–84. doi: 10.1038/sj.icb.7100035. [DOI] [PubMed] [Google Scholar]
- (12).Xue ML, Zhu H, Thakur A, Willcox M. 1 alpha,25-Dihydroxyvitamin D3 inhibits proinflammatory cytokine and chemokine expression in human corneal epithelial cells colonized with Pseudomonas aeruginosa. Immunol Cell Biol. 2002;80(4):340–345. doi: 10.1046/j.1440-1711.80.4august.1.x. [DOI] [PubMed] [Google Scholar]
- (13).Laragione T, Shah A, Gulko PS. The vitamin D receptor regulates rheumatoid arthritis synovial fibroblast invasion and morphology. Mol Med. 2012;18:194–200. doi: 10.2119/molmed.2011.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ascherio A, Munger KL, Simon KC. Vitamin D and multiple sclerosis. The Lancet Neurology. 2010;9(6):599–612. doi: 10.1016/S1474-4422(10)70086-7. [DOI] [PubMed] [Google Scholar]
- (15).Mathieu C, Gysemans C, Giulietti A, Bouillon R. Vitamin D and diabetes. Diabetologia. 2005;48(7):1247–1257. doi: 10.1007/s00125-005-1802-7. [DOI] [PubMed] [Google Scholar]
- (16).Pappa HM, Grand RJ, Gordon CM. Report on the vitamin D status of adult and pediatric patients with inflammatory bowel disease and its significance for bone health and disease. Inflamm Bowel Dis. 2006;12(12):1162–1174. doi: 10.1097/01.mib.0000236929.74040.b0. [DOI] [PubMed] [Google Scholar]
- (17).Als OS, Riis B, Christiansen C. Serum concentration of vitamin D metabolites in rheumatoid arthritis. Clin Rheumatol. 1987;6(2):238–243. doi: 10.1007/BF02201030. [DOI] [PubMed] [Google Scholar]
- (18).Merlino LA, Curtis J, Mikuls TR, Cerhan JR, Criswell LA, Saag KG. Vitamin D intake is inversely associated with rheumatoid arthritis: results from the Iowa Women’s Health Study. Arthritis Rheum. 2004;50(1):72–77. doi: 10.1002/art.11434. [DOI] [PubMed] [Google Scholar]
- (19).Costenbader KH, Feskanich D, Holmes M, Karlson EW, ito-Garcia E. Vitamin D intake and risks of systemic lupus erythematosus and rheumatoid arthritis in women. Ann Rheum Dis. 2008;67(4):530–535. doi: 10.1136/ard.2007.072736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Haga HJ, Schmedes A, Naderi Y, Moreno AM, Peen E. Severe deficiency of 25-hydroxyvitamin D(3) (25-OH-D (3)) is associated with high disease activity of rheumatoid arthritis. Clin Rheumatol. 2013 doi: 10.1007/s10067-012-2154-6. [DOI] [PubMed] [Google Scholar]
- (21).Kostoglou-Athanassiou I, Athanassiou P, Lyraki A, Raftakis I, Antoniadis C. Vitamin D and rheumatoid arthritis. Ther Adv Endocrinol Metab. 2012;3(6):181–187. doi: 10.1177/2042018812471070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Baykal T, Senel K, Alp F, Erdal A, Ugur M. Is there an association between serum 25-hydroxyvitamin D concentrations and disease activity in rheumatoid arthritis? Bratisl Lek Listy. 2012;113(10):610–611. doi: 10.4149/bll_2012_137. [DOI] [PubMed] [Google Scholar]
- (23).Higgins MJ, Mackie SL, Thalayasingam N, Bingham SJ, Hamilton J, Kelly CA. The effect of vitamin D levels on the assessment of disease activity in rheumatoid arthritis. Clin Rheumatol. 2013 doi: 10.1007/s10067-013-2174-x. [DOI] [PubMed] [Google Scholar]
- (24).Song GG, Bae SC, Lee YH. Association between vitamin D intake and the risk of rheumatoid arthritis: a meta-analysis. Clin Rheumatol. 2012;31(12):1733–1739. doi: 10.1007/s10067-012-2080-7. [DOI] [PubMed] [Google Scholar]
- (25).Rossini M, Maddali BS, La MG, Minisola G, Malavolta N, Bernini L, et al. Vitamin D deficiency in rheumatoid arthritis: prevalence, determinants and associations with disease activity and disability. Arthritis Res Ther. 2010;12(6):R216. doi: 10.1186/ar3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: Associations with disease and evolution. Genome Res. 2010;20(10):1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Eyre S, Bowes J, Diogo D, Lee A, Barton A, Martin P, et al. High-density genetic mapping identifies new susceptibility loci for rheumatoid arthritis. Nat Genet. 2012;44(12):1336–1340. doi: 10.1038/ng.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Cooper JD, Smyth DJ, Walker NM, Stevens H, Burren OS, Wallace C, et al. Inherited variation in vitamin D genes is associated with predisposition to autoimmune disease type 1 diabetes. Diabetes. 2011;60(5):1624–1631. doi: 10.2337/db10-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Hingorani A, Humphries S. Nature’s randomised trials. The Lancet. 2003;366(9501):1906–1908. doi: 10.1016/S0140-6736(05)67767-7. [DOI] [PubMed] [Google Scholar]
- (30).Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41(8):882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376(9736):180–188. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Martin P, Barton A, Eyre S. ASSIMILATOR: a new tool to inform selection of associated genetic variants for functional studies. Bioinformatics. 2011;27(1):144–146. doi: 10.1093/bioinformatics/btq611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4(10):e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5(6):e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.