Abstract
Sagittal craniosynostosis is the most common form of craniosynostosis, affecting approximately one of 5,000 newborns. We conducted the first genome-wide association study (GWAS) for non-syndromic sagittal craniosynostosis (sNSC) using 130 non-Hispanic white (NHW) case-parent trios. Robust associations were observed in a 120 kb region downstream of BMP2, flanked by rs1884302 (P = 1.13 × 10−14; odds ratio [OR] = 4.58) and rs6140226 (P = 3.40 × 10−11; OR = 0.24) and within a 167 kb region of BBS9 between rs10262453 (P = 1.61 × 10−10; OR=0.19) and rs17724206 (P = 1.50 × 10−8; OR = 0.22). We replicated the associations to both loci [rs1884302 (P = 4.39 × 10−31); rs10262453 (P = 3.50 × 10−14)] in an independent NHW population of 172 unrelated sNSC probands and 548 controls. Both BMP2 and BBS9 are genes with a role in skeletal development warranting functional studies to further understand the etiology of sNSC.
Keywords: genome-wide association study, non-syndromic sagittal craniosynostosis, BMP2, BBS9, meta-analysis, nonsyndromic
Skull development is a complex process that involves ongoing interaction between the bones of the skull and cranial soft tissues. The cranial vault is comprised of intramembranous bones joined by sutures of dense fibrous tissue that accommodate the growing brain. Bone is added at these sutures during growth, and the skull eventually ossifies completely. Craniosynostosis (CS), the premature closure of one or more of the cranial vault sutures is a common congenital anomaly. Approximately 80% of CS occurs as an isolated anomaly, non-syndromic craniosynostosis (NSC), without major associated malformations (1). Rare mutations in the FGFR2, TWIST1, FREM1, LRIT3, EFNA4, and RUNX2 duplications have been reported in a minor fraction of NSC cases (2-9). The remainder (approximately 20%) of CS cases are syndromic, occurring with one or more additional major malformations, due to single-gene mutations in one of at least eight genes (FGFR1, FGFR2, FGFR3, TWIST1, EFNB1, POR, MSX2 and RAB23) primarily involving the coronal sutures (10-12).
The most common type of NSC is synostosis of the sagittal suture (sNSC), accounting for 40 to 58% of all cases (13-14). The etiology of sNSC is not well understood, although the published literature suggests that it is a multifactorial condition, where both genetic and environmental factors play a role, as indicated by a higher rate of concordance in monozygotic as compared to dizygotic twins (30% vs. 0% in sNSC) (15), increased male to female ratio (3:1) (1, 16), and a high recurrence risk in affected families (17). Several environmental factors have also been associated with sNSC, including parity, prematurity, intrauterine constraint, and maternal tobacco or nitrosatable drug use (18-21).
In an attempt to identify susceptibility loci for sNSC, we recruited 201 case-parent trios and 13 nuclear families with at least one individual affected with sNSC (68% non-Hispanic white [NHW], 32% mixed ethnicity families). These participants were enrolled and evaluated through the collaborative effort of the International Craniosynostosis Consortium (genetics.ucdmc.ucdavis.edu/icc.cfm). Samples were genotyped on the Illumina 1M Human Omni1-Quad array. Final criteria for the current analyses were based on strict sNSC phenotype (no additional synostoses, congenital anomalies or developmental delay present in the proband or affected sibling) and NHW ethnicity as determined by self-reported ancestry and confirmed by principal-component analysis (PCA) using EIGENSOFT 3.0 (22). Application of these criteria reduced our discovery population to 130 case-parent trios.
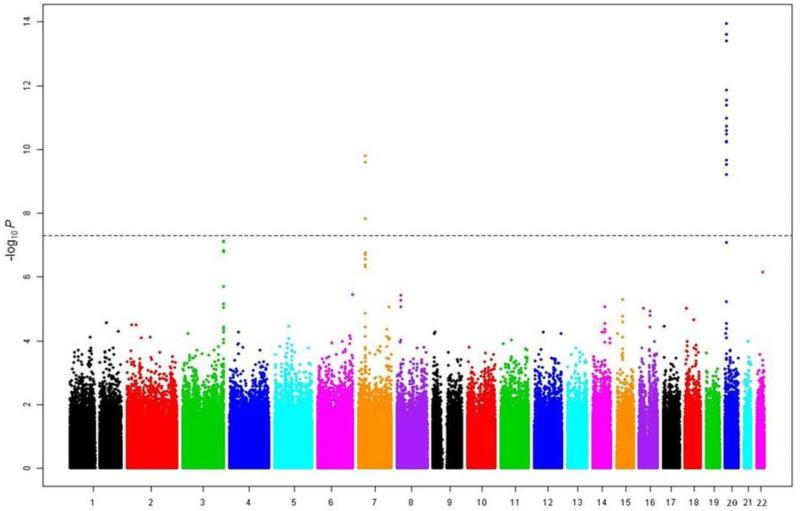
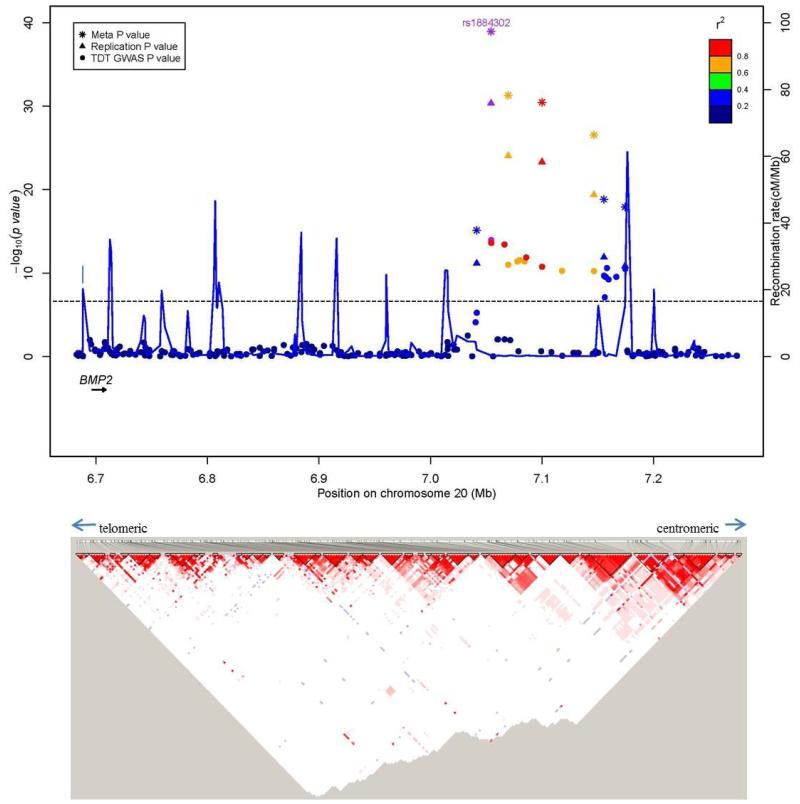
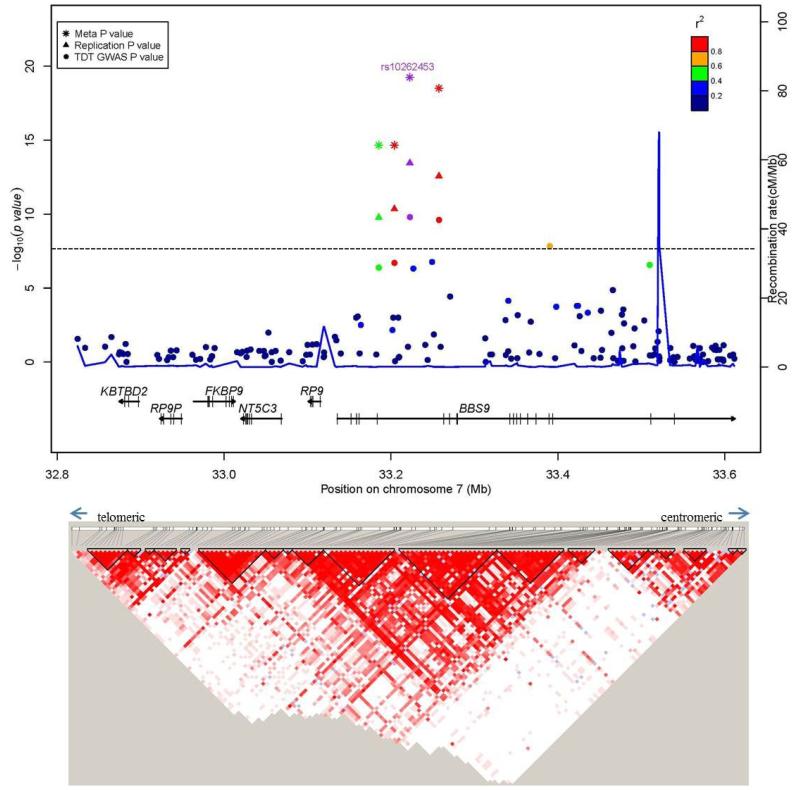
After stringent quality control procedures, we retained genotypes for 915,307 SNPs (914,402 autosomal and 905 pseudo-autosomal) which were analyzed using the transmission disequilibrium test (TDT) as implemented in PLINK v1.07 (23). This resulted in the identification of 21 SNPs , 18 on 20p12.3 and 3 on 7p14.3, which reached the genome-wide significance threshold of P < 5 × 10−8 (Fig. 1, Supplementary Fig. 1). The 18 chromosome 20 SNPs were in two LD blocks (92 kb and 19 kb, on chromosome 20p12.3; Fig. 2a, Supplementary Fig. 2, Table 1), with the most significant SNP, rs1884302 (P = 1.13 × 10−14), located approximately 345 kb downstream of BMP2. Conditional analysis using extended TDT (ETDT) in UNPHASED (24) confirmed that rs1884302 is leading these signals (data not shown). Except for a predicted mRNA, BC038533, no other known genes, CNVs, transcripts, miRNA or regulatory elements were identified in this region. However, at least four highly conserved genome regions with multiple predicted sites for transcription factor binding were identified by bioinformatic analysis in the vicinity of rs1884302 (Supplementary Fig. 3). Exonic resequencing of BMP2 in our cohort did not identify any variants with obvious deleterious effects. The three SNPs on chromosome 7p14.3 span a region of 167 kb within BBS9 introns 4 and 15 (Fig. 2b, Supplementary Fig. 4). RT-PCR sequencing of cDNA from eight sNSC calvarial osteoblast cell lines discovered six BBS9 variants, but three were determined to be previously reported SNPs and all were benign based on bioinformatic protein prediction tools and conservation analysis (see online Methods). We also searched for two-way gene-gene interactions using the 100 most significant TDT GWAS SNPs using a conditional logistic regression model (25). No results were significant after correcting for multiple testing (1247 tests, Bonferroni P ≤ 4 × 10−5, Supplementary Table 1).
Figure 1. Manhattan plot of P-values obtained from the genome-wide TDT of 130 trios (N=914,402).
The x-axis corresponds to the genomic position of the autosomes, while the y-axis is the −log10 of the P-value. Horizontal dashed line corresponds to the genome-wide significance threshold of P ≤ 5 × 10−8.
Figure 2. Regional association plots for associations with sNSC at genome-wide significance.
Upper panel: P-values (-log10) of the GWAS (solid circle), replication (triangle) and meta-analysis (asterisk) on the y-axis are plotted against the genomic positions of each SNP on the x-axis on (a) chromosome 20 and on (b) chromosome 7. Horizontal dashed line corresponds to the genome-wide significance threshold of P ≤ 5 × 10−8. Genes in region are depicted below (not to scale). The LD (r2) between the lead SNP and other SNPs are indicated in different colors. The blue lines indicate the recombination rates in cM per Mb using HapMap controls. Lower panel: LD plots (based on r2 value) using genotyped subjects from the GWAS study.
Table 1.
GWAS, replication study and meta-analysis results for most significant SNP on chromosomes 20 and 7
| GWAS | Replication | Meta-analysis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chr. | SNP | Allelesa | RAFb | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | P het c |
| 20 | rs1884302 | C/T | 0.3 | 4.58 (2.99–6.97) | 1.1 × 10−14 | 4.30 (3.33–5.56) | 4.4 × 10−31 | 4.38 (3.51–5.45) | 1.1 × 10−39 | 0.81 |
| 7 | rs10262453 | C/A | 0.69 | 0.20 (0.11–0.34) | 1.6 × 10−10 | 0.26 (0.18–0.37) | 3.5 × 10−14 | 0.24 (0.17–0.32) | 5.6 × 10−20 | 0.44 |
Minor allele/major allele.
Risk allele frequency (RAF) calculated from the replication study controls.
P value for heterogeneity between the discovery and replication cohorts. Underlined bases are overtransmitted alleles. Chr., chromosome.
Considering the male predominance of sNSC, we performed a separate TDT analysis for 104 NHW trios with a male proband. The 21 SNPs on chromosomes 20 and 7 remained genome-wide significant, with an additional pseudo-autosomal marker reaching a low, but not genome–wide statistically significant P-value (rs2522623, P = 5.75 × 10−5) and thus did not meet our criteria for replication. This marker is located in protocadherin 11 X-linked (PCDH11X), which to date has not been reported to play a functional role in sNSC. Analysis of a larger cohort may demonstrate a stronger association to this locus.
From a total of 46 autosomal SNPs that showed significant and suggestive associations with P < 1 × 10−5, 25 SNPs were prioritized for the replication study (Supplementary Table 2). In order to confirm the observed signals, we genotyped a NHW replication population of 172 unrelated cases with sNSC and 548 unaffected controls. We considered our data as a replication when the direction of effect of the alleles surpassing nominal significance was consistent between the discovery and replication datasets. All seven genotyped markers on chromosome 20 and four out of five markers on 7p14.3, with one SNP failing genotyping, were successfully replicated with meta-analysis P-values <10−9 (Supplementary Table 2, Supplementary Table 3). There was little or no evidence of heterogeneity of the odds ratio in the two different sample sets for the eleven replicated SNPs. Additionally, genotypic TDT (gTDT) analysis indicated the effect of SNPs was consistent with an additive model (Supplementary Table 4). We detected that, in most cases, transmitted minor alleles conferred a 2.36-4.38 fold risk of sNSC at the chromosome 20 markers, whereas the chromosome 7 transmitted minor alleles were associated with a decreased risk of sNSC (Supplementary Table 3). We were unable to replicate other signals from the GWAS TDT with P < 1 × 10−5. These associations could be false positives or were missed due to differences between the study design and statistical methods used for the discovery and replication stages. We also relied on self-reported maternal ethnicity instead of ancestry informative markers for the replication cohort, which limited our ability to precisely assign ethnicity to the case-control study. The effect of gene-environment interaction or the relatively small sample of the replication cohort may also have affected the findings.
Imputation on borderline significant regions on chromosomes 3, 7, 8, 15, 22 was performed using BEAGLE v3.3.2 (26) to identify additional genome-wide significant SNPs. Fifty-two intronic SNPs in DLG1 reached genome-wide significance (Supplementary Table 5), but we previously failed to replicate this association in our case/control sample (Supplementary Table 2). The lack of replication of a common variant may be due to our small sample size, differences in study design between our GWAS and replication datasets, or rare variants that contribute to the risk. Imputation was carried out using the 1000Genomes data for the reference panel, which does include some sequence variants with minor allele frequencies < 1%. Given that sNSC has a prevalence of 1 in 5000, we suspect that rare functional variants not identified by 1000Genomes may be involved and sequencing of the associated region will be necessary. No statistically significant signals were observed after the imputation at the other loci.
Using these modestly-sized populations we discovered and replicated common genetic variants in at least two loci that contribute to sNSC. The chromosome 20 locus is located in the vicinity of the BMP2 gene that plays a role in skeletal development. BMP2 is a member of the TGFβ superfamily that activates SMAD1, 5, and 8, and together with other osteogenic transcription factors such as RUNX2, ultimately regulating osteoblast development (27).
An interesting observation in humans and animal models is that mutations with opposite effects in the same gene result in craniosynostosis (accelerated ossification) or parietal foramina/large fontanelles (deficient ossification) phenotypes. This holds true for mutations in at least 4 genes: MSX2 (28-30), TWIST1 (31-32), RUNX2 (6-7, 33) and Nell1 (34-35) Consistent with this, gain-of-function mutations in ALX4, a gene associated with parietal foramina, have been identified in sNSC probands by our group (36). Similarly, a variant in BMP2, p.S37A, has been associated with osteoporosis (37): thus, if p.S37A results in loss-of-function, one can speculate that gain-of-function variants causing overexpression of BMP2 would result in a phenotype of accelerated ossification and potentially CS. Additionally of interest, a limb specific regulatory element located 110 kb downstream of BMP2 is mutated in brachydactyly type A2 (38). Therefore, we predict that a skull-specific BMP2 regulatory element is located within or near the association region. Also, non-coding SNPs can affect gene expression variation of one or many genes. Even though none of our top hits were found in the eQTL databases, this may be due to the fact that no human eQTL studies have been conducted in the skull suture tissues.
The GWAS and the replication analysis also showed evidence for association on chromosome 7p14 within an intronic region of BBS9. BBS9 is a component of the multiprotein complex BBSome which plays critical roles in intraflagellar transport, or moving cargo molecules in and out of cilia. Loss-of-function mutations in BBS9 have been found in patients with Bardet-Biedl syndrome (BBS) (39). BBS arises from defects of primary cilia that function as a platform for many important signaling events involving at least PDGFRα, hedgehog, epidermal growth factor, and 5-HT6 serotonin receptor (40-44). Although BBS does not present with suture phenotypes, it presents with a skeletal phenotype (polydactyly) and orofacial anomalies (39). It is noteworthy that aberrant signaling of PDGFRα and epidermal growth factor or hedgehog has been implicated in craniofacial development (45-49). In addition, it has been shown that FGF-signaling regulates cilia length and function during development of zebrafish and Xenopus (50). Thus, it is possible that CS-causing FGFR gain-of-function mutations also work via aberrant assembly and/or signaling of cilia. It remains to be seen whether gain-of-function mutations or overexpression of BBS9 would influence suture fusion. Three of the genes in the vicinity of the association on chromosome 7 (RP9P, KBTBD2, FKBP9) are pseudogenes and the remaining two, RP9 and NT5C3, have no known role in skeletal development and cause retinitis pigmentosa and hemolytic anemia, respectively (51-52). This region is located approximately 14 Mb telomeric of TWIST1 and thus is unlikely to impact its function.
Our findings implicate variants within or near genes with relevant function in skeletal development, suggesting a major genetic effect for sNSC. A larger sample size will be needed to detect additional susceptibility genes with moderate genetic effects. The lack of Mendelian segregation suggests a more complex pattern of inheritance for sNSC. The causative variants may be of low frequency and in linkage disequilibrium with the common alleles associated with sNSC in this study. Another possibility is that one or more environmental factors interacting with genetic variants contribute to sNSC. Less likely, but still possible are oligogenic inheritance, gene-gene interactions, imprinting, epigenetic changes, or incomplete penetrance of the variants.
Biologically plausible candidate genes, BMP2 and BBS9, both involved in skeletal development, were identified in our study. Functional studies of these genes, such as RNA sequencing and possibly tiling arrays of the regions to look for non-coding or microRNAs, are warranted. The potential contribution of these loci to other types of NSC is worthy of investigation. Our results represent the first major step toward deciphering the genetic etiology of the sNSC.
Methods
Subjects
Informed consents were obtained from all patients and/or their parents. This study was approved by the Institutional Review Boards of the University of California, Davis, and other participating institutions and was conducted in accordance with institutional guidelines. Craniosynostosis (CS) was documented by computerized tomography of the skull and/or the protocol of the surgical correction. All patients with sagittal non-syndromic craniosynostosis (sNSC) were clinically assessed by a clinical geneticist and those with synostosis of additional sutures, associated extracranial congenital anomalies or developmental delays were excluded. Patients for both the initial GWAS study and the replication study were recruited within the USA.
The case-control replication sample was comprised of archived, residual newborn blood spots from non-Hispanic white (NHW) infants diagnosed with sNSC (n=172) and control infants without a congenital malformation (n=548). Case infants were delivered by New York State residents from January 1, 1998 – December 31, 2005 and identified from the New York State Congenital Malformations Registry, a population-based birth defects surveillance program. Control infants were a random sample of live births in New York State delivered during the same time frame with the control-to-case ratio of approximately 3:1. Records for cases and controls were linked to records of the New York State Newborn Screening Program to obtain the respective blood spots and provided anonymously to study investigators. Use of the blood spots was approved by the Institutional Review Board of the New York State Department of Health and Mount Sinai School of Medicine.
SNP genotyping and quality control in GWAS
Genomic DNA was extracted from whole blood or oral samples (468 whole blood, 139 mouth wash, 2 buccal and 24 Oragene). A total of 680 samples, including 16 blind duplicates and 13 HapMap controls (11 Utah residents of European ancestry, CEU and 2 Yoruban controls, YRI) were genotyped on the Illumina 1M Human Omni1-Quad array at the Center for Inherited Disease Research (http://www.cidr.jhmi.edu). Genotypes for 1,006,422 SNPs and 133,997 CNV probes were released. The missing data rate was 0.39%, the blind duplicate rate was 99.99%, and the HapMap sample concordance rate was 99.7%. To check for cryptic relatedness and population stratification, 92,867 independent SNPs were selected based on a pair-wise inter-marker linkage disequilibrium r2 < 0.4, minor allele frequency (MAF) > 0.1 and genotype call rates > 0.99. Pairwise identity-by-descent (IBD) calculations were performed using PLINK v1.0.7 (53) and inconsistent samples were dropped, reducing the sample size to 646 individuals (213 trios). To confirm the self-reported ancestry of samples, principal components analysis (PCA) was performed using EIGENSOFT 3.0 (22)]. A total of 145 families (451 individuals) were found to cluster with the 11 HapMap CEU controls, with no other well-defined clusters noted. A strict sNSC phenotype (no additional synostosis, congenital anomalies or developmental delay present in proband or affected sibling) further reduced our number of NHW trios to 130. SNPs were excluded from analysis if the call rate was less than 98%, they were discordant in more than one duplicate, had a Mendelian inconsistency in more than one trio, were missing in more than 10% of the samples, had a MAF of 0.0, and/or had a Hardy-Weinberg equilibrium P-value ≤ 1 × 10-5 with no heterozygotes. This reduced the number of SNPs available for analysis to 914,402 autosomal SNPs and 905 pseudo-autosomal SNPs.
SNP selection and quality control in replication study
Genomic DNA was extracted from 3 mm dried blood spot punches using a sodium hydroxide precipitation protocol described previously (54). SNPs from the discovery stage were selected if P-value for association was ≤ 1.0 × 10-5 in the discovery stage. When multiple SNPs from the same region passed the P-value cutoff for replication, SNPs with the lowest P-value and/or lowest pairwise LD were selected. Using this screening process, from 46 autosomal SNPs with P-values ≤ 1.0 × 10-5, we selected to genotype 25 SNPs (Supplementary Table 2) that tagged all 11 significant regions of interest (1 region each on 10 different chromosomes and 2 regions on chromosome 7). SNPs for the replication phase were genotyped using the ABI TaqMan allelic discrimination assay according to the manufacturer’s protocol (Applied Biosystems). SNP genotypes were called and discriminated using software S.D.S v3.4.4. Genotyping was conducted in duplicates and the concordance rate was 100%. Case and control samples were randomly distributed across the DNA plates. Disease status was unknown at the time of the genotyping experiments. Of the 25 SNPs from the original discovery phase that were selected for replication, two failed multiple TaqMan assay attempts even when conditions were modified (chr.7: rs17724206, failed and chr.22: rs4634034, call rate <90%). For the remaining SNPs, the genotype call rates were >95%.
Sequencing
BMP2 sequencing was carried out on both exons for 149 sagittal NSC probands, of which 102 were part of the 130 proband cohort used in the initial discovery phase. Primer sequences for BMP2 are available in Supplementary Table 6A. Three variants were identified. Two of them, c.109T>G (p.S37A) and c.570A>T (p.R190S), were previously reported (rs2273073 and rs23576, respectively) and the allelic frequencies for the CEU population (obtained from HapMap Rel. 28, Phase II + III) was not significantly different than that found in our sequencing sample. The other SNP, c.393A>T (p.R131S) was novel, present in the proband’s father and characterized as non-damaging based on bioinformatic predictions from SIFT (Sorting Tolerant from Intolerant, Fred Hutchinson Cancer Research Center, Seattle, web software, http://blocks.fhcrc.org/sift/SIFT.html) and PPH (PolyPhen, Bork Group & Sunyaev Lab, Harvard, web software, http://genetics.bwh.harvard.edu/pph).
Reverse transcriptase-PCR using control fetal calvarial osteoblast cDNA was attempted with all possible exonic primer combinations for predicted mRNA BC038533, but did not produce PCR amplicons. BBS9 sequencing was performed on cDNA from eight NSC fetal calvarial osteoblast lines. cDNA was created via RT-PCR (using Invitrogen SuperScript protocol and kit) on RNA extracted from available patients. BBS9 sequencing primers are available in Supplementary Table 6B. Six variants were identified; three (c.1029A>G G343G, c.2100G>A L700L and c.1729A>C I577L) were determined to be previously reported (rs25195153, rs115809567 and rs34209904 respectively) while the remaining three (BBS9_3′UTR +108 T>A, BBS93′UTR +115 C>T and c.1284C>T) were not found in any searched databases (1000Genomes, HapMap, NCBI or MapBack). All six variants were indicated as non-damaging or benign by SIFT and PPH respectively.
ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/) alignment of BBS9 protein sequence for conservation analysis was performed in twelve available species - NP_001028776.1 (Homo sapiens), NP_848502.1 (Mus musculus), XP_002664838.1 (Danio rerio), XP_235942.4 (Rattus norvegicus), NP_001179782.1 (Bos taurus), XP_418843.3 (Gallus gallus), NP_001016821.1 (Xenopus tropicalis), XP_002957514.1 (Volvox carteri f. nagariensis), XP_001107028.1 (Macaca mulatta), XP_852259.1 (Canis lupis familiaris), XP_001168457.1 (Pan troglodytes), NP_001087412.1 (Xenopus laevis). Parental DNA was not available for sequencing analysis.
Imputation
Ungenotyped markers 50 kb upstream and downstream from borderline significant regions from the TDT GWAS (chromosomes 3, 7, 8, 15 and 22) were imputed for the discovery sample using a trio design as implemented in BEAGLE v3.3.2 (26). A reference panel of 379 European individuals (GBR, FIN, IBS, CEU and TWI) from the 1000 Genomes Project database was used. We first pre-phased the trio GWAS data and then imputed SNPs using a phased reference panel. There were 58, 19, 75, 37, and 30 genotyped SNPs and 1074, 234, 613, 276, and 431 imputed variants on chromosomes 3, 7, 8, 15, and 22 respectively. We carried out TDT tests on imputed variants of high quality (r2 ≥ 0.3).
Statistical analysis
GWAS association tests were carried out using TDT analysis as implemented in PLINK v1.07 (53). Manhattan plots were generated using the R script, and LD plots and r2 measures were obtained using HaploView v4.06 (55). All SNP positioning information was gathered from human genome version hg18 (March 2006; NCBI36).
Association mapping in the replication panel was performed by the χ2 test for allelic association using SAS/Genetics software version 9.2 (SAS Institute, Inc., Cary, NC). Direction of effect of markers surpassing nominal significance in the replication dataset was compared between both the discovery and replication datasets and three markers had opposite effects (chr.6: rs4716412; chr.7: rs7457112; and chr.16: rs4522429).
We searched for 2-way gene-gene interactions using the 100 most significant SNPs from the 130 NHW trio TDT analysis. We performed a genotypic TDT (gTDT) for the interaction of each pair of SNPs, excluding results from SNP pairs on the same chromosome due to long range LD (1247 tests). For each case-parents trio, we generated 15 pseudo-controls matched to each proband as a function of parental genotypes of the two considered SNPs. Pseudo-controls for each case are made of one of the two-locus genotypes that could have been, but were not transmitted to the case. Using these matched case and 15 pseudo-controls, we fitted a conditional logistic regression model (25) to test for epistatic interactions that incorporate additive and dominance effects at the two loci. The ‘trio’ R package v1.4.24 was used to perform these tests (56).
Conditional analysis was performed to identify additional SNPs potentially masked by the primary significant SNP using the conditional extended transmission disequilibrium test (ETDT) for trio data (57) as implemented in UNPHASED v3.0.9 (24). Meta-analysis was performed by combining the trait-marker associations of the discovery TDT and replication case-control study using a fixed-effects model after testing for inter-study heterogeneity (58).
Supplementary Material
Acknowledgements
The authors are highly indebted to all families who contributed to this study. S.A.B. is partially funded through a Children’s Miracle Network Endowed Chair and through grants K23 DE00462, R03 DE016342, and R01 DE016886 from NIDCR/NIH and M01-RR00052 from NCRR/NIH and fully supported by Zlatka, Anton and Alec Boyadjiev. Partial funding was also obtained from grants to E.W.J. (CDC, 5 R01 DD000350), M.L.C. (R01 DE018227), A.O.M.W. (Wellcome Trust; #093329), P.A.R. (Centers for Disease Control and Prevention; #5U01DD000492), J.K. (National Institute Of Dental & Craniofacial Research, NIH; # R21DE022419), J.L.M. (Intramural Research Program of the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development ; IRP # HHSN267200703431C; NICHD # N01-DK-7-3431), P.A.S-L. (Robert Wood Johnson Foundation; #3R37DE012711-13S1 and CHLA-USC Child Health Research Career Development Program; #NIH K12-HD05954), J.T.R. (NIDCR/NIH and the American Recovery and Reinvestment Act: #R01 DE018500 and 3R01 DE018500-02S1) and I.P. (National Center for Advancing Translational Sciences, NIH; #UL1TR000067). This project was also supported, in part, by the Division of Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (C.M.J., Y.K. and A.F.W.). Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. We are indebted to Bridget Wilson, Nisha Issac, Christopher Nauta, Erica Goude, Elijah Cherkez, Linda Peters, and Jeanette Harrison for patient recruitment, Corrine Boehm and Alan Scott for coordination of discovery phase genotyping, Christina Stevens, Armineh Stoner, Jia Lie Liu, Adam Gearhart, April Atkins and Emily McGrath for bench work, Douglas Mortlock for bioinformatic analysis and informative discussion and John Graham, Jonathan Bernstein, Jeffrey Marsh, Jayesh Panchal, Travis Tollefson and Maria Passos-Bueno for contributing clinical information and biospecimens for this project.
Footnotes
Author Contributions C. M. J. – Co-first author with G. Y., performed statistical analysis, analyzed data and wrote the manuscript
G. Y. – Co-first author with C. M. J., performed experiments, analyzed data and wrote the manuscript
Y. K. – Performed statistical analyses, analyzed the data and contributed to writing of manuscript.
I. P. – Performed statistical analyses, analyzed the data and contributed to writing of manuscript
E. W. J. – Helped design the experiments, contributed materials and reagents and contributed to editing of manuscript
M. E. – Performed experiments
X. Y. – Performed experiments
E. A. – Performed experiments
L.S. – Performed experiments
M. L. C. – Analyzed the data and contributed materials and reagents
V. K. – Analyzed the data and contributed materials and reagents.
T. R. – Analyzed the data
S. A. W. – Contributed materials and reagents
A. O. M. W. – Analyzed the data and contributed to editing of manuscript
J. S. – Analyzed the data and contributed materials and reagents
J. T. R. – Analyzed the data and contributed to writing of manuscript.
Y. H. – Analyzed the data and contributed to writing of manuscript.
P. A. S-L. – Analyzed the data and contributed materials and reagents
M. F. B. – Analyzed the data, contributed materials and reagents, contributed to editing of the manuscript
C. M. D. – Contributed materials and reagents and helped with experimental design
J. L. M. – Contributed materials and reagents and helped with experimental design
M. C. – Contributed materials and reagents and helped with experimental design
P. A. R. – Contributed materials and reagents, helped with experimental design and contributed to writing of manuscript
D. M. K. – Contributed materials and reagents, helped with experimental design, and helped with editing of the manuscript
C. S. – Contributed materials and reagents and helped with experimental design
P. J. T. – Contributed materials and reagents, helped with experimental design, and helped with editing of the manuscript
O. D. K. – Contributed materials and reagents and helped with experimental design
J. B. – Contributed materials and reagents and helped with experimental design
M. Z-L. – Contributed materials and reagents and helped with experimental design
C. N. – Contributed materials and reagentsand helped with experimental design
J. K. – Supervised research and helped analyze data
A. F. W. – Supervised research, analyzed the data, and helped with writing of manuscript
S. A. B – Principal investigator, designed and supervised research, recruited and evaluated participants, analyzed the data, and contributed to writing of manuscript
References
- 1.Cohen MM. Craniosynostosis: Diagnosis, Evaluation, and Management. Oxford University Press; New York: 2000. [Google Scholar]
- 2.Johnson D, et al. A novel mutation, Ala315Ser, in FGFR2: a gene-environment interaction leading to craniosynostosis? Eur J Hum Genet. 2000;8(8):571–577. doi: 10.1038/sj.ejhg.5200499. [DOI] [PubMed] [Google Scholar]
- 3.Seto ML, et al. Isolated sagittal and coronal craniosynostosis associated with TWIST box mutations. Am J Med Genet A. 2007;143(7):678–86. doi: 10.1002/ajmg.a.31630. [DOI] [PubMed] [Google Scholar]
- 4.Weber I, et al. Molecular analyisis of 74 patients with craniosynostosis. Eur J Hum Genet. 2001;9(Sup.1):P0409, 179. [Google Scholar]
- 5.Merrill AE, et al. Cell mixing at a neural crest-mesoderm boundary and deficient ephrin-Eph signaling in the pathogenesis of craniosynostosis. Hum Mol Genet. 2006;15(8):1319–28. doi: 10.1093/hmg/ddl052. [DOI] [PubMed] [Google Scholar]
- 6.Wilkie AO, et al. Clinical dividends from the molecular genetic diagnosis of craniosynostosis. Am J Med Genet A. 2007;143A(16):1941–9. doi: 10.1002/ajmg.a.31905. [DOI] [PubMed] [Google Scholar]
- 7.Mefford HC, et al. Copy number variation analysis in single-suture craniosynostosis: multiple rare variants including RUNX2 duplication in two cousins with metopic craniosynostosis. Am J Med Genet A. 2010;152A(9):2203–10. doi: 10.1002/ajmg.a.33557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vissers LE, et al. Heterozygous mutations of FREM1 are associated with an increased risk of isolated metopic craniosynostosis in humans and mice. PLoS Genet. 2011;7(9):e1002278. doi: 10.1371/journal.pgen.1002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S-D, et al. Leucine-rich repeat, immunoglobulin-like and transmembrane domain 3 (LRIT3) is a modulator of FGFR1. Federation of European Biochemical Societies. 2012;586(10):1516–1521. doi: 10.1016/j.febslet.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melville H, et al. Genetic basis of potential therapeutic strategies for craniosynostosis. Am J Med Genet A. 2010;152A(12):3007–15. doi: 10.1002/ajmg.a.33703. [DOI] [PubMed] [Google Scholar]
- 11.Wilkie AO, et al. Prevalence and complications of single-gene and chromosomal disorders in craniosynostosis. Pediatrics. 2010;126(2):e391–400. doi: 10.1542/peds.2009-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Passos-Bueno MR, et al. Genetics of craniosynostosis: genes, syndromes, mutations and genotype-phenotype correlations. Front Oral Biol. 2008;12:107–43. doi: 10.1159/000115035. [DOI] [PubMed] [Google Scholar]
- 13.Kimonis V, et al. Genetics of craniosynostosis. Semin Pediatr Neurol. 2007;14(3):150–61. doi: 10.1016/j.spen.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Kolar JC. An epidemiological study of nonsyndromal craniosynostoses. J Craniofac Surg. 2011;22(1):47–9. doi: 10.1097/SCS.0b013e3181f6c2fb. [DOI] [PubMed] [Google Scholar]
- 15.Lajeunie E, et al. Genetic considerations in nonsyndromic midline craniosynostoses: a study of twins and their families. J Neurosurg. 2005;103(4 Suppl):353–6. doi: 10.3171/ped.2005.103.4.0353. [DOI] [PubMed] [Google Scholar]
- 16.Lajeunie E, et al. Syndromal and nonsyndromal primary trigonocephaly: analysis of a series of 237 patients. Am J Med Genet. 1998;75(2):211–215. doi: 10.1002/(sici)1096-8628(19980113)75:2<211::aid-ajmg19>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 17.Lajeunie E, et al. Genetic study of scaphocephaly. Am J Med Genet. 1996;62(3):282–5. doi: 10.1002/(SICI)1096-8628(19960329)62:3<282::AID-AJMG15>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Lara PA, et al. Fetal constraint as a potential risk factor for craniosynostosis. Am J Med Genet A. 2010;152A(2):394–400. doi: 10.1002/ajmg.a.33246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeiger JS, et al. Genetic and environmental risk factors for sagittal craniosynostosis. J Craniofac Surg. 2002;13(5):602–6. doi: 10.1097/00001665-200209000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Gardner JS, et al. Maternal exposure to prescription and non-prescription pharmaceuticals or drugs of abuse and risk of craniosynostosis. Int J Epidemiol. 1998;27(1):64–7. doi: 10.1093/ije/27.1.64. [DOI] [PubMed] [Google Scholar]
- 21.Kallen K. Maternal smoking and craniosynostosis. Teratology. 1999;60(3):146–150. doi: 10.1002/(sici)1096-9926(199909)60:3<146::aid-tera10>3.3.co;2-v. [DOI] [PubMed] [Google Scholar]
- 22.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25(2):115–21. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 25.Cordell HJ. Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11(20):2463–8. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- 26.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84(2):210–23. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosen V. BMP2 signaling in bone development and repair. Cytokine Growth Factor Rev. 2009;20(5-6):475–80. doi: 10.1016/j.cytogfr.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Jabs E, et al. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75:443–450. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- 29.Spruijt L, et al. A novel mutation in the MSX2 gene in a family with foramina parietalia permagna (FPP) Am J Med Genet A. 2005;139(1):45–7. doi: 10.1002/ajmg.a.30923. [DOI] [PubMed] [Google Scholar]
- 30.Wilkie A, et al. Functional haploinsufficiency of the human homeobox gene MSX2 causes defects in skull ossification. Nat Genet. 2000;24(4):330–331. doi: 10.1038/74224. [DOI] [PubMed] [Google Scholar]
- 31.Howard T, et al. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat Genet. 1997;15(1):36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- 32.Stankiewicz P, et al. Phenotypic findings due to trisomy 7p15.3-pter including the TWIST locus. Am J Med Genet. 2001;103(1):56–62. doi: 10.1002/ajmg.1512. [DOI] [PubMed] [Google Scholar]
- 33.Mundlos S, et al. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 34.Desai J, et al. Nell1-deficient mice have reduced expression of extracellular matrix proteins causing cranial and vertebral defects. Hum Mol Genet. 2006;15(8):1329–41. doi: 10.1093/hmg/ddl053. [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, et al. The role of NELL-1, a growth factor associated with craniosynostosis, in promoting bone regeneration. J Dent Res. 2010;89(9):865–78. doi: 10.1177/0022034510376401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yagnik G, et al. ALX4 gain-of-function mutations in nonsyndromic craniosynostosis. Hum Mutat. 2012 doi: 10.1002/humu.22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Styrkarsdottir U, et al. Linkage of osteoporosis to chromosome 20p12 and association to BMP2. PLoS Biol. 2003;1(3):E69. doi: 10.1371/journal.pbio.0000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dathe K, et al. Duplications involving a conserved regulatory element downstream of BMP2 are associated with brachydactyly type A2. Am J Hum Genet. 2009;84(4):483–92. doi: 10.1016/j.ajhg.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tobin JL, Beales PL. Bardet-Biedl syndrome: beyond the cilium. Pediatr Nephrol. 2007;22(7):926–36. doi: 10.1007/s00467-007-0435-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marshall WF, Nonaka S. Cilia: tuning in to the cell’s antenna. Curr Biol. 2006;16(15):R604–14. doi: 10.1016/j.cub.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 41.Schneider L, et al. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15(20):1861–6. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Haycraft CJ, et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1(4):e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma R, et al. PKD2 functions as an epidermal growth factor-activated plasma membrane channel. Mol Cell Biol. 2005;25(18):8285–98. doi: 10.1128/MCB.25.18.8285-8298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brailov I, et al. Localization of 5-HT(6) receptors at the plasma membrane of neuronal cilia in the rat brain. Brain Res. 2000;872(1-2):271–5. doi: 10.1016/s0006-8993(00)02519-1. [DOI] [PubMed] [Google Scholar]
- 45.Jenkins D, et al. RAB23 mutations in Carpenter syndrome imply an unexpected role for hedgehog signaling in cranial-suture development and obesity. Am J Hum Genet. 2007;80(6):1162–70. doi: 10.1086/518047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ehlen HW, Buelens LA, Vortkamp A. Hedgehog signaling in skeletal development. Birth Defects Res C Embryo Today. 2006;78(3):267–79. doi: 10.1002/bdrc.20076. [DOI] [PubMed] [Google Scholar]
- 47.Miettinen PJ, et al. Epidermal growth factor receptor function is necessary for normal craniofacial development and palate closure. Nat Genet. 1999;22(1):69–73. doi: 10.1038/8773. [DOI] [PubMed] [Google Scholar]
- 48.Ding H, et al. A specific requirement for PDGF-C in palate formation and PDGFR-alpha signaling. Nat Genet. 2004;36(10):1111–6. doi: 10.1038/ng1415. [DOI] [PubMed] [Google Scholar]
- 49.Choi DS, et al. 5-HT2B receptor-mediated serotonin morphogenetic functions in mouse cranial neural crest and myocardiac cells. Development. 1997;124(9):1745–55. doi: 10.1242/dev.124.9.1745. [DOI] [PubMed] [Google Scholar]
- 50.Neugebauer JM, et al. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458(7238):651–4. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keen TJ, et al. Mutations in a protein target of the Pim-1 kinase associated with the RP9 form of autosomal dominant retinitis pigmentosa. Eur J Hum Genet. 2002;10(4):245–9. doi: 10.1038/sj.ejhg.5200797. [DOI] [PubMed] [Google Scholar]
- 52.Marinaki AM, et al. Genetic basis of hemolytic anemia caused by pyrimidine 5′ nucleotidase deficiency. Blood. 2001;97(11):3327–32. doi: 10.1182/blood.v97.11.3327. [DOI] [PubMed] [Google Scholar]
- 53.McDonald-McGinn D, et al. Molecular characterization of Pfeiffer syndrome: implications for prognosis and genetic counseling. Am J Hum Genet. 2003;73(5):212, A253. [Google Scholar]
- 54.Mills JL, et al. Folate and vitamin B12-related genes and risk for omphalocele. Hum Genet. 2012;131(5):739–46. doi: 10.1007/s00439-011-1117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 56.Schwender H, et al. Rapid Testing of SNPs and Gene-Environment Interactions in Case-Parent Trio Data Based on Exact Analytic Parameter Estimation. Biometrics. 2011 doi: 10.1111/j.1541-0420.2011.01713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koeleman BP, et al. Adaptation of the extended transmission/disequilibrium test to distinguish disease associations of multiple loci: the Conditional Extended Transmission/Disequilibrium Test. Annals of Human Genetics. 2000;64(Pt 3):207–13. doi: 10.1017/S0003480000008095. [DOI] [PubMed] [Google Scholar]
- 58.Kazeem GR, Farrall M. Integrating case-control and TDT studies. Annals of Human Genetics. 2005;69:329–335. doi: 10.1046/j.1529-8817.2005.00156.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.