Abstract
Vibrio cholerae is a globally important pathogen that is endemic in many areas of the world and causes 3-5 million reported cases of cholera every year (http://www.who.int/wer). Historically there have been seven acknowledged cholera pandemics; included in the 7th and ongoing pandemic are the recent outbreaks in Zimbabwe and Haiti1. Only serogroup O1 isolates (consisting of two biotypes known as ‘classical’ and ‘El Tor’) and the derivative O1392,3 can cause epidemic cholera2. It is believed that the first six cholera pandemics were caused by the classical biotype but El Tor has subsequently spread globally and replaced the classical biotype in the current pandemic1. Detailed molecular epidemiological mapping of cholera has been compromised by a reliance on sub-genomic regions such as mobile elements to infer relationships, making El Tor isolates associated with the 7th pandemic appear superficially diverse. To understand the underlying phylogeny of the lineage responsible for the current pandemic we identified high resolution markers (single nucleotide polymorphisms; SNPs) in 154 whole genome sequences of globally and temporally representative V. cholerae isolates. Using this phylogeny we show that the 7th pandemic has spread from the Bay of Bengal in at least three independent but overlapping waves with a common ancestor in the 1950’s and identify multiple transcontinental transmission events. Additionally, we show how the acquisition of the SXT family of antibiotic resistance elements has shaped the pandemic spread and show that it was first acquired at least 10 years prior to its discovery in V. cholerae.
Whole genome analysis is perhaps the ultimate approach to building a robust phylogeny within recently emerged pathogens through the identification of SNPs and other rare genetic variants4. Consequently, we sequenced the genomes of 136 V. cholerae (including 113 isolates from the 7th pandemic) and added to these 18 previously published genomes1,2,5 to produce a global genomic database from isolates collected over a century. We included representative El Tor isolates collected over the past four decades and compared these to previously reported and novel classical and non-O1 genome sequences1,2.
The sequence reads were mapped to the reference sequence of El Tor N169616, a 7th pandemic V. cholerae isolated in Bangladesh in 1975(see footnote Table S1) and the resulting consensus tree identified 8 distinct phyletic lineages (L1-L8, see Fig S1 and Table S1 for strain and lineage information), 6 of which incorporated O1 clinical isolates. The classical isolates formed a distinct highly clustered group (L1), distant from the El Tor isolates of the 7th pandemic (L2). It is clear from Fig S1 that the classical and El Tor clades did not originate from a recent common ancestor and instead appear to be independent derivatives with distinct phylogenetic histories, consistent with previous proposals2. Isolates of L4 share a common ancestor with previously reported non-conventional O1 isolates2 (Fig S2) and are likely to have acquired the O1 antigen genes by a recombination event onto a genetically distinct genomic backbone. Isolates of L7 also have a distinct backbone, while L2, L3 (US Gulf Coast strains), L5, L6 and L8 share a more ‘El Tor-like’ genome backbone, and the L1 backbone is of the ‘classical type’.
Genome-wide SNP analysis showed that the 123 El Tor isolates within the L2 cluster (Fig S1) differed from the reference by only 50-250 SNPs. With this large sample size we were able to construct a high-resolution phylogeny that shows unequivocally that the current pandemic is monophyletic and originated from a single source, providing a framework for future epidemiological and phenotypic analysis of V. cholerae including transmission tracking and typing.
Predicted recombined regions were identified, and along with genomic islands and mobile genetic elements, these were initially excluded from the phylogenetic analysis of 7th pandemic isolates, to determine the underlying phylogeny. Interestingly, analysis of the tree (Fig 1, see Fig S3 for tree with strain names) provides clear evidence of a clonal expansion of the lineage with a strong temporal signature. This is most clearly illustrated by the fact that the most divergent isolates from the N16961 reference are represented by the oldest 7th pandemic isolate in our collection, A6 collected in 1957, together with the most recent Haitian isolates5 from late 2010. We performed a linear regression analysis on all the L2 isolates to calculate the rate of SNP accumulation based on the date of isolation and the root to tip distance. The shape of the tree and temporal signatures in Fig 1 show that there is a very consistent rate of SNP accumulation, 3.3 SNPs per year (R2 = 0.73, Fig S4) in the core genome, emphasizing its robustness and utility for transmission studies. The only exception to this is V. cholerae A4, a repeatedly passaged laboratory strain that was originally isolated in 1973 (Fig S3, Fig S4). The estimated rate of mutation for our 7th pandemic V. cholerae collection was 8.3 × 10−7 SNPs/site/year, which is between 5 and 2.5 times slower than that estimated for recent clonal expansions of some other human pathogenic bacteria4,7.
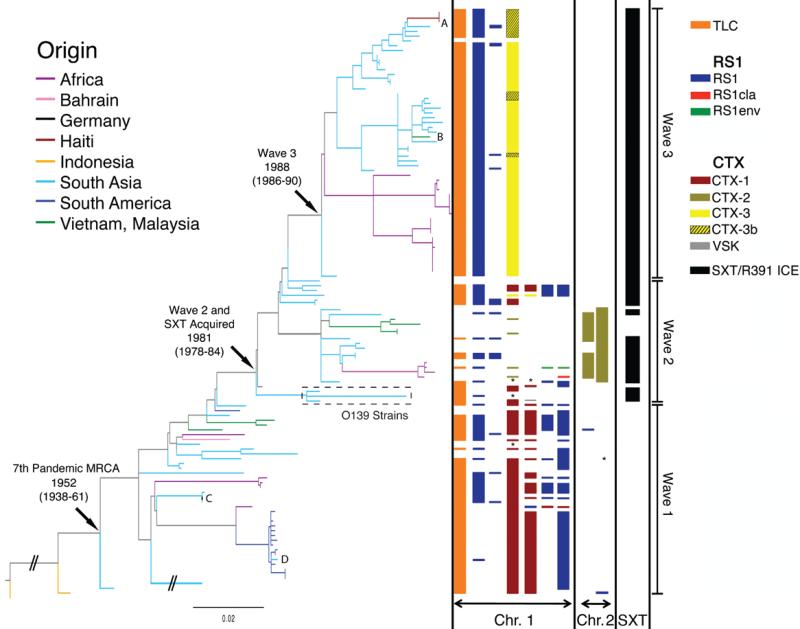
Fig 1.
A maximum likelihood phylogenetic tree of the 7th pandemic lineage of V. cholerae based on the SNP differences across the whole core genome, excluding likely recombination events. The pre-7th pandemic isolate M66 was used as an outgroup to root the tree. Branches are colored based on the region of isolation of the strains. The branches representing the three major waves are indicated on the far right, and the nodes representing the most recent common ancestors (MRCA) of the 7th pandemic, and subsequent waves 2 and 3, are indicated with arrows, and labeled with inferred dates. The presence and type of CTX and SXT elements in each strain are shown to the right of the tree. The presence of TLC and RS1 elements are shown but their number and position, respectively, are arbitrarily assigned. A-D mark cases of sporadic intercontinental transmission. The dates shown are the median estimates for the indicated nodes, taken from the results of the BEAST analysis. * indicates where no data was available and the scale is given as the number of substitutions per variable site.
Significantly, the 7th pandemic tree can be subdivided into three major groups or clades by clustering analysis using Bayesian Analysis of Population Structure8,9 (shown as waves 1-3 in Fig 1), mostly consistent with their cholera toxin (CTX) type, representing independent waves of transmission. Although examples of genetic determinants differentiating these three CTX types have previously been published10, they have not previously been put into a phylogenetic context, undermining efforts to investigate the evolutionary aspects of their emergence. Perhaps consequently, there has been substantial uncertainty in naming new CTX-types, as they have been discovered. Our data shows that the first CTX type is canonical CTX-El Tor and we propose that it is re-named CTX-1, while for the other two we propose a new expandable nomenclature and class them as CTX-2 and CTX-3 (Table S2).
Isolates spanning A18 to PRL5 (the lower clade in Fig 1) represent wave 1 covering ~16 years (1977-1992). All isolates within this group lack an SXT/R391 family Integrative and Conjugative Element (ICE) encoding multiple antibiotic resistance3,11,12. It is within this time period that 7th pandemic cholera occurred in South America6. Our data shows that the South American isolates form a discrete cluster, which also includes a single Angolan isolate collected in 1989. The position of the Angolan isolate at the base of the South American group suggests that transmission to South America may have been via Africa as also proposed by Lam et al13. We used BEAST14 to translate evolutionary distance in SNPs into time (Fig S5), and this indicated that transmission to South America is likely to have occurred between 1981 and 1985. The branch harboring this West African-South American (WASA) clade is distinguished from all other V. cholerae by the acquisition of novel VSP-215 genes and a novel genomic island we have denoted WASA1 (Table S3). Strikingly, the Angolan isolate A5 and all the South American isolates are discriminated by just 10 SNPs. Based on the accumulation rate of 3.3 SNPs per year (Fig S4), the 3 year time period between the isolation of A5 and the oldest South American isolate A32 included in this study is consistent with previous studies suggesting that cholera spread as a single epidemic13.
The first acquisition of an SXT/R391 ICE, encoding multiple antibiotic resistances, lies at the point of transition from wave 1 strains being the dominant clinical isolates to those of wave 2. Using our dated phylogeny (Fig S5)14, we were able to date this transition and the first acquisition of SXT/R391 ICE to 1978-84, 10 years prior to its discovery in O139 strains, which also fits with the otherwise surprising discovery of SXT in a Vietnamese strain isolated before 199216. This date would also correspond to the most recent common ancestor (MRCA) of the O1 and O139 serogroup isolates. Analysis of the diversity of the common regions of SXT/R391 ICEs in our 7th pandemic collection (Fig S6) shows that they are discriminated by 3161 SNPs, compared to only 1757 SNPs, which were used to define the core whole genome phylogeny in Fig 1. This indicates that there have either been multiple recombination events within these ICEs, or that they have been independently acquired multiple times on the tree11. Isolates from wave 2 represent a discrete cluster that shows a complex pattern of accessory elements within the CTX locus (Fig 1) and a wide phylogeographic distribution. It is also important to note that isolates collected in Vietnam between 1995-2004 and strain A109 are the only wave 2 isolates we studied from this time period that lack an SXT/R391 ICE. We examined the genomic locus in these clones that marks the point of insertion of SXT/R391 ICE in all other V. cholerae isolates and found no remnants of this conjugative element, which may have been lost from this lineage (no DNA sequence ‘scar’ is expected following the precise excision of SXT/R391 ICE).
Interestingly, ignoring the CTX related genomic regions, the 7th pandemic L2 isolates show relatively little evidence of recombination either within or from outside of the tree. Based on the SNP distribution, 1930 out of 2027 SNPs (Table S4) are congruent with the tree, leaving 97 homoplasies that could be due to selection or homologous recombination amongst the L2 isolates. Just 270 SNPs were predicted to be due to homologous recombination from outside the tree. The only two branches where the SNP distribution suggested significant recombination were those leading to the WASA cluster (Fig S7) and the O139 serogroup. Aside from the acquisitions of CTX and the SXT/R391 ICEs, we found evidence of gene flux affecting only a further 155 genes (Fig S8, Table S3 and Fig S9).
Also represented within our collection are two isolates of serogroup O139, which are known to have arisen from a homologous replacement of their O-antigen determinant into an El Tor genomic backbone2,3,13. CTX types different from El Tor, classical, CTX-2 and 3 have been reported for the O139 serogroup17-20, however phylogenetic position of the two strains included in this study shows that O139 was derived from O1 El Tor and therefore represents another distinct, but spatially restricted wave from the common source.
We were also able to date the ancestor of the El Tor 7th pandemic lineage, L2, as having existed between 1827-1936 (Fig S5), which is consistent with the predicted date of origin from the linear regression plot (1910, Fig S4). This also corresponds well with the date of isolation of the first El Tor biotype strain in 190521.
It is apparent from Fig 1 that V. cholerae wave 1, which spread globally was later replaced by the more geographically restricted wave 2 and wave 3, a phenomenon supported by local clinical observations and phage analysis10. This also reflects the fact that V. cholerae epidemics since 2003 to 2010 have been restricted to South Asia and Africa. Interestingly, the rates of SNP accumulation calculated independently for wave 1, wave 3 and wave 2 (2.3, 2.6 and 3.5 SNPs/year respectively) are consistent with the rate calculated over the whole collection period (Fig S4).
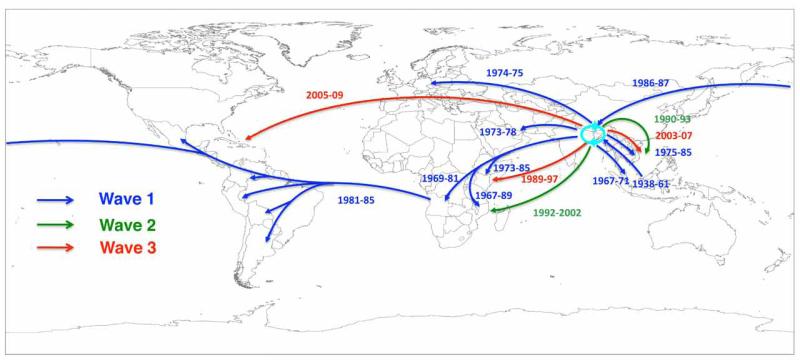
The clonal clustering of L2 isolates, the constant rate of SNP accumulation and the temporal and geographic distribution support the concept that the 7th pandemic has spread by periodic radiation from a single source population located in the Bay of Bengal, followed by local evolution and ultimately local extinction in non-endemic areas. This is evidenced by the disappearance of wave 1 isolates followed by the independent expansion of wave 2 and wave 3, both derived from the same original population, occurring within 7 years of each other. These two waves are clearly distinguished from the first by the acquisition of SXT/R391 ICEs (Fig 1). Plotting the intercontinental spread of each wave onto the world map (Fig 2) shows clearly that the V. cholerae 7th pandemic is sourced from a restricted single geographical location but has spread in overlapping waves. Within these ancestral waves, there are at least four recent long-range transmission events (A-D in Fig 1), where isolates clearly share a common ancestor with recent strains at a distant location indicating that such events are not uncommon. The most recent example of this is the Haitian outbreak, whose strains share a very recent common ancestor with South Asian strains at the tip of wave 3, and where the number of SNP differences even at whole genome resolution between the Haitian and most closely related Indian and Bangladesh strains is very low. This result demonstrates clearly that the Haitian strains must have come from South East Asia, at most within the last 6 years. However, the limited discrimination means that it may prove challenging to make country-specific inferences as to the origins of the Haitian strains based on DNA sequence alone, and in order for such conclusions to be robust, great care must be taken in the selection of samples for the analysis.
Fig 2.
Transmission events inferred for the 7th Pandemic phylogenetic tree drawn on a global map. The date ranges shown for transmission events are taken from the BEAST analysis, and represent the median values for the MRCA of the transmitted strains (later bound), and the MRCA of the transmitted strains and their closest relative from the source location (earlier bound).
Despite clear evidence of sporadic long-range transmission events likely to be associated with direct human carriage, the overall pattern seen in our data is one of continued local evolution of V. cholerae in the Bay of Bengal, with multiple independent waves of global transmission resulting in short term epidemics in non-endemic countries. Although our sample set is substantial, there are clearly areas where geographic coverage is limited. However, the structure of the tree, with deep branches between the major waves, means that further increasing the number of strains and the resolution should only indentify further independent waves of transmission. Indeed, we cannot rule out the possibility of an El Tor population persisting or evolving as a new wave of the 7th pandemic, for example in areas such as China that were not sampled in this study.
One significant factor in the ongoing evolution of pandemic cholera was the acquisition of the SXT/R391-family antibiotic resistance element. Interestingly, the clinical use of the antibiotics tetracycline and furazolidone for cholera treatment started in 1963 and 1968 respectively, ~15 years before our prediction of the first acquisition of an SXT/R391 ICE (1978-1984). Our analysis provides a robust framework for further elucidating the evolution of the 7th pandemic and for studying local evolution, particularly in the Bay of Bengal, which plays such a key role in cholera.
Methods
Genomic library creation and multiplex sequencing
Unique index-tagged libraries for each sample were created, and up to 12 separate libraries sequenced in each of 8 channels in Illumina Genome Analyzer GAII cell with 54bp paired-end reads. The index tag sequence information was used for downstream processing to assign reads to the individual samples4.
Detection of SNPs in core genome
54-base paired-end reads were mapped against the N16961 El Tor reference (accession numbers AE003852 and AE003853) and SNPs were identified using methods followed by Croucher et al7. The unmapped reads and the sequences that were not present in all the genomes were not considered a part of the core genome and therefore SNPs from these regions were not included in the analysis. Appropriate SNP cut offs were chosen to minimize the number of false positive and negative calls; SNPs were filtered to remove those at sites with a SNP quality score below 30, and SNPs at sites with heterogeneous mappings were filtered out if the SNP was present in less than 75% of reads at that site. From the 7th pandemic dataset, high-density SNP clusters indicating possible recombination were excluded7. In total 2027 SNPs were detected in the core genome of the El Tor lineage. Of these, 270 SNPs are predicted to be due to recombination. Removing these provides a dataset characterized by 1757 SNPs that were used to produce the final phylogeny.
Comparative Genomics
Raw Illumina data was split to generate paired end reads and was assembled using a de-novo genome assembly program Velvet v0.7.0322 to generate a multi-contig draft genome for each of 133 V. cholerae strains4. The parameters were optimized to give the longest kmer size and at least 20X kmer coverage. As 7th pandemic Vibrio cholerae strains are closely related in the core, Abacas23 was used to order the contigs using N16961 El Tor strain as reference followed by annotation transfer from the reference strain to each draft genome4. Using the N16961 sequence as a database to perform a TBLASTX24 for each draft genome, a genome comparison file was generated that was subsequently used in Artemis Comparison Tool25 to manually compare the genomes and search for novel genomic islands.
Phylogenetic Analysis
A phylogeny was drawn for V. cholerae using RAxML v0.7.426 to estimate the trees for all the SNPs called from core genome and the general time reversible (GTR) model with gamma correction was used for among site rate variation for ten initial trees4. US gulf coast strains A215 and A325, which have substantially different core genome from all other strains in our collection, were used as an outgroup to root the global phylogeny (Fig S1), whereas a pre-7th pandemic strain M66 (accession numbers CP001233 and CP001234) and strain A6 (from our collection) was used to root the 7th pandemic phylogenetic tree (Fig 1).
CTX prophage analysis
For each strain, the CTX structure and the sequence of rstA, rstR and ctxB was determined using approach same as Lee et al27 and Nguyen et al28.
Linear Regression and Bayesian Analysis
The phylogram for the 7th pandemic was exported to Path-O-Gen v1.3 (http://tree.bio.ed.ac.uk/software/pathogen) and a linear regression plot for isolation date vs. root to tip distance was generated. The same plot was also constructed individually for the three waves but A4, being a laboratory strain, was excluded from the latter analysis.
The presence of three waves was checked, and their makeup determined using a BAPS analysis performed on the SNP alignment containing the unique SNP patterns from the 7th Pandemic isolates. The program was run using the BAPS individual mixture model and three independent iterations were performed using an upper limit for the number of populations of 20, 21 and 22 to obtain the most optimal partitioning of the sample. The dates for the acquisition of SXT and the ancestors of the three waves were inferred using the Bayesian Markov Chain Monte Carlo framework BEAST29. We used the final SNP alignment with recombinant sites removed and fixed the tree topology to the phylogeny produced by RAxML, as described above. We used BEAST to estimate the rates of evolution on the branches of the tree using a relaxed molecular clock14, which allows rates of evolution to vary amongst the branches of the tree. BEAST produced estimates for the dates of branching events on the tree by sampling dates of divergence between isolates from their joint posterior distribution in which the sequences are constrained by their known date of isolation. The data were analyzed using a coalescent constant population size and a GTR model with gamma correction. The results are produced from three independent chains of 50 million steps each, sampled every 10,000 steps to ensure good mixing. The first 5 million steps of each chain were discarded as a burn-in. The results were combined using Log Combiner, and the maximum clade credibility tree was generated using Tree Annotator, both parts of the BEAST package (http://tree.bio.ed.ac.uk/software/beast/). Convergence and the effective sample size values were checked using Tracer 1.5 (available from http://tree.bio.ed.ac.uk/software/tracer) ESS values in excess of 200 were obtained for all parameters.
Nomenclature
The 7th cholera pandemic strains were clearly distinguished by three waves and we therefore propose their CTX types to be CTX-1, CTX-2 and CTX-3 under the new nomenclature scheme (see Table S2). Our nomenclature system is expandable and would be suitable for naming any new 7th pandemic Vibrio cholerae strains. With CTX-1 representing canonical El Tor, we followed the rationale defined below:
For CTX-1 to CTX-2, as there was a shift of rstREl Tor to rstRClassical, rstA El Tor to rstAClassical+El Tor and ctxBEl Tor to ctxBClassical, we called it CTX-2.
For CTX-1 to CTX-3, as there was a shift of ctxBEl Tor to ctxBClassical, we called it CTX-3.
For CTX-3 to CTX-3b, as there was only one SNP mutation in ctxBClassical from CTX-2 and rest was identical, we call it the next variant of CTX-3, which is CTX-3b.
In summary, if there is a shift of any gene from one biotype to another, the new CTX will be called CTX-‘n’ and so will be the strains e.g. the next strains fitting this criteria will be called CTX-4. However, if there is mutation(s) in the gene that does not lead to a shift of the gene to another biotype gene, CTX-1b, CTX-1c or CTX-2b, CTX-2c or CTX-3b, CTX-3c and so on should be followed as appropriate.
Supplementary Material
Acknowledgements
This work was supported by The Wellcome Trust and The IVI is supported by the Governments of Korea, Sweden, and Kuwait. JP was supported by Wellcome Trust grant 076964, DWK was partially supported by grant RTI05-01-01 from the Ministry of Knowledge and Economy (MKE), Korea and R01-2006-000-10255-0 from the Korea Science and Engineering Foundation and JLNW was supported by the Alborada Trust and the RAPIDD program of the Science & Technology Directorate, Department of Homeland Security. Special thanks to Andrew Camilli at Tufts University Medical School, Boston, USA for providing the corrected N16961 sequence, to BM Nguyen at NIHE, Vietnam and M Ansaruzzaman at ICDDR, Bangladesh for providing strains and to Maria Fookes at WTSI for the training support.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information All the genomic sequences have been submitted to the Genbank with the accession numbers listed in Table S1. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Chin CS, et al. The origin of the Haitian cholera outbreak strain. N Engl J Med. 2011;364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chun J, et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A. 2009;106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hochhut B, Waldor MK. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol. 1999;32:99–110. doi: 10.1046/j.1365-2958.1999.01330.x. [DOI] [PubMed] [Google Scholar]
- 4.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Update: cholera outbreak --- Haiti, 2010. MMWR Morb Mortal Wkly Rep. 2010;59:1473–1479. [PubMed] [Google Scholar]
- 6.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corander J, Marttinen P, Siren J, Tang J. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics. 2008;9:539. doi: 10.1186/1471-2105-9-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corander J, Waldmann P, Sillanpaa MJ. Bayesian analysis of genetic differentiation between populations. Genetics. 2003;163:367–374. doi: 10.1093/genetics/163.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safa A, Nair GB, Kong RY. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 2010;18:46–54. doi: 10.1016/j.tim.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Garriss G, Waldor MK, Burrus V. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet. 2009;5:e1000775. doi: 10.1371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wozniak RA, et al. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 2009;5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam C, Octavia S, Reeves P, Wang L, Lan R. Evolution of seventh cholera pandemic and origin of 1991 epidemic, Latin America. Emerg Infect Dis. 2010;16:1130–1132. doi: 10.3201/eid1607.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Shea YA, et al. The Vibrio seventh pandemic island-II is a 26.9 kb genomic island present in Vibrio cholerae El Tor and O139 serogroup isolates that shows homology to a 43.4 kb genomic island in V. vulnificus. Microbiology. 2004;150:4053–4063. doi: 10.1099/mic.0.27172-0. [DOI] [PubMed] [Google Scholar]
- 16.Bani S, et al. Molecular characterization of ICEVchVie0 and its disappearance in Vibrio cholerae O1 strains isolated in 2003 in Vietnam. FEMS Microbiol Lett. 2007;266:42–48. doi: 10.1111/j.1574-6968.2006.00518.x. [DOI] [PubMed] [Google Scholar]
- 17.Basu A, et al. Vibrio cholerae O139 in Calcutta, 1992-1998: incidence, antibiograms, and genotypes. Emerg Infect Dis. 2000;6:139–147. doi: 10.3201/eid0602.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faruque SM, Mekalanos JJ. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 2003;11:505–510. doi: 10.1016/j.tim.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Faruque SM, et al. The O139 serogroup of Vibrio cholerae comprises diverse clones of epidemic and nonepidemic strains derived from multiple V. cholerae O1 or non-O1 progenitors. J Infect Dis. 2000;182:1161–1168. doi: 10.1086/315807. [DOI] [PubMed] [Google Scholar]
- 20.Nair GB, Bhattacharya SK, Deb BC. Vibrio cholerae O139 Bengal: the eighth pandemic strain of cholera. Indian J Public Health. 1994;38:33–36. [PubMed] [Google Scholar]
- 21.Cvjetanovic B, Barua D. The seventh pandemic of cholera. Nature. 1972;239:137–138. doi: 10.1038/239137a0. [DOI] [PubMed] [Google Scholar]
- 22.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics. 2009;25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Carver T, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 27.Lee JH, et al. Classification of hybrid and altered Vibrio cholerae strains by CTX prophage and RS1 element structure. J Microbiol. 2009;47:783–788. doi: 10.1007/s12275-009-0292-6. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen BM, et al. Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J Clin Microbiol. 2009;47:1568–1571. doi: 10.1128/JCM.02040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.