Abstract
The main malaria vectors of sub-Saharan Africa, Anopheles gambiae sensu stricto and Anopheles arabiensis are morphologically indistinguishable, but often occur in sympatry and differ in feeding preference and vector competence. It is important to assess vector species identity for understanding the vectorial system and establishing appropriate vector control measures. The currently available species diagnosis methods for An. gambiae sensu latu require equipment to which public health practitioners in many African countries may not have access. This report describes a loop-mediated isothermal amplification technique (LAMP) for An. gambiae species diagnosis. The LAMP method was tested in single mosquito legs and whole body. The sensitivity and specificity of the LAMP method, in reference to the conventional rDNA-polymerse chain reaction (PCR) method, ranged from 0.93 to 1.00. The LAMP-based species identification method can be performed in a water bath and completed within 65 minutes, representing an alternative method for rapid and field applicable vector species diagnosis.
INTRODUCTION
With more than one million deaths per year, malaria is one of the most fatal infectious diseases in Africa.1 The majority of malaria-caused deaths occur in children less than 5 years of age. Vector control, mainly through insecticide-treated bed nets (ITNs) and indoor residual sprays, is one of the most important measures for malaria prevention. In sub-Saharan Africa, the principal malaria vector species are Anopheles gambiae sensu stricto (hereafter referred to as An. gambiae) and Anopheles arabiensis, both in the An. gambiae species complex.2 They are morphologically indistinguishable and often occur in sympatry, but they profoundly differ in their ability to vector malaria parasites, in host feeding preferences, resistance to desiccation, larval habitat requirement, and responses to the application of ITNs. Assessment of malaria risks, deployment of vector control techniques, and evaluation of the impact of control measures require information on the identity and abundance of vector species. Therefore, a simple and rapid identification method for An. gambiae and An. arabiensis is particularly valuable.
To date, several techniques have been developed to discriminate between the member species of An. gambiae species complex, including polytene chromosome binding patterns,3 isoenzyme electrophoresis,4 high-performance liquid chromatography of cuticular hydrocarbons,5 and polymerase chain reaction (PCR) methods targeting ribosomal DNA (rDNA) species-specific polymorphism.6–8 All these methods require expensive and delicate laboratory equipment to which public health practitioners in many African countries may not have access. As such, these techniques are not readily applicable in field settings in many developing countries.
Here, we describe a novel diagnostic method for discrimination between An. gambiae and An. arabiensis that can be performed rapidly with limited equipment requirements. This novel method takes advantage of the peculiarities of the loop-mediated isothermal amplification (LAMP) technique.9 The LAMP is a one-step nucleic acid amplification that relies on autocycling strand-displacement DNA synthesis. It is performed under isothermal conditions using a DNA polymerase with strand displacement activity. The LAMP technique uses one forward outer primer (F3), one backward outer primer (B3), one forward internal primer (FIP), and one backward internal primer (BIP). Specifically, the two external primers initiate the synthesis, and two internal primers have both sense and antisense sequences in such a way that a loop with a free 3′ end is generated as the amplification takes place.9 Moreover, the amplification products can be visualized directly.10 The LAMP method offers a high sensitivity, but requires simple equipment (e.g., a water bath) and no electrophoresis, rendering the method particularly suitable for field settings where sophisticated equipment is lacking.
MATERIAL AND METHODS
Mosquitoes
Anopheles gambiae G3 strain and An. arabiensis Dongola strain, obtained from the Malaria Research and Reference Reagent Resource Center (MR4), were used for LAMP method development. The LAMP method validation against the conventional rDNA-PCR method used morphologically identified An. gambiae sensu latu adult mosquitoes collected in two sites in western Kenya. The first was Asembo Bay (34°22′0E, 0°–14°60′N, elevation about 1,200 m above sea level), Siaya District, Nyanza Province, collected between June and September 2008. The second was from Iguhu subdistrict (34°–35°E, 0°010″S, elevation 1480–1580 m), Kakamega district, western province, collected in May–June 2008. Anopheles arabiensis is known to be the predominant species in Asembo Bay after the use of insecticide-treated bednets,11 whereas An. gambiae is the major malaria vector in Iguhu.12 Mosquitoes were collected by aspiration and pyrethrum indoor spray catch methods. Mosquito samples were stored individually at −20°C and sent to the University of California at Irvine for molecular analyses.
Preparation of the DNA template
We used two methods to extract mosquito DNA. The first method used the Promega Wizard Genomic DNA purification kit (Promega, Madison, WI), and the extracted DNA from individual mosquitoes was used for LAMP method development. The second method (termed as cheap method) involved simple procedures, and was intended for applications under field settings. Extracted DNA from the second method was used for comparison between the LAMP method and rDNA-PCR method for An. gambiae and An. arabiensis species identification. Briefly, mosquito legs or the remaining carcasses were ground in a freshly prepared extraction buffer (0.5% SDS, 0.2 M NaCl, 25 mM EDTA, 10 mM tris HCl ph 8, RNasi 20 mg/mL). Following incubation at 37°C for 60 min, proteinase K (20 mg/mL) was added, and then incubated at 50°C for 60 min. The tube was centrifugated at 13,000 rpm for 10 min, and the supernatant was recovered and precipitated in cold 100% ethanol. The DNA was washed with 70% ethanol, dried, and re-suspended in molecular grade water.
LAMP assay
The LAMP primers were designed using the program PrimerExplorer V413 based on the intergenic spacer region (IGS) of An. gambiae and An. arabiensis 28S rDNA sequences. The LAMP reactions were carried out using the loopamp DNA amplification kit, following manufacturer’s recommended procedures (Eiken Chemical Co., Tokyo, Japan). Briefly, 2 µL of genomic DNA (about 5 to 10 ng) were placed in a reaction tube along with 12.5 µL of reaction buffer, 5 pmol of F3 and B3 primer, 40 pmol of BIP and FIP primers, 1 µL of Bst DNA polymerase, and 1 µL of fluorescent reagent calcein. Molecular grade water was added to reach a final volume of 25 µL. Reaction tubes were placed under isothermal conditions, at 63°C for 1 hr, followed by 5 min at 80°C to inactivate the enzyme. The color of the final amplification product was inspected visually: a positive amplification shows a turbid yellow liquid, whereas in the absence of amplification, the reaction mixture retains a clear orange-like color.9 We first tested the specimen using An. gambiae specific primers, and then repeated the reaction using An. arabiensis -specific primers. If a specimen were amplified by both An. gambiae -specific primers and An. arabiensis -specific primers, the specimen was recorded as “hybrid.” When a sample was identified as hybrid, the LAMP reaction was repeated two other times with each set of primers to confirm the result.
To reduce the costs associated with LAMP amplification, we performed the LAMP assays using reduced reaction volumes: reduction by half (12.5 µL final volume) and by two-thirds (8.3 µL final volume) from the original 25 µL reaction. The concentration of the primers was maintained at 40 pmol for FIP/BIP and 5 pmol for F3/B3.
rDNA-PCR for species identification
To determine the specificity and the sensitivity of the LAMP method, we compared the species identity results from the LAMP method with the rDNA-PCR method6 for mosquito samples collected in Asembo (N = 23) and Iguhu (N = 124). The rDNA-PCR method is the most frequently used method for species diagnosis of mosquitoes of the An. gambiae complex. It is based on a PCR reaction involving primers designed on the IGS 28S rDNA sequence: one primer is species-specific, the other is based on a sequence conserved among the species of the complex.6 Gel electrophoresis of the resulting PCR product allows the identification of the species based on a different size for the PCR fragments: An. gambiae DNA generates a PCR fragment of 390 bp, An. arabiensis a fragment of 310 bp.6 The original rDNA-PCR protocol did not include An. bwambae, which was shown later to harbor enough polymorphism in the same IGS 28S rDNA region to allow the design of a species-specific primer.14 The hybrids identified by the rDNA-PCR method were repeated three times to confirm the diagnosis result. The specificity and the sensitivity of the LAMP method were calculated using the rDNA-PCR method as the “gold standard.”15,16
RESULTS
LAMP primer design and amplification using laboratory-reared mosquitoes
Using the PrimerExplorer V4 program, we designed LAMP primers to encompass the IGS region of the 28S rDNA gene of An. gambiae and An. arabiensis, previously shown to harbor species-specific polymorphism6 (Table 1). The forward outer primer (F3) and backward outer primer (B3) were designed to contain two point mutations each. The FIP and the BIP contain seven point mutations and an insertion/deletion of three bases. The LAMP amplification of An. gambiae and An. arabiensis uses two separate sets of primers (Table 1).
Table 1.
Primer sequences of the LAMP-based Anopheles gambiae and Anopheles arabiensis species identification method
| Species | Primer | Sequence (5′-3′) |
|---|---|---|
| An. gambiae | F3 | ACGTAACACTAGTGAGCTTGTC |
| B3 | CCACCTCGACACACGACG | |
| FIP | GGTGTGTAAGCTTACTGGTTTGGTGCGTGCCTCGTTCTCGA | |
| BIP | ATAAGTTAATCCGTTTGGGCCGGTAACCGAACATGGTCAACAACA | |
| An. arabiensis | F3 | AGGACACTTAACACTAATGAGC |
| B3 | CTCGACACACGACCTGTT | |
| FIP | CGAGCATGTGTAAGCTTACTGGTTCTCGACTTGATTGTCTTGATG | |
| BIP | AGTTGTATAAGTTGACCCGTTTGGCAACCGAACATGGTCAACACC |
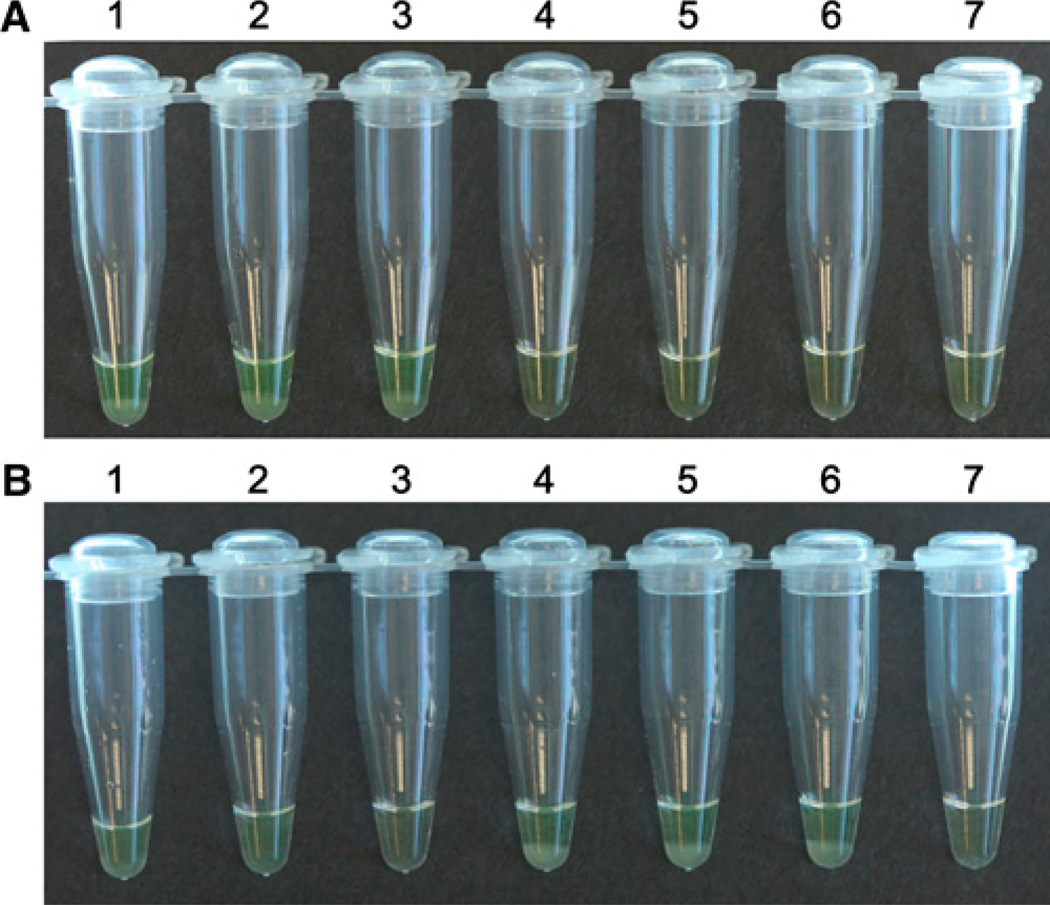
We tested the utility of the designed LAMP primers and amplification protocol using DNA samples from laboratory-reared mosquitoes (An. gambiae G3 and An. arabiensis Dongola strains). Results of this test confirmed that the primers designed on the IGS 28S rDNA sequence of An. gambiae work exclusively on An. gambiae DNA, and did not amplify An. arabiensis DNA (Figure 1B). Similarly, the primers designed for An. arabiensis specifically amplified An. arabiensis DNA, but not An. gambiae (Figure 1A).
Figure 1.
LAMP method for Anopheles gambiae and Anopheles arabiensis species diagnosis. A, LAMP reaction using An. arabiensis -specific primers; and B, LAMP reaction using An. gambiae -specific primers. Tubes 1–3: DNA samples from three An. arabiensis Dongola individuals; Tubes 4–6: DNA samples from three An. gambiae G3 individuals. Tube 7: negative control with water as a template. Positive samples are shown with a turbid yellow liquid, with a white precipitate (tubes 1–3 in panel A and tubes 4–6 in panel B) that results from amplification success. A clear orange-like liquid (tubes 4–7 in panel A and tubes 1–3 and tube 7 in panel B) indicates lack of LAMP amplification. This figure appears in color at www.ajtmh.org.
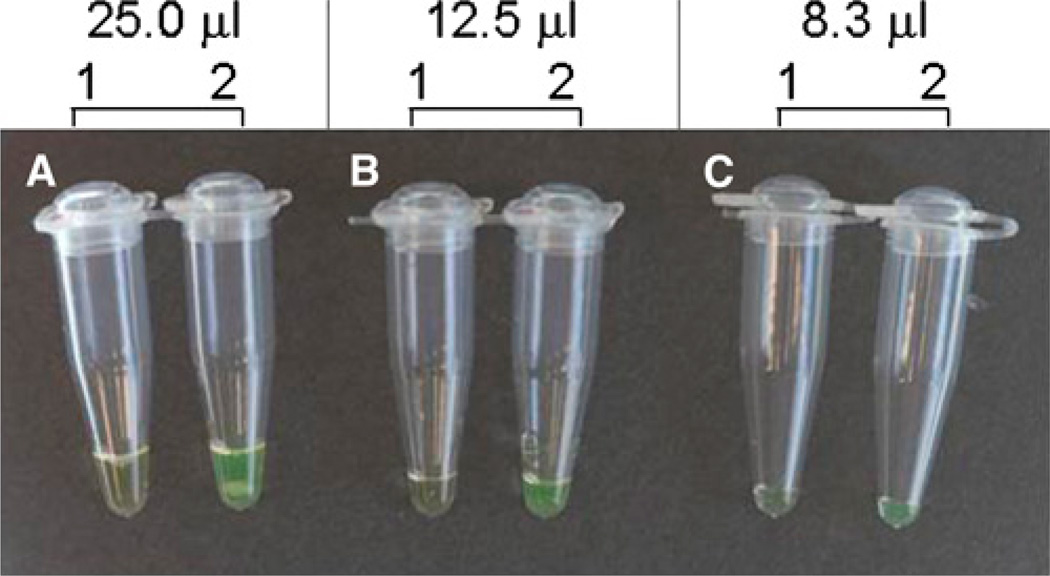
To reduce the amount of reagents for LAMP reaction, we compared three volumes of LAMP reactions (25, 12.5, and 8.3 µL final volumes). We found that reducing LAMP reaction volume has no impact on the ability to distinguish the two species (Figure 2). Consequently, the lowest volume (8.3 µL) was adopted for LAMP method validation with field collected An. gambiae s.l. mosquitoes.
Figure 2.
Effects of LAMP reaction volumes on the ability of Anopheles gambiae species diagnosis. One An. gambiae mosquito collected from Iguhu, Kakamega was used as template for all reactions. A, 25 µL final reaction volume; B, 12.5 µL final reaction volume; and C, 8.3 µL final reaction volume. Tube 1: LAMP reaction using An. arabiensis- specific primers; and tube 2: LAMP reaction using An. gambiae -specific primers. A turbid yellow liquid (tube 2) indicates positive amplification, whereas a clear orange-like liquid (tube 1) shows lack of application. No difference in the ability to diagnose An. gambiae species was detected among the LAMP reactions with different volumes. Please note the white precipitate (pyrophosphate ions) generated by the LAMP reaction in the bottom of the tube. This figure appears in color at www.ajtmh.org.
LAMP method specificity and sensitivity on field-collected mosquitoes
A total of 147 An. gambiae s.l. mosquitoes, 23 from Asembo Bay and 124 from Iguhu were tested. Results of the LAMP-based species identification assay are summarized in Table 2. Out of the 147 mosquitoes tested, 18 mosquitoes were identified as An. arabiensis (14 from Asembo Bay, 4 from Iguhu); 108 as An. gambiae (1 from Asembo Bay, 107 from Iguhu); and 12 as hybrid between An. gambiae and An. arabiensis (1 from Asembo Bay, 11 from Iguhu). Nine mosquitoes (7 from Asembo Bay, 2 from Iguhu) showed no amplification with either primer sets. The number of mosquitoes that were not amplified by the LAMP method was the same as that with the conventional rDNA-PCR species identification method, with the exception of three samples from Asembo Bay that were identified as hybrids by the rDNA-PCR method (Table 2).
Table 2.
Sensitivity and specificity of the LAMP-based Anopheles gambiae and Anopheles arabiensis species identification method, in comparison to the conventional rDNA-polymerase chain reaction (PCR) method
| No. mosquitoes (Asembo Bay, Iguhu) | Sensitivity | Specificity | ||
|---|---|---|---|---|
| Species | LAMP method | rDNA-PCR | (95% CI)* | (95% CI)* |
| An. gambiae | 108 (1, 107) | 116 (1, 115) | 0.93 (0.87–0.96) | 1.0 (0.89–1.00) |
| An. arabiensis | 18 (14, 4) | 18 (14, 4) | 1.0 (0.82–1.00) | 1.0 (0.97–1.00) |
| Hybrid | 12 (1, 11) | 7 (4, 3) | ||
| Not amplified | 9 (7, 2) | 6 (4, 2) | ||
| Total | 147 (23, 124) | 147 (23, 124) | ||
95% CI refers to 95% confidence interval.
To assess the specificity and the sensitivity of the LAMP-based species identification method, all 147 field caught mosquitoes were subjected to the conventional rDNA-PCR species identification method.6 For samples from Asembo Bay, results from the two methods were congruent, with the exception that three specimens identified as hybrids between An. gambiae and An. arabiensis by the rDNA-PCR method showed no amplification by the LAMP method. In Iguhu, results were consistent between the two methods except that 8 out of 118 samples, which scored as An. gambiae by the rDNA-PCR method, were identified as An. gambiae / An. arabiensis hybrid by the LAMP. In comparison to the rDNA-PCR method, the LAMP method showed a sensitivity between 0.93 and 1.00, and a specificity of 1.00 (Table 2). Overall, the two methods yielded similar species composition for the two sites (G = 4.80, degrees of freedom [df] = 3, two-tailed test, P = 0.19).
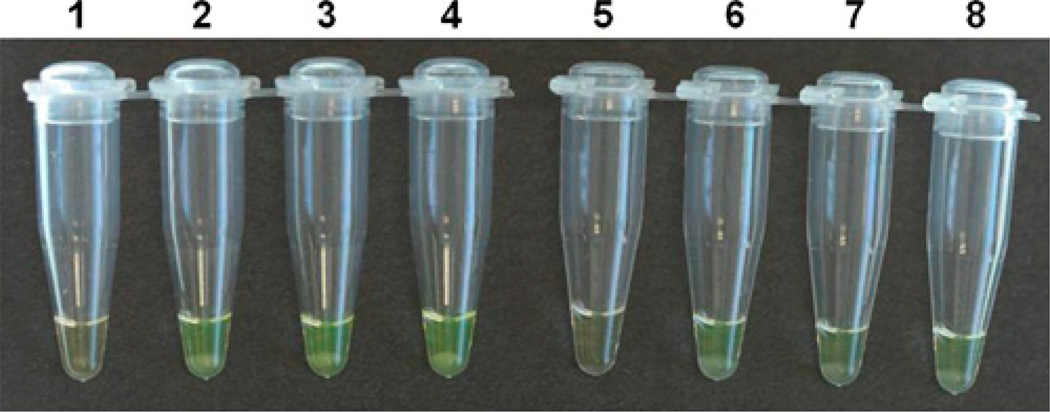
Because An. gambiae s.l. mosquito head is often used for Plasmodium falciparum sporozoite detection and abdomen for bloodmeal analysis, mosquito leg is the preferred body part for species identification. We therefore tested whether mosquito leg is sufficient for species diagnosis by the LAMP method. The DNA from a single mosquito leg and from the remaining body carcass was extracted using the second method described previously. Even though the LAMP products from the single mosquito leg were slightly fainter than the ones from the remaining body carcass, mosquito species can be determined with certainty (Figure 3). Therefore, a single mosquito leg is sufficient for LAMP-based An. gambiae and An. arabiensis species diagnosis.
Figure 3.
LAMP-based Anopheles gambiae and Anopheles arabiensis identification using template DNA from a single mosquito leg and whole body. LAMP assay using An. arabiensis -specific primers (tubes 1–4) and An. gambiae -specific primers (tubes 5–8). Tubes 1 and 5: negative controls using water as template; tubes 2 and 6: the positive controls using DNA extracted by Promega Wizard Genomic DNA purification kit (Promega, Madison, WI) from the whole body of An. arabiensis Dongola and An. gambiae G3, respectively; tube 3: single leg of An. arabiensis Dongola; tube 4: whole body of An. arabiensis Dongola; tube 7: single leg of An. gambiae G3; and tube 8: whole body of An. gambiae G3. Tubes with a turbid yellow liquid, with a white precipitate, indicate positive amplification (tubes 2–4 and 6–8), and those showing a clear orange-like liquid indicate lack of LAMP amplification (tubes 1 and 5). This figure appears in color at www.ajtmh.org.
DISCUSSION
The An. gambiae species complex comprises seven morphologically indistinguishable species, of which only An. gambiae and An. arabiensis are the principal malaria vectors in sub-Saharan Africa; the other five species (Anopheles melas, Anopheles merus, Anopheles bwambae, and Anopheles quadriannulatus species A and B) have limited geographic distribution or are not competent malaria vectors.2,7,8,17–20 It is on this basis that the present study focused on developing a rapid and field applicable diagnostic method for An. gambiae and An. arabiensis. The LAMP method was based on the 28S rDNA gene sequences previously shown to harbor point mutations and insertions/deletions among An. gambiae, An. arabiensis, An. melas, An. merus, An. quadriannulatus, and An. bwambae.6,14 Because the LAMP method requires four primers that recognize six different regions of the target DNA, it is supposed to be extremely specific.9 Consequently, based on the expected specificity of the LAMP amplification and the fact that the IGS region of the 28S rDNA was shown to contain enough polymorphism to allow the PCR-based identification of the members of the An. gambiae species complex,6–8,14 it is very unlikely that the LAMP primers here described for the identification of An. gambiae and An. arabiensis will also amplify other members of the An. gambiae species complex.
The major difference between the LAMP and the rDNA-PCR methods is that the LAMP method does not require a thermocycler and does not require gel electrophoresis. The LAMP-based diagnostic method is performed under isothermal conditions (e.g., in a water bath), and is completed within 65 minutes. Furthermore, the species identity results can be determined visually based on the turbidity and the color of the reaction mixture. During the LAMP amplification, large amounts of pyrophosphate ions are generated leading to a white precipitant that can be directly visualized.9 To facilitate the discrimination between positive and negative LAMP results, the fluorescent dye calcein can be used as in the present study.10 When calcein is added to the reaction mixture, the positive LAMP products appear turbid yellow-green, whereas the negative retain a clear orange-like color.
We showed a high sensitivity of the LAMP-based species diagnosis method. In comparison to the rDNA-PCR method for An. gambiae s.l. species diagnosis, the LAMP method exhibited a sensitivity of 0.93 and a specificity of 1.0 for An. gambiae; for An. arabiensis sensitivity and specificity were both 1.0. The discrepancy between the LAMP method and rDNA-PCR method was in the detection of An. gambiae / An. arabiensis hybrids, with the number of hydrids detected by the two methods being substantially different for the Iguhu samples (Table 2). In nature, hybrids have been observed at a very low rate.21 However, the present study found a quite high proportion of hybrids by the two methods (8.2% by LAMP method and 4.8% by rDNA-PCR method). Because the hybrids were confirmed by repeating both methods three times, the high proportion of hybrids in the population is not likely caused by contaminations or misidentification. Overall, the two methods detected a similar species composition in the two study sites. Similar to the rDNA-PCR method, the LAMP assay can detect species identity when DNA from a single mosquito leg is used. Thus, other body parts of a mosquito can be used for other tests, such as head for sporozoite infection and abdomen for bloodmeal source test.
We estimate that the material cost for one LAMP reaction based on the current price for the loopamp DNA amplification kit by the manufacturer (Eiken Chemical Co.) is about 1.6 US dollars per reaction if 8.3 µL final reaction volume is used. Additionally, it has been suggested that purchasing the chemicals for the LAMP reaction separately, rather than in kit form, will reduce the overall cost of the LAMP reaction to less than one dollar.22 Recently, the LAMP method was successfully applied to the identification of oocysts and sporozoites of Plasmodium berghei in Anopheles stephensi mosquitoes.23 This report, along with modern advancements of the LAMP technique toward the simultaneous detection of two amplicons in a single reaction24 further enhance the feasibility and convenience for field application of the LAMP method for mosquito species diagnosis, and detection of P. falciparum sporozoites simultaneously.
Acknowledgments
We thank the two anonymous reviewers for their comments, which helped to improve the manuscript.
Financial support: This research is supported by grants from the National Institutes of Health (R01 AI050243 and D43 TW001505).
REFERENCES
- 1.World Health Organization. Malaria Control Today: Current WHO Recommendations. Geneva: Roll Back Malaria Department; 2005. [Google Scholar]
- 2.Coetzee M, Craig M, le Sueur D. Distribution of African malaria mosquitoes belonging to the Anopheles gambiae complex. Parasitol Today. 2000;16:74–77. doi: 10.1016/s0169-4758(99)01563-x. [DOI] [PubMed] [Google Scholar]
- 3.Coluzzi MS, Petrarca V, de Deco MA. Chromosomal differentiation and adaptation to human environment in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979;73:479–483. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- 4.Coosemans M, Smits A, Roelants P. Intraspecific isozyme polymorphism of Anopheles gambiae in relation to environment, behavior, and malaria transmission in southwestern Burkina Faso. Am J Trop Med Hyg. 1998;58:70–74. doi: 10.4269/ajtmh.1998.58.70. [DOI] [PubMed] [Google Scholar]
- 5.Carlson DA, Service M. Differentiation between species of the Anopheles gambiae Giles complex (Diptera: Culicidae) by analysis of cuticular hydrocarbons. Ann Trop Med Parasitol. 1979;73:589–592. doi: 10.1080/00034983.1979.11687301. [DOI] [PubMed] [Google Scholar]
- 6.Scott JA, Brogdon W, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 7.Walker ED, Thelen AP, Bullard BA, Huang J, Odiere MR, Bayoh NM, Wilkins EE, Vulule JM. Identification of field caught Anopheles gambiae s.s. and Anopheles arabiensis by TaqMan single nucleotide polymorphism genotyping. Malar J. 2007;6:23–30. doi: 10.1186/1475-2875-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bass CW, Field LM. Development of a multiplex real-time PCR assay for identification of members of the Anopheles gambiae species comples. Acta Trop. 2008;107:50–53. doi: 10.1016/j.actatropica.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Parida M, Sannarangaiah S, Dash PK, Rao PV, Morita K. Loop mediated isothermal amplification (LAMP): a new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev Med Virol. 2008;18:407–421. doi: 10.1002/rmv.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomita N, Mori YY, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 11.Lindblade KA, Gimnig JE, Kamau L, Hawley WA, Odhiambo F, Olang G, Ter Kuile FO, Vulule JM, Slutsker L. Impact of sustained use of insecticide-treated bednets on malaria vector species distribution and culicine mosquitoes. J Med Entomol. 2006;43:428–432. doi: 10.1603/0022-2585(2006)043[0428:iosuoi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 12.Minakawa N, Munga S, Atieli F, Mushinzimana E, Zhou G, Githeko AK, Yan G. Spatial distribution of anopheline larval habitats in western Kenyan highlands: effects of land cover types and topography. Am J Trop Med Hyg. 2005;73:157–165. [PubMed] [Google Scholar]
- 13.Explorer P. PrimerExplorer V.4. [Accessed June 16, 2009];2005 Available at: http://primerexplorer.jp/elamp4.0.0/index.html. [Google Scholar]
- 14.Towson HOA. Identification by rDNA-PCR of Anopheles bwambae, a geothermal spring species of the An. gambaie complex. Insect Mol Biol. 1994;3:279–282. doi: 10.1111/j.1365-2583.1994.tb00178.x. [DOI] [PubMed] [Google Scholar]
- 15.Banoo S, Bell D, Bossuyt P, Herring A, Mabey D, Poole F, Smith PG, Sriram N, Wongsrichanalai C, Linke R, O’Brien R, Perkins M, Cunningham J, Matsoso P, Nathanson CM, Olliaro P, Peeling RW, Ramsay A. Evaluation of diagnostic tests for infectious diseases: general principles. Nat Rev Microbiol. 2006;4:S20–S32. doi: 10.1038/nrmicro1570. [DOI] [PubMed] [Google Scholar]
- 16.Confidence Interval Calculator. Confidence Interval Calculator V4. [Accessed June 16, 2009];2002 Available at: http://vl.academicdirect.org/applied_statistics/binomial_distribution/ref/CIcalculator.xls. [Google Scholar]
- 17.Coetzee M. Distribution of the African malaria vectors of the Anopheles gambiae complex. Am J Trop Med Hyg. 2004;70:103–104. [PubMed] [Google Scholar]
- 18.Moreno MC, Nzambo S, Bobuasaki L, Buatiche JN, Ondo M, Micha F, Benito A. Malaria Panel Assay versus PCR: detection of naturally infected Anopheles melas in a coastal village of Equatorial Guinea. Malar J. 2004;3:20–25. doi: 10.1186/1475-2875-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsy LD, Marrama L, Rabarison P, Le Goff G, Rajaonarivelo V, Robert V. Distribution of the species of the Anopheles gambiae complex and first evidence of Anopheles merus as a malaria vector in Madagascar. Malar J. 2003;2:33–39. doi: 10.1186/1475-2875-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White GB. Anopheles bwambae sp.n., a malaria vector in Semliki Valley, Uganda, and its relationships with other sibling species of the An. gambiae complex. Syst Entomol. 1985;10:501–522. [Google Scholar]
- 21.Temu EA, Coetzee M, Minjas JN, Schiff CJ. Detection of hybrids in natural populations of the Anopheles gambiae complex by rDNA-based, PCR method. Ann Trop Med Parasitol. 1997;8:963–965. doi: 10.1080/00034989760383. [DOI] [PubMed] [Google Scholar]
- 22.Poon LL, Ma EH, Chan KH, Chow LM, Abeyewickreme W, Tangpukdee N, Yuen KY, Guan Y, Looareesuwan S, Peiris JS. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem. 2006;52:303–306. doi: 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- 23.Aonuma H, Suzuki M, Iseki H, Perera N, Nelson B, Igarashi I, Yagi T, Kanuka H, Fukumoto S. Rapid identification of Plasmodium-carrying mosquitoes using loop-mediated isothermal amplification. Biochem Biophys Res Commun. 2008;376:671–676. doi: 10.1016/j.bbrc.2008.09.061. [DOI] [PubMed] [Google Scholar]
- 24.Mori Y, Hirano T, Notomi T. Sequence specific visual detection of LAMP reactions by addition of cationic polymers. BMC Biotechnol. 2006;6:3–12. doi: 10.1186/1472-6750-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]