Abstract
Although larvae feeding and food source are vital to the development, survival and population regulation of African malaria vectors, the prey organisms of Anopheles gambiae larvae in the natural environment have not been well studied. This study used a molecular barcoding approach to investigate the natural diets of Anopheles gambiae s.l. larvae in Western Kenya. Gut contents from third- and fourth-instar larvae from natural habitats were dissected and DNA was extracted. The 18S ribosomal DNA gene was amplified, the resulting clones were screened using a RFLP method and non-mosquito clones were sequenced. Homology search and phylogenetic analyses were then conducted using the sequences of non-mosquito clones to identify the putative microorganisms ingested. The phylogenetic analyses clustered ingested microorganisms in four clades, including two clades of green algae (Chlorophyta, Chlorophyceae Class, Chlamydomonadales and Chlorococcales families), one fungal clade, and one unknown eukaryote clade. In parallel, using the same approach, an analysis of the biodiversity present in the larval habitats was done. This present study demonstrated the feasibility of the barcoding approach to infer the natural diets of Anopheles gambiae larvae. Our analysis suggests that despite the wide range of microorganisms available in natural habitats, mosquito larvae fed on specific groups of algae. The novel tools developed from this study can be used to improve our understanding of the larval ecology of African malaria vectors and to facilitate the development of new mosquito control tools.
Keywords: food source, barcoding, Anopheles gambiae s.l., Kenya, larval ecology
Introduction
Larval ecology of Anopheles gambiae mosquitoes has been a neglected area. Despite their role as vectors of malarial parasites in Africa, important ecological characteristics such as feeding behaviors, natural diets along the larval development and population regulation are poorly understood (Merritt et al. 1992b). Understanding the factors that regulate larval mosquito populations is considered fundamental to predict transmission rates (Okech et al. 2007) and to control vector population (Molineaux 1997). Moreover, identification of food sources and their importance to larval development and survival is particularly relevant to the development of novel vector control methods such as identification of productive habitats for targeted control, encapsulation of bacterial toxins into larval food items, or genetic modification of bacteria or algae to spread parasite-inhibiting genes (Federici et al. 2003; Thiery et al. 1991).
Although predation is vital to the dynamics and regulation of anopheline larval populations (Service 1976), the prey organisms of the anopheles larvae in the natural environment have not been determined. Are they generalists or do they select their food source? What type of food do they ingest? Anopheles larvae utilize “mouth brushes” or lateral palatal brushes in the labrum of the head to generate currents for ingestion of food particles approaching the mouth, an active filter-feeding behavior that involves energy expenditure for creation of feeding currents (Merritt et al. 1992a,b). The mechanisms by which Anopheles larvae capture and retain food particles are poorly understood, but videotape recording and microscopic observations of the larval mouthparts of An. quadrimaculatus have suggested that the mouth brushes act as paddles to create currents or flows, thereby collecting particles from the surface microlayers and bringing them to the cibarium (Merritt et al. 1992a). Further, Merritt et al. (1992a) suggested that algae and microorganisms are the main food source of Anopheles quadrimaculatus larvae, and mosquito larvae are not discriminatory in the type of food they ingest. Heterotrophic bacteria are thought to be important in larval diet (Wallace, Merritt 2004; Wotton et al. 1997), but they have been shown to be a minor constituent in An. albimanus larval diet (Rejmankova et al. 1996).
Anopheles gambiae sensu stricto and An. arabiensis, two sibling species responsible for most of the malaria transmission in sub-Saharan Africa, are common in temporary, small, sunlit, turbid aquatic habitats (Gimnig et al. 2001). The importance of algae as the main food source for An. gambiae s.s. has been demonstrated in western Kenya (Gimnig et al. 2001; 2002). Tuno et al. (2006) established a strong, positive association between the presence of epizoic algae and An. gambiae s.l. larval abundance, emphasizing the importance of the algal communities in the regulation of mosquito larval populations. In laboratory experiments, the size of the particles ingested by An. gambiae s.s. varies from 0.1 to 165μm in diameter and the physiological state of the blue-green algae strain appears to be critical (Thiery et al. 1991). Interestingly, Ye-Ebiyo et al. (2000) found that larvae ingesting maize pollen exhibited higher fitness. Attempts have been made to analyze gut contents through epifluorescence microscopy with DAPI staining in Asian anopheline species (An. culicifacies s.l. and An. varuna) (Piyaratne et al. 2005) and in An. quadrimaculatus in North America (Wallace, Merritt 2004). These studies identified detritus, bacteria, filamentous algae, diatoms and desmids in the guts of larvae. However, this method did not identify biotic materials at a specific level. Further, gut contents in the larvae of An. gambiae s.l. have not been published.
A new DNA fingerprinting technology termed barcoding has been recently developed to investigate biodiversity (Miller 2007). This technique is based on the idea that a short DNA sequence can be used to identify organisms. For example, cytochrome oxidase I (COI) sequencing, which is the reference barcoding sequence, has been used to catalog Indian mosquito biodiversity (Kumar et al. 2007). This DNA-based approach has also been applied to study diet and prey organisms of several marine invertebrates or vertebrates (Blankeship, Yayanos 2005; Suzuki et al. 2006). If DNA from consumed organisms is not completely degraded by digestion, certain genes could be amplified via PCR and sequenced (Jarman et al. 2004). Subsequently, DNA sequences recovered from gut contents can be compared against DNA databases to provide taxonomic insight of the consumed organisms. In the present study, we investigated prey organisms of An. gambiae and An. arabiensis larvae using the barcoding approach with universal primers targeting the variable central domain of the 18S rDNA gene.
Material and methods
Sample collection and DNA extraction
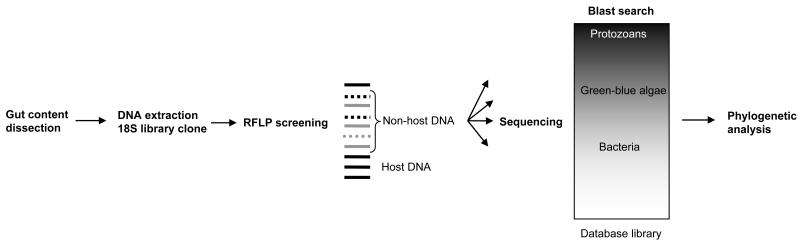
The flow chart of the experimental protocol is presented in Fig. 1. Larvae and water samples were collected in larval habitats around the Vector Biology Control and Research Center near Kisumu, in western Kenya where Anopheles gambiae s.s. and An. arabiensis are dominant malaria vector species (Gimnig et al. 2001). Collections were made from July to September 2006. Larvae were collected using a standard 350-ml dipper. All stages of anopheline larvae were kept in ethanol. Water samples were collected in the larval habitats where larvae were collected and transported to the laboratory and stored at 4 °C until centrifugation. Third and 4th instars were randomly selected to be dissected under a binocular microscope using sterilized dissection devices. Prior to dissection, the surface of the larvae was washed and rinsed three times with sterile distilled water to remove contaminants from the body surface. Gut contents were dissected and added into a 1.5-ml tube with 100uL of DNA extraction buffer (0.5% SDS, 0.2M NaCl, 25mM EDTA and 10mM Tris pH=8.0 in final concentrations). Gut contents were individually homogenized and ground in tubes with a pestle, and then DNA of the gut contents was extracted following the method of Linton et al. (2001). The remaining carcasses were kept in individual tubes, and the DNA was extracted following Linton et al. (2001). The allele-specific PCR assay of Scott et al. (1993) was used to identify all specimens to An. gambiae s.s. and An. arabiensis. Out of 73 DNA-extracted larvae, 5 were identified as An. gambiae s.s. and 68 as An. arabiensis.
Figure 1.
A diagram of experimental procedure.
Fifty milliliters of the surface water was collected from the same larval habitats, and the water was centrifuged three times at 13,000 rpm for 15 minutes. The pellets were preserved in 100% ethanol and then the DNA was extracted, using Ultraclean™ Soil DNA extraction kit (Mo Bio Laboratories, Carlsbad, CA).
18S rDNA amplification
The 18S rDNA, using both gut content DNA and water sample 0 DNA as templates, was amplified by PCR using the universal primers 5′-ATCCAAGGAAGGCAGCAGGC-3′ [18SU467F] and 5′-CTCCACCAACTAAGAACGGC-3′ [18SU1310R], primers that can amplify a wide range of eukaryotes (Suzuki et al. 2006). The PCR product size was ~800bp. PCR reactions were performed in a 25-ul reaction volume containing 200uM dNTPS, 10× buffer, 10uM of each primer, and 0.5 units of Taq polymerase. Thermal cycling conditions were initial denaturation at 95 °C for 2 min, followed by 30 cycles at 95 °C for 2 min, 55°C for 30 sec, 72 °C for 1.5 min, and then a final elongation at 72 °C for 10min. PCR products were electrophoresed through a 2% agarose gel in TBE buffer, and stained with ethidium bromide.
Cloning, screening and sequencing of the amplified fragments
PCR products were purified using ExoSAP-It (Amersham Pharmacia Biotech, Piscataway, NJ) following the manufacturer’s instructions. PCR products were ligated into pGEMTr-T Easy Vector System (Promega, Madison, WI) and transformed into competent One Shot cells (Invitrogen, Carlsbad, CA) following manufacturer’s instructions. The transformed clones were first screened by PCR with the 18S primer set. During the process of gut contents dissection, it was inevitable that mosquito host cells inside the food bolus were collected. Thus, mosquito DNA may also be cloned. To differentiate mosquito DNA clones from non-mosquito DNA clones, all clones were screened with a restriction enzyme PshA I (New England Biolabs, Inc., Ipswich, MA) that digests mosquito 18S DNA but not algal 18S DNA based on known DNA sequences. This endonuclease was selected based on the unique sequence difference between algal and mosquito 18S sequences (data not shown). Minipreps were prepared from non-mosquito clones and purified using the QIAgen Spin Miniprep Kit (Qiagen, Valencia, CA). Clones were then sequenced twice using the forward primer.
Similarity search and sequence analysis
The DNA sequences from non-mosquito clones were subjected to similarity searches against GenBank to investigate sequence similarities to those deposited in the Genbank. We kept only the top-five sequences with nucleotide homology >93%. Sequences were incorporated in the alignment with the non-mosquito sequences, using the ClustalW algorithm (Thompson et al. 1994). Phylogenetic reconstructions were implemented using MEGA software (Kumar et al. 2004) with Neighbor-Joining (NJ) reconstruction under Kimura substitution model (two parameters). Trees were unrooted.
Results
Sequence analysis of mosquito gut content DNA
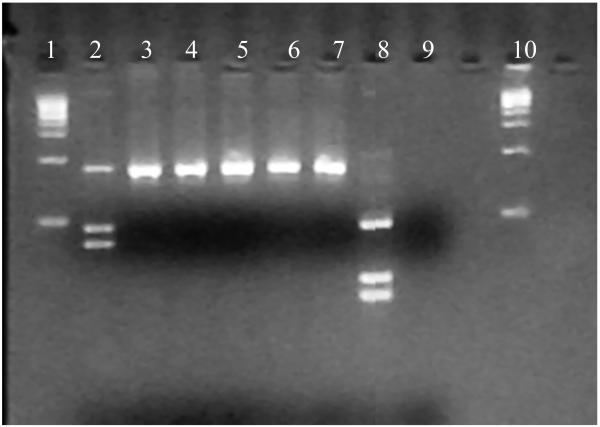
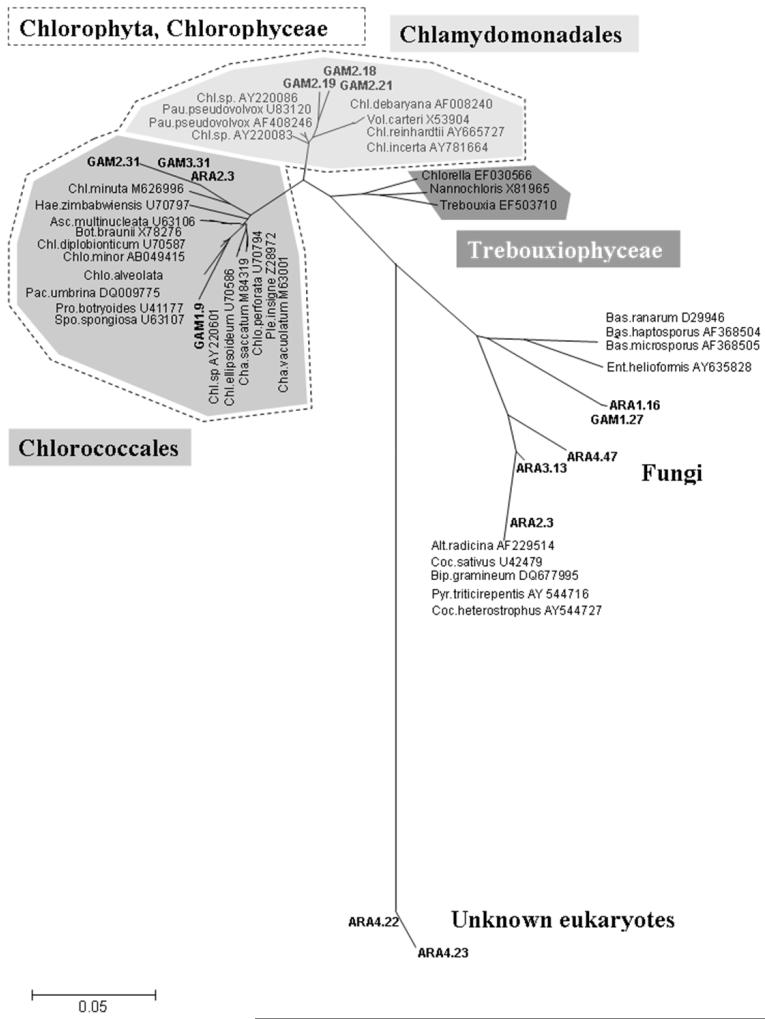
Among the eight larvae (5 An. arabiensis and 3 An. gambiae s.s.) randomly selected for dissection of gut contents and cloning of 18S rDNA PCR products, a total of 355 clones were screened by RFLP (from 36 to 64 clones per gut), and 14 unique non-mosquito sequences were identified (Fig 2). The proportion of clones showing non-mosquito restriction profiles varied among the samples ranging from 1.6 to 13.9%. The unique 14 non-mosquito sequences of 708bp were blasted against GenBank, and the 36 sequences with the highest homology hits were selected for phylogenetic analysis (total number of sequences was 50). Some taxa were hit only several times, which reflects the poor diversity sampling in the Genbank for this marker and these groups. The 18S non-mosquito sequences were clustered into three main clades, unknown eukaryotes (2), green algae (7), and fungi (5) (Fig 3). Fungi sequences belonged to Ascomyceta and Zygomyceta groups. Within the green algae clade (Chlorophyceae Class, Chlorophyta), the sequences were clustered into two main families, the Chloroccocales and Chlamydomonadales families.
Figure 2.
Agarose gel electrophoresis showing the restriction profiles for 18S gut contents clones. Lane 2, 3, 4, 5, 6, 7: 18S non-mosquito DNA restriction pattern; Lane 8: 18S mosquito restriction pattern; Lane 9: negative control, Lane 1 and 10: molecular size marker.
Figure 3.
Neighbor-joining tree of 18S rDNA sequences obtained from the larvae gut content and the organisms of blast search with the highest homology. The clones are showed as bold and named from the host species and an arbitrary clone number sequenced. The hit blast search clones are designed by their Genbank accession numbers. Species genus abbreviation: Alt, Alternacia; Asc, Ascochloris; Bas, Basidiobolus; Bip, Bipolaris; Bot, Botryococcus; Coc, Cochliobolus; Chl, Chlamydomonas; Chlo, Chlorosarcinopsis; Cha, Characium; Ent, Entophyctis; Hae, Haematococcus; Pac, Pachycladella; Pau, Paulschulzia; Pro, Protosiphon; Pyr, Pyrenophora; Spo, Spongiochloris; Vol, Volvox.
Sequence analysis of water samples
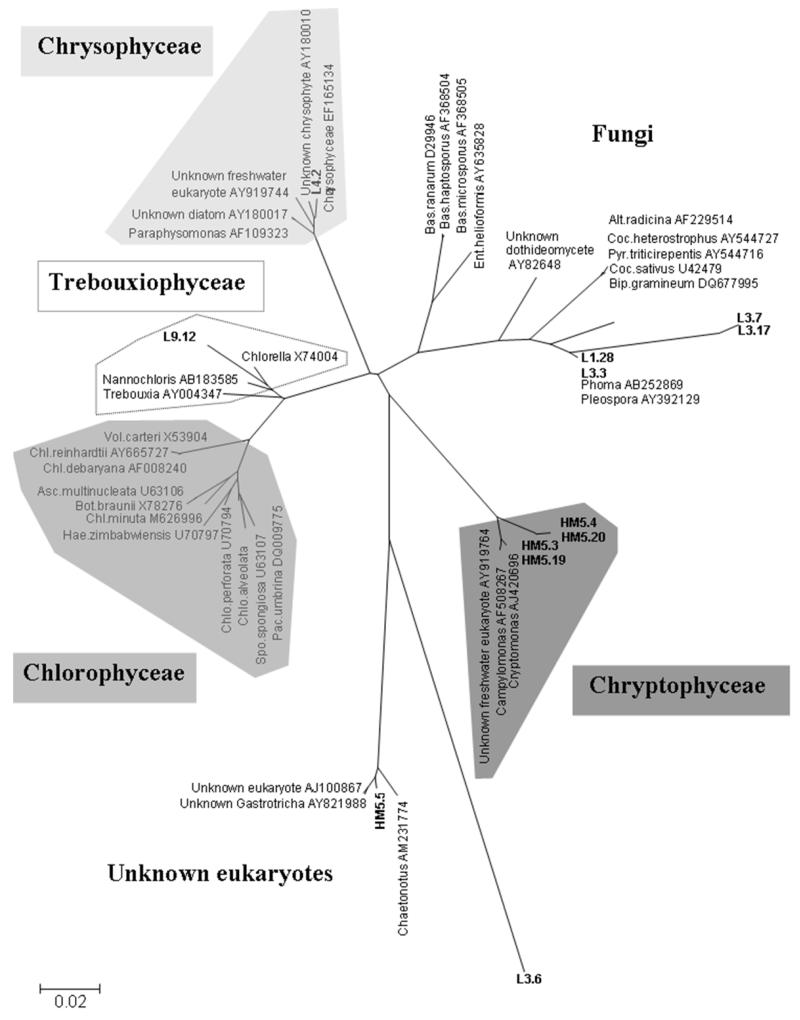
A total of 5 water samples were analyzed, which represented 160 clones (32 clones per water samples). Twelve unique sequences were identified. The phylogenetic reconstruction found five main clades. Four groups were representative of microalgae, Chlorophyceae (green algae), Chryptophyceae, Chrysophyceae (golden algae) and Trebouxiophyceae. Metazoaires and fungi were also found (Fig 4).
Figure 4.
Neighbor-joining tree of 18S rDNA sequences obtained from water samples content and the organisms of blast search with the highest homology. The clones are showed as bold and named from the larval habitat number and an arbitrary clone number sequenced. The hit blast search clones are designed by their GenBank accession numbers. Species genus abbreviation: Alt, Alternacia; Asc, Ascochloris; Bas, Basidiobolus; Bip, Bipolaris; Bot, Botryococcus; Coc, Cochliobolus; Chl, Chlamydomonas; Chlo, Chlorosarcinopsis; Cha, Characium; Ent, Entophyctis; Hae, Haematococcus; Pac, Pachycladella; Pau, Paulschulzia; Pro, Protosiphon; Pyr, Pyrenophora; Spo, Spongiochloris; Vol, Volvox.
Discussion
The successful detection of DNAs from gut contents of An. gambiae s.l. larvae indicates that the method presented in this study may be a promising tool to investigate natural diets of larvae in natural habitats. The large amount of 18S rDNA data over a wide range of eukaryote taxa available in the GenBank database allows the possibility to investigate taxonomic affinity of mosquito gut contents from natural habitats with little information on the biotic community. The barcoding technique should be applied to earlier stages of Anopheles larvae in order to investigate not only the changes of diet among different larval stages and seasonal changes in diet, but also variation in diet in different habitat types.
Using universal 18S rDNA sequences may be advantageous in investigating gut contents because the PCR primers can amplify homologous regions in almost all eukaryotes, even the unknown ones. Because the rDNA is constituted of multiple copy sequences (Hillis, Dixon 1991), degraded DNA or small amounts of DNA could be amplified. We amplified the 18S rDNA region because we targeted only eukaryotic prey organisms, such as microalgae, which have not been well studied. The choice of 18S rDNA eliminates the possibility of amplifying prokaryotic organisms such as bacteria. We recognize some drawbacks with using 18S rDNA nucleotide databases to identify prey organisms. First, the number of 18S rDNA sequences in the GenBank is limited, particularly in organisms from remote African regions, and in microorganisms and freshwater plankton. Thus, a sequence homology search could lead to inaccuracies or the inability to identify the prey species. In our case, two unique sequences were identified as unknown eukaryotes. Second, the prey organisms that have been well digested may not be amplified. Controlled experiments on the threshold duration beyond which the DNA of prey organisms can no longer be amplified by PCR would reveal the critical period needed for feeding experiments and gut content digestion.
The present study sheds light on prey organisms of Anopheles larvae during their late development. We confirmed that microalgae appeared as a food source for An. gambiae s.l. larvae, as suggested by previous works (Gimnig et al. 2002; Tuno et al. 2006). Fungi may be also a food source as well. Tuno et al. (2006) found a positive association between larval abundance and a Chlorophyceae microalga, Chlorococcaeceae family (Characium anopheloides, Tuno N., personal communication). It is noteworthy that the algal food source identified in our study belonged to the same taxonomic family. The sample size used in the present study did not allow investigation of differences between An. gambiae s.s and An. arabiensis. Future studies should focus on this group of microalgae using microalgae-specific primers (Jarman et al. 2004) with a large sample size. Chloroccocales and Chlamydomonales are freshwater microalgae that are widely distributed over the world. Although we found a large range of microalgae in the larval habitats, only two types of microalgae were identified in the gut contents. Anopheline larvae also feed off submerged plant surfaces and at the bottom of shallow pools. This may explain the difference in the types of microalgae found in the water samples and gut samples. Therefore, further investigation on the microorganisms present at different depths of natural larval habitats is required. The fact that we found these algal DNAs in the gut contents does not necessarily indicate that they were digested. Whether the ingested microalgae are being digested is an objective of our on-going study.
In summary, this study used a novel approach to molecularly investigate the prey organisms of An. gambiae s.l. larvae. Such studies will not only improve our understanding of Anopheles larval ecology, but also provide fundamental information to facilitate the development of novel larval control tools.
Acknowledgments
This study is published with the permission of the Director, Kenya Medical Research Institute. We thank Cynthia Spurgeon and two anonymous reviewers for valuable comments on the manuscript. This work was supported by grants from the National Institutes of Health (NIH) D43 TW01505 and R01 A150243.
Bibliography
- Blankeship LE, Yayanos AA. Universal primers and PCR of gut content to study marine invertebrate diets. Molecular Ecology. 2005;14:891–899. doi: 10.1111/j.1365-294X.2005.02448.x. [DOI] [PubMed] [Google Scholar]
- Federici BA, Park H-W, Bideshi DK, Wirth MC, Johnson JJ. Recombinant bacteria for mosquito control. The Journal of Experimental Biology. 2003;206:3877–3885. doi: 10.1242/jeb.00643. [DOI] [PubMed] [Google Scholar]
- Gimnig JE, Ombok M, Kamau L, Hawley WA. Characteristics of larval anopheline (Diptera: Culicidae) habitats in western Kenya. Journal of Medical Entomology. 2001;38:282–288. doi: 10.1603/0022-2585-38.2.282. [DOI] [PubMed] [Google Scholar]
- Gimnig JE, Ombok M, Otieno S, et al. Density-dependant development of Anopheles gambiae (Diptera: Culicidae) larvae in artificial habitats. Journal of Medical Entomology. 2002;39:162–172. doi: 10.1603/0022-2585-39.1.162. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Dixon MT. Ribosomal DNA: molecular evolution and phylogenetic inference. Quaterly Review of Biology. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- Jarman SN, Deagle BE, Gales NJ. Group-specific polymerase chain reaction for DNA-based analysis of species diveristy and identity in dietary samples. Molecular Ecology. 2004;13:1313–1322. doi: 10.1111/j.1365-294X.2004.02109.x. [DOI] [PubMed] [Google Scholar]
- Kumar NP, Rajavel AR, Natarajan R, Jambulingam P. DNA barcodes can distinguish species of Indian mosquitoes (Diptera: Culicidae) Journal of Medical Entomology. 2007;44:1–7. doi: 10.1603/0022-2585(2007)44[1:dbcdso]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Linton YM, Harbach RE, Chang Moh S, Anthony TG, Matusop A. Morphological and molecular identity of Anopheles (Cellia) sundaicus (Diptera: Culicidae), the nominotypical member of a malaria vector species complex in Southeast Asia. Systematic Entomology. 2001;26:357–366. [Google Scholar]
- Merritt RW, Craig DA, Walker ED, Vanderploeg HA, Wotton RS. Interfacial feeding behavior and particle flow patterns of Anopheles quadrimaculatus larvae (Diptera: Culicidae) Journal of Insect Behavior. 1992a;5:741–761. [Google Scholar]
- Merritt RW, Craig DA, Wotton RS, Walker ED. Feeding behavior of aquatic insects: case studies on black fly and mosquito larvae. Invertebrate Biology. 1996;115:206–217. [Google Scholar]
- Merritt RW, Dadd RH, Walker ED. Feeding behavior, natural food, and nutritional relationships of larval mosquitoes. Annual Review of Entomology. 1992b;37:349–376. doi: 10.1146/annurev.en.37.010192.002025. [DOI] [PubMed] [Google Scholar]
- Miller SE. DNA barcoding and the renaissance of taxonomy. Proceedings of the National Academy of Sciences of USA. 2007;104:4775–4776. doi: 10.1073/pnas.0700466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineaux L. Malaria and mortality: some epidemiological considerations. Annales of Tropical Medicine and Parasitology. 1997;91:811–825. doi: 10.1080/00034989760572. [DOI] [PubMed] [Google Scholar]
- Okech BA, Gouagna LC, Yan G, Githure JI, Beier JC. Larval habitats of Anopheles gambiae s.s. (Diptera: Culicidae) influences vector competence to Plasmodium falciparum parasites. Malaria Journal. 2007:6. doi: 10.1186/1475-2875-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piyaratne MK, Amerasinghe PH, Amerasinghe FP, F K. Food of larval Anopheles culicifacies and Anopheles varuna in a stream habitat in Sri Lanka. Journal of the American Mosquito Control Association. 2005;21:387–394. doi: 10.2987/8756-971X(2006)21[387:FOLACA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Rejmankova E, Roberts DR, Manguin S, et al. Anopheles albimanus (Diptera: Culicidae) and cyanobacteria: an example of larval habitat selection. Environmental Entomology. 1996;25:1058–1067. doi: 10.1093/ee/25.5.1058. [DOI] [PubMed] [Google Scholar]
- Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. American Journal of Tropical Medecine and Hygiene. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- Service MW. Mosquito ecology. John Wiley & Sons; New York: 1976. [Google Scholar]
- Suzuki N, Murakami K, Takeyama H, Chow S. Molecular attempt to identify prey organisms of lobster phyllosoma larvae. Fisheries Science. 2006;72:342–349. [Google Scholar]
- Thiery I, Nicolas L, Rippka R, Tandeau de Marsac N. Selection of cyanobacteria isolated from mosquito breeding sites as a potential food source for mosquito larvae. Applied and Environmental Microbiology. 1991;57:1354–1359. doi: 10.1128/aem.57.5.1354-1359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuno N, Githeko AK, Nakayama T, et al. The association between the phytoplankton, Rhopalosolen species (Chlorophyta; Chlorophyceae), and Anopheles gambiae sensu lato (Diptera: Culicidae) larval abundance in western Kenya. Ecological Research. 2006;21:476–482. [Google Scholar]
- Wallace JR, Merritt RW. Diel feeding periodicity of larval anopheline mosquitoes on microorganisms and microinvertebrates: A spatial and temporal comparison of Anopheles quadrimaculatus (Diptera: Culicidae) diets in a Michigan pond. Journal of Medical Entomology. 2004;41:853–860. doi: 10.1603/0022-2585-41.5.853. [DOI] [PubMed] [Google Scholar]
- Wotton RS, Chaloner DT, Yardley CA, Merritt RW. Growth of Anopheles mosquito larvae on dietary microbiota in aquatic surface microlayers. Medical and Veterinary Entomology. 1997;11:65–70. doi: 10.1111/j.1365-2915.1997.tb00291.x. [DOI] [PubMed] [Google Scholar]
- Ye-Ebiyo Y, Pollack RJ, Spielman A. Enhanced development in nature of Anopheles arabiensis mosquitoes feeding on maize pollen. American Journal of Tropical Mediceine and Hygiene. 2000;63:90–93. doi: 10.4269/ajtmh.2000.63.90. [DOI] [PubMed] [Google Scholar]