Abstract
The present work focuses on a local survey of free-living amoebae (FLA) that cause opportunistic and nonopportunistic infections in humans. Determining the prevalence of FLA in water sources can shine a light on the need to prevent FLA related illnesses. A total of 150 samples of tap water were collected from six districts of Sivas province. The samples were filtered and seeded on nonnutrient agar containing Escherichia coli spread. Thirty-three (22%) out of 150 samples were found to be positive for FLA. The FLA were identified by morphology and by PCR using 18S rDNA gene. The morphological analysis and partial sequencing of the 18S rDNA gene revealed the presence of three different species, Acanthamoeba castellanii, Acanthamoeba polyphaga, and Hartmannella vermiformis. Naegleria fowleri, Balamuthia mandrillaris, or Sappinia sp. was not isolated during the study. All A. castellanii and A. polyphaga sequence types were found to be genotype T4 that contains most of the pathogenic Acanthamoeba strains. The results indicated the occurrence and distribution of FLA species in tap water in these localities of Sivas, Turkey. Furthermore, the presence of temperature tolerant Acanthamoeba genotype T4 in tap water in the region must be taken into account for health risks.
1. Introduction
Free-living amoebae (FLA), ubiquitous and widely distributed protozoa, feed on bacteria, algae, fungi, and small organic particles and are adaptable to their environment [1]. They can be found in dust, air, seawater, dental treatment units, sewage, eyewash solutions, contact lenses, and dialysis units and are particularly abundant in soil and water [2, 3]. Among them, only four genera including Acanthamoeba, Naegleria, Balamuthia, and Sappinia cause opportunistic and nonopportunistic infections in humans and in animals, but infections are not commonly reported with the exception of Acanthamoeba keratitis which is reported in over 1 to 2 cases per million contact lens wearers in the USA annually [4–6]. Acanthamoeba spp. and Balamuthia mandrillaris cause granulomatous amoebic encephalitis (GAE) while Naegleria fowleri causes primary amoebic meningoencephalitis (PAM). Both GAE and PAM are central nervous system infections. Some Acanthamoeba spp., commonly Acanthamoeba castellanii, cause amoebickeratitis (AK), a vision-threatening corneal infection. In humans Acanthamoeba spp. may also affect the skin and lungs [3, 7]. Hartmannella spp. invade animal tissues and have been found in nasal mucosa of humans, the bronchial system of dogs, and the intestines of turkeys [8]. Sappinia diploidea have been reported, only once, from a brain infection in a healthy man [9]. This amoeba was identified later as Sappinia pedata, by using real-time PCR tests based on 18S rRNA gene sequences [10].
The presence of FLA in tap water may represent a health risk to both immunocompromised and immunocompetent individuals [11] and they are resistant to extreme conditions of temperature, pH, and exposure to various chemicals [2, 5]. In addition to their pathogenicity, FLA serve as hosts for a large number of pathogenic bacteria and viruses for humans including Legionella spp., Vibrio cholerae, Burkholderia cepacia, Listeria monocytogenes, Escherichia coli O157, Mycobacterium spp., Coxsackievirus, Adenovirus, and Echovirus [2, 5, 11]. Furthermore, FLA can increase virulence of some of the bacteria called amoeba-resisting microorganisms (ARMs) including Mycobacterium spp., Pseudomonas aeruginosa, Legionella spp., Cryptococcus neoformans, and Histoplasma capsulatum [11].
An increase in the number of intracerebral infections caused by FLA in the USA and worldwide has been reported [6].
FLA human infections are documented [12–14], but limited information is available in the literature concerning FLA from the environmental samples in Turkey [15–17]. Therefore, the aim of this study is to isolate FLA from tap water samples collected from various districts in the province of Sivas by employing morphological and molecular methods in order to contribute to the study of understanding their ecology and to identify any potential health risks.
2. Subjects and Methods
2.1. Study Location, Sampling, Isolation, and Identification of Amoebae and DNA Extraction
A total of 150 tap water samples were collected between March and August 2011 from the districts of Divriği, Kangal, Suşehri, Ulaş, Gölova, and Gemerek of Sivas province, located in the central Anatolia region of Turkey. The total surface area of Sivas province is 28500 km2 and the study area is about 10000 km2 (Figure 1). Sivas is located at the junction of different regions and reflects typical climates of Turkey's various regions. Therefore, a prevalence study in a transition region like Sivas may reflect overall Turkish FLA distribution.
Figure 1.
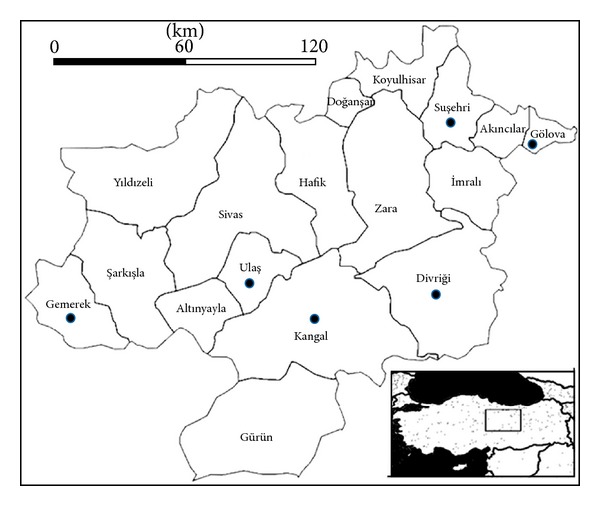
Districts of Sivas province and water sample locations (dots), Turkey.
All samples (except two of them, see Table 2) in this study were chlorinated by the city officials in drinking water plant according to world health organization criteria (0.2–0.7 ppm).
Table 2.
Species, genotypes, GenBank accession numbers, and isolation sources of free living amoebae in districts of Sivas, Turkey.
| No. | Species, identified | Genotype | Gene Bank ref. no. | Source of tap water |
|---|---|---|---|---|
| 1 | Acanthamoeba castellanii | T4 | U07403 | Divriği village |
| 2 | Acanthamoeba castellanii | T4 | U07413 | Kangal village |
| 3 | Hartmannella vermiformis | AF426157 | Suşehri village | |
| 4 | Hartmannella vermiformis | AF426157 | Kangal village | |
| 5 | Hartmannella vermiformis | AF426157 | Kangal village | |
| 6 | Hartmannella vermiformis | AF426157 | Kangal village | |
| 7 | Hartmannella vermiformis | AF426157 | Kangal city center* | |
| 8 | Hartmannella vermiformis | AF426157 | Suşehri village | |
| 9 | Hartmannella vermiformis | AF426157 | Suşehri village | |
| 10 | Hartmannella vermiformis | AF426157 | Kangal village | |
| 11 | Hartmannella vermiformis | AF426157 | Suşehri village | |
| 12 | Hartmannella vermiformis | AF426157 | Divriği village | |
| 13 | Acanthamoeba castellanii | T4 | U07413 | Kangal village |
| 14 | Hartmannella vermiformis | AF426157 | Ulaş village | |
| 15 | Hartmannella vermiformis | AF426157 | Suşehri village | |
| 16 | Hartmannella vermiformis | AF426157 | Divriği village | |
| 17 | Hartmannella vermiformis | AF426157 | Gölova village | |
| 18 | Hartmannella vermiformis | AF426157 | Divriği village | |
| 19 | Acanthamoeba castellanii | T4 | U07413 | Kangal village |
| 20 | Hartmannella vermiformis | AF426157 | Kangal village** | |
| 21 | Acanthamoeba castellanii | T4 | U07413 | Kangal village |
| 22 | Acanthamoeba castellanii | T4 | U07413 | Divriği hospital |
| 23 | Hartmannella vermiformis | AF426157 | Suşehri village | |
| 24 | Hartmannella vermiformis | AF426157 | Ulaş healthcare center | |
| 25 | Hartmannella vermiformis | AF426157 | Gemerek village | |
| 26 | Hartmannella vermiformis | AF426157 | Kangal village | |
| 27 | Hartmannella vermiformis | AF426157 | Kangal village | |
| 28 | Hartmannella vermiformis | AF426157 | Suşehri village | |
| 29 | Hartmannella vermiformis | AF426157 | Ulaş village | |
| 30 | Acanthamoeba castellanii | T4 | U07413 | Kangal village |
| 31 | Hartmannella vermiformis | AF426157 | Suşehri village | |
| 32 | Acanthamoeba polyphaga | T4 | U07407 | Ulaş village |
| 33 | Acanthamoeba castellanii | T4 | U07413 | Kangal village |
*Spring water.
**Fountain water.
A total of 500 mL of water sample was collected from each tap focus in a sterile plastic container from different villages and districts. They were then immediately transferred to the laboratory. FLA were isolated from the samples as previously described elsewhere [7, 18]. Briefly, water samples were filtered through 0.45 μm pore size cellulose nitrate membrane filter (47 mm in diameter) under a vacuum. The membrane filters for each water sample were scraped and collected materials were placed in 15 mL of sterile cover tubes containing 10 mL phosphate buffered saline (PBS). Tubes were incubated at room temperature overnight and then centrifuged for ten minutes at 1500 rpm to collect particles on filters. After centrifugation, the supernatant solution was discarded and the pellet was inoculated onto 1.5% nonnutrient agar (NNA) plates. A dense suspension of heat inactivated Escherichia coli, prepared in Page Saline, was seeded onto NNA plates to grow FLA. After the inoculation of the samples, all plates were incubated at 30°C and examined daily for the presence of FLA for up to 10 days using a light microscope (100x). Once a growth was observed, a piece of NNA containing the amoebae was excised to inoculate a fresh NNA plate to subculture and incubated until trophozoites were grown. Then, the trophozoites were scraped to isolate genomic DNA (QIAmp DNA Mini Kit, QIAGEN).
Amoebae were isolated and identified by morphologic features as well as PCR based sequence analysis. Smirnov and Goodkov's basic morphotyping list was used to identify amoebae [19]. Acanthamoeba was identified both by acanthopodia in the trophozoite form and by double-layered polygonal walls in the cysts form (Figure 2(a)). The Hartmannella was identified by smooth, spherical appearance (Figure 2(b)) [16, 20]. A temperature tolerance test was also performed for Acanthamoeba: three sets of subculture plates (NNA-E. coli) for each sample were incubated at 37, 42, and 52°C, respectively. All plates were examined daily for amoebal growth by phase contrast microscopy for seven days [21]. When an FLA strain was isolated, the flagellate transformation test was applied for the identification of N. fowleri [20]. All FLA strains were transferred to a fresh NNA-E. coli plate every month to check their viability and each of them were used in the experiments.
Figure 2.
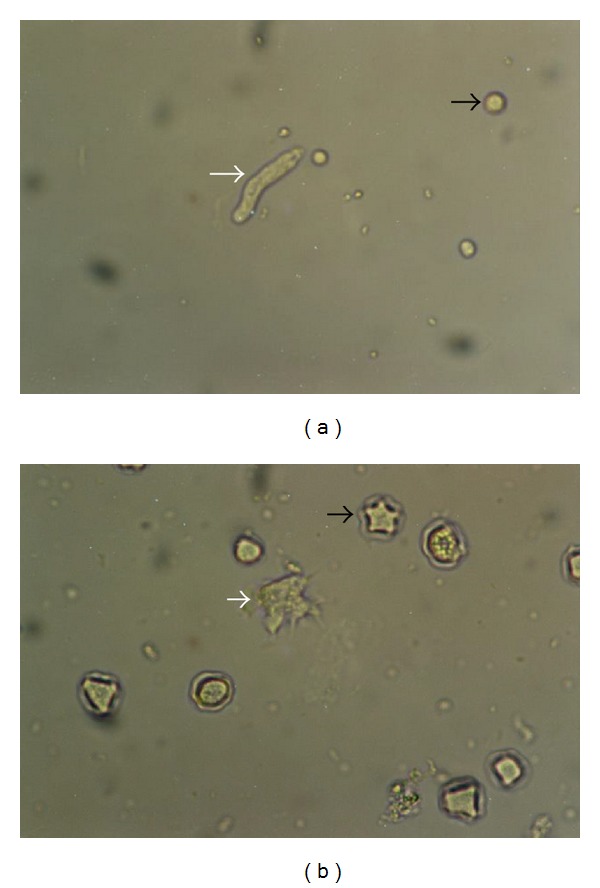
Hartmannella (a) and Acanthamoeba (b) trophozoites (white arrow), cyst forms (black arrow) (original magnification; 40x).
2.2. PCR Amplification, Sequencing, Blast Search of Sequenced Amplicons, and Cluster Analysis of Amoebae
Acanthamoeba, N. fowleri, B. mandrillaris,and Sappinia genus specific primer pairs along with common amoebae specific primers (Table 1) were employed in molecular detection of the amoebae species [7, 10, 22–24]. Fifty µL of PCR mixture contained 1 ng DNA, 5 µL 10x Taq buffer, 5 µL 2 mM dNTP, 4 µL 25 mM MgCl2, 0.5 µL 100 mM primer, 0.5 µL Taq DNA Polymerase. The thermal cycling conditions were an initial incubation of 94°C for 7 min and 45 cycles of 94°C for 60 s (95°C for Acanthamoeba), X°C for 60 s, and 72°C for 60 s with a terminal extension of 72°C for 10 min (X = 55°C for Naegleria, Sappina, and Balamuthia; 60°C for Acanthamoeba; and 65°C for common primers). PCR reactions were performed to amplify 18S rDNA sequences. These primers yielded 750–1000 bp fragments. The PCR amplicons were separated by 1% agarose gel (data not shown but available on request). Amplicon sizes were estimated using a DNA ladder. Amplicons were gel purified prior to sequencing. Sequencing was unidirectional (5-GTCAGAGGTGAAATTCTTGG-3′). Nucleotide similarity search was performed by blast search (basic local alignment search tool) of sequenced amplicons against amoeba species. Acanthamoeba has been classified into genotypes based on 18S ribosomal RNA nucleotide sequencing recently [25, 26]. GenBank accession numbers of these species are given in Table 2. Morphological observations and sequencing yielded three different species: A. castellanii, Hartmannella vermiformis, and A. polyphaga.
Table 1.
Primers used in this study (5′ → 3′).
| Species | Forward primer | Reverse primer | Reference |
|---|---|---|---|
| Common for FLA | CGCGGTAATTCCAGCTCCAATAGC | CAGGTTAAGGTCTCGTTCGTTAAC | Tsvetkova et al., 2004 [7] |
| Acanthamoeba spp. | GGCCCAGATCGTTTACCGTG | TCTCACAAGCTGCTAGGGAGTCA | Schroeder et al., 2001 [22] |
| N. fowleri | CAAACACCGTTATGACAGGG | CTGGTTTCCCTCACCTTACG | Schild et al., 2007 [23] |
| B. mandrillaris | CGCATGTATGAAGAAGACCA | TTACCTATATAATTGTCGATACCA | Booton et al., 2003 [24] |
| Sappinia | TCT GGT CGC AAG GCT GAA AC | GCA CCA CCA CCC TTG AAA TC | Qvarnstrom et al., 2009 [10] |
FLA: Free-living amoebae.
Cluster analysis was performed for FLA by using the latest version of MEGA 5.05 software [27]. Phylogenetic construct was produced by employing 18S rRNA gene sequences and amoebae genus 18S rDNA gene sequences accession numbers listed in Table 2. These were aligned with the corresponding sequences from 33 reference Acanthamoeba isolates, and a neighbor-joining tree was obtained using MEGA 5.05 (Figure 3) [27].
Figure 3.

Phylogenetic analysis of FLA isolates. Neighbor-joining tree based on 18S rDNA sequences. The sequences from Sivas isolates were aligned by MEGA software using reference isolates from GenBank. Bar is index of evolutionary distances (0.2) among the different sequences. The numbers on the branch nodes of phylogenetic tree corresponded to bootstrap value.
3. Results
FLA were detected in 33 (22%) out of 150 water samples in six Sivas districts. All Hartmannella isolates (n = 24) were identified as H. vermiformis and except one isolate, which was identified as A. polyphaga, all Acanthamoeba isolates (n = 8) were identified as A. castellanii. No representative of any of the other three genera of FLA of clinical relevance, Balamuthia, Naegleria, and Sappinia, was present in any of the samples (Table 2). It was observed that all Acanthamoeba isolates were capable of growth at all incubation temperatures and they grew easily and fast at 37°C incubation but the amoeba trophozoites grew slower at either 42°C or 52°C of incubations. After two days of incubation, respective Acanthamoeba isolates stayed alive at 42°C (samples numbers, 2, 13, 19, 27, 30, 32, and 33) and at 52°C (samples numbers, 2, 13, 27, 30, 32, and 33). The flagellate transforming test was found to be negative for all the 33 isolates.
Morphological identification of amoebae revealed Acanthamoeba and Hartmannella trophozoites. Acanthamoeba strains belong to the T4 genotype as confirmed by genus specific primer pair (Table 1).
The main target of the current study was to apply and evaluate molecular methods to recognize FLA along with classical microscopic determination method. We used PCR primers to diagnose FLA species by employing a general primer set and four genus specific primer sets (Table 1). In this study FLA isolates were compared to GenBank and the reference strains (Table 2) to determine species. PCR amplicon lengths varied from 500 to 1000 base pairs.
Acanthamoeba isolates were further examined by phylogenetic analyses by comparison of sequenced amplicons to Acanthamoeba strains. This included all representative sequences available from GenBank. All Acanthamoeba isolates were clustered in sequence type group T4.
A homology search was performed with BLAST from NCBI. The deduced sequences were aligned by ClustalW with FLA sequences and the phylogenetic tree was then displayed by neighbor-joining analysis conducted with MEGA (Figure 3). All sequences gave 100% similarity except sample number 2 (99%) with accession numbers being given in Figure 3. In the phylogenetic tree Acanthamoeba and Hartmannella species clustered in different branches but sequences of the same genus clustered together in the same branch. This shows their genetic similarity to a certain degree and consistency with identification of isolated samples.
Neighbor-joining gene tree analysis identifies individual strains of the isolates obtained in this study as revealed by reference numbers given at Table 2. The phylogenetic analysis illustrated that Acanthamoeba isolates were clustered to pathogenic genotype T4 and closest to U07413 reference number. However, two Acanthamoeba isolates belong to the U7413 and U07407 reference numbers. Similarly, Hartmannella isolates belong to the AF426157 reference number.
The phylogenetic analysis confirms clinically relevant amphizoic amoebae in tap water which may present a risk to people's health.
4. Discussion
FLA were distributed worldwide and the composition of these species at certain locations depends on the surroundings. Also, the spreading of FLA species depends on its tolerance to survive under adverse conditions. Therefore, the ecological importance of FLA must be adequately studied to prevent fatal human diseases. In this present comprehensive study, waterborne amoebae were isolated in various districts of Sivas province. We found FLA nearly in one out of five water sources in Sivas districts. Results revealed that Acanthamoeba and Hartmannella have a high distribution in the samples compared to other FLA. Although specific primers were designed, we did not isolate any of N. fowleri, B. mandrillaris, and Sappinia sp. during the study. This could be due to a lower prevalence of such FLA in the environment. Our findings are in agreement with a study from Bulgaria [7]. This Bulgarian study group determined Acanthamoeba and Hartmannella abundantly in water and soil samples compared to other species in Bulgaria. The high prevalence of the above FLA in human-related habitats was observed in environmental freshwaters of several countries as reported by the studies [28, 29].
Acanthamoebae have been isolated previously from bottled drinking water, tap water, soil, and dust in Burdur and İstanbul provinces in Turkey [15]. Furthermore both Acanthamoebae and Naegleria have also been isolated from soil and thermal water specimens in our region [16]. However, in that study, the amoebae have not been identified below the genus level. In our study except Naegleria we have also shown the presence of Acanthamoeba in the same region and identified those species. Acanthamoeba isolates belonging to T2, T3, T4, and T7 genotypes from Ankara [17] and T4 and T9 genotypes from Aydin province [13] have been found in environmental samples in Turkey.
To our knowledge the presence of FLA in the environment does not mean a risk factor for illness. However, many species of Acanthamoeba are potentially pathogenic for animals and for humans. A. castellanii, A. polyphaga, and A. culbertsoni are the most common species to infect humans [30, 31]. Acanthamoeba spp. are the main cause of AK associated with contact lenses [2, 3], although cases involving Hartmannella sp. have also been described [32]. H. vermiformis has been suggested as a cause of AK but this is still under debate by other investigators [33]. In a study performed in Aydin province, in the western part of Turkey, a case of AK caused by a genotype T4 Acanthamoeba strain related possibly with source of tap water was reported [13]. Another genotype, T4 Acanthamoeba strain, was also reported in an AK case in İzmir province, a neighbor province of Aydin. Tap water has been assumed to be the most important source of AK caused by genotype T4 Acanthamoeba [7] and worldwide the most important AK-causing strains are associated with genotype T4 [34, 35].
Acanthamoeba can tolerate extreme temperatures and thus become cold resistant, A. polyphaga may survive below 4°C. Some strains of Hartmannella can tolerate up to 48°C [11, 36]. Since Sivas province is a place of extreme hot and cold temperatures during the year, we conclude that determined FLA species are tolerant to ecological conditions. Air temperature ranges between winter and summer have been reported as −34.6 to 40°C during the years of 1972 to 2011 in Sivas (Turkish State Meteorological Service at http://www.dmi.gov.tr). In addition to environmental temperature conditions, several other factors such as cyst structure and surface availability of the organism, pH alterations, and osmolarity changes in water also determine the FLA life [37].
Acanthamoeba spp. and Hartmannella spp. can harbor pathogenic microorganism which indicates the importance of these amoebae to public health. Acanthamoeba T4 genotype and H. vermiformis may be infected naturally by pathogenic ARMs. Acanthamoeba T4 genotype may be infected by Legionella sp. and Neochlamydia sp., whereas H. vermiformis may be infected by Neochlamydia sp. and Legionella donaldsonii [37]. FLA can facilitate growth and transportation of waterborne pathogens. Therefore, they are used by pathogenic waterborne ARMs to proliferate in drinking water systems. We have limited data about host FLA in drinking water. It was reported previously that the infected FLA rate by ARM in drinking water system was 16% [5]. It is likely that several other infected FLA by unidentified pathogenic ARMs are yet to be determined.
Interestingly, FLA detection reported the presence of several genera of FLA, namely, Acanthamoeba, Naegleria, and Hartmannella, at different stages of the water treatment in drinking water treatment plants [38, 39]. Furthermore, Hartmannella spp. have been reported to resist disinfection in treatment plants [38, 39]. Acanthamoeba spp. can resist water treatment for sanitizing drinking water as well [28]. This data is consistent with our findings since 24 out of 33 detected FLA were H. vermiformis and 9 out of 33 detected FLA were Acanthamoeba spp. (Table 2). One other reason is that the high H. vermiformis prevalence in our region might be due to high levels of active biomass and natural organic matter [40] since agriculture and animal husbandry is common in Sivas province.
5. Conclusions
The results indicated the presence of A. castellanii, H. vermiformis, and A. polyphaga in tap water in Sivas localities. Furthermore, presence of temperature tolerant Acanthamoeba T4 genotypes in the region must be taken into account for AK.
FLA were underestimated by medical community until some species of amebae caused systemic infections in immunocompromised individuals. The amebic diseases are difficult to determine clinically and a patient may suffer from delay in treatment. Further, this delay may lead to deadly infections and cause fatal cases. Therefore, an investigation of the connection between environment and the patient's infections is essential. An individual history including interaction with amebic water and inhalation of cysts during dust storm may help a physician to diagnose the infection. Knowledge of prior prevalence of amebae in the region may help physicians to diagnose and treat either healthy or immunocompromised individuals. Clinicians may benefit from the reported data in the treatment of amebae related infections. Presence of FLA can lead to take precautions and reported FLA distribution can help understand the potential threat to the health of individuals.
The data obtained from the study may be beneficial for the clinicians and the environmental professionals in the region and regions around the world that have similar ecological conditions.
Conflict of Interests
The authors declare no conflict of interests.
Authors' Contribution
Y. Tutar was the principal investigator and takes primary responsibility for the paper. K. A. Çoşkun and L. Tutar performed the experiments. N. Elaldı and S. Özçelik coordinated the research. Y. Tutar wrote the paper.
Acknowledgment
This work was funded by a seed grant from the Turkish National Academy of Sciences (TUBA-GEBIP).
References
- 1.Stockman LJ, Wright CJ, Visvesvara GS, Fields BS, Beach MJ. Prevalence of Acanthamoeba spp. and other free-living amoebae in household water, Ohio, USA—1990–1992. Parasitology Research. 2011;108(3):621–627. doi: 10.1007/s00436-010-2120-7. [DOI] [PubMed] [Google Scholar]
- 2.Trabelsi H, Dendana F, Sellami A, et al. Pathogenic free-living amoebae: epidemiology and clinical review. Pathologie Biologie. 2012;60(6):399–405. doi: 10.1016/j.patbio.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Martinez AJ, Visvesvara GS. Free-living, amphizoic and opportunistic amebas. Brain Pathology. 1997;7(1):583–598. doi: 10.1111/j.1750-3639.1997.tb01076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea . FEMS Immunology and Medical Microbiology. 2007;50(1):1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- 5.Thomas V, Loret J-F, Jousset M, Greub G. Biodiversity of amoebae and amoebae-resisting bacteria in a drinking water treatment plant. Environmental Microbiology. 2008;10(10):2728–2745. doi: 10.1111/j.1462-2920.2008.01693.x. [DOI] [PubMed] [Google Scholar]
- 6.Diaz JH. Increasing intracerebral infections caused by free-living amebae in the United States and worldwide. Journal of Neuroparasitology. 2010;1(1):1–10. [Google Scholar]
- 7.Tsvetkova N, Schild M, Panaiotov S, et al. The identification of free-living environmental isolates of amoebae from Bulgaria. Parasitology Research. 2004;92(5):405–413. doi: 10.1007/s00436-003-1052-x. [DOI] [PubMed] [Google Scholar]
- 8.Visvesvara GS, Stehr-Green JK. Epidemiology of free-living ameba infections. Journal of Protozoology. 1990;37(4):25S–33S. doi: 10.1111/j.1550-7408.1990.tb01142.x. [DOI] [PubMed] [Google Scholar]
- 9.Gelman BB, Rauf SJ, Nader R, et al. Amoebic encephalitis due to sappinia diploidea. Journal of the American Medical Association. 2001;285(19):2450–2451. doi: 10.1001/jama.285.19.2450. [DOI] [PubMed] [Google Scholar]
- 10.Qvarnstrom Y, Da Silva AJ, Schuster FL, Gelman BB, Visvesvara GS. Molecular confirmation of sappinia pedata as a causative agent of amoebic encephalitis. Journal of Infectious Diseases. 2009;199(8):1139–1142. doi: 10.1086/597473. [DOI] [PubMed] [Google Scholar]
- 11.Thomas V, McDonnell G, Denyer SP, Maillard J-Y. Free-living amoebae and their intracellular pathogenic microorganisms: Risks for water quality. FEMS Microbiology Reviews. 2010;34(3):231–259. doi: 10.1111/j.1574-6976.2009.00190.x. [DOI] [PubMed] [Google Scholar]
- 12.Sarica FB, Tufan K, Çekinmez M, Erdoğan B, Altinörs MN. A rare but fatal case of granulomatous amebic encephalitis with brain abscess: the first case reported from Turkey. Turkish Neurosurgery. 2009;19(3):256–259. [PubMed] [Google Scholar]
- 13.Ertabaklar H, Türk M, Dayanir V, Ertuğ S, Walochnik J. Acanthamoeba keratitis due to Acanthamoeba genotype T4 in a non-contact-lens wearer in Turkey. Parasitology Research. 2007;100(2):241–246. doi: 10.1007/s00436-006-0274-0. [DOI] [PubMed] [Google Scholar]
- 14.Ozkoc S, Tuncay S, Delibas SB, et al. Identification of Acanthamoeba genotype T4 and Paravahlkampfia sp. from two clinical samples. Journal of Medical Microbiology. 2008;57(3):392–396. doi: 10.1099/jmm.0.47650-0. [DOI] [PubMed] [Google Scholar]
- 15.Mergeryan H. The prevalence of Acanthamoeba in the human environment. Reviews of Infectious Diseases. 1991;13(5):S390–S391. doi: 10.1093/clind/13.supplement_5.s390. [DOI] [PubMed] [Google Scholar]
- 16.Saygi G, Akin Z, Tecer H. Isolation of Acanthamoeba and Naegleria spp. from soil and thermal water samples in Sivas. Acta Parasitologica Turcica. 2000;24(3):237–242. [Google Scholar]
- 17.Kilic A, Tanyuksel M, Sissons J, Jayasekera S, Khan NA. Isolation of Acanthamoeba isolates belonging to T2, T3, T4 and T7 genotypes from environmental samples in Ankara, Turkey. Acta Parasitologica. 2004;49(3):246–252. [Google Scholar]
- 18.Zeybek Z, Ustunturk M, Binay AR. Morphological characteristics and growth abilities of free living amoeba isolated from domestic tap water samples in İstanbul. IUFS Journal of Biology. 2010;69(1):17–23. [Google Scholar]
- 19.Smirnov AV, Goodkov AV. An illustrated list of basic morphotypes of Gymnamoebia (Rhizopoda, Lobosea) Protistology. 1999;1(1):20–29. [Google Scholar]
- 20.Visvesvara GS. Pathogenic and opportunistic amebae. In: Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, editors. Manual of Clinical Microbiology. Washington, DC, USA: ASM Press; 2007. pp. 2082–2091. [Google Scholar]
- 21.Winck MAT, Caumo K, Rott MB. Prevalence of Acanthamoeba from tap water in Rio Grande do Sul, Brazil. Current Microbiology. 2011;63(5):464–469. doi: 10.1007/s00284-011-0003-5. [DOI] [PubMed] [Google Scholar]
- 22.Schroeder JM, Booton GC, Hay J, et al. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of Acanthamoebae from humans with keratitis and from sewage sludge. Journal of Clinical Microbiology. 2001;39(5):1903–1911. doi: 10.1128/JCM.39.5.1903-1911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schild M, Gianinazzi C, Gottstein B, Müller N. PCR-based diagnosis of Naegleria sp. infection in formalin-fixed and paraffin-embedded brain sections. Journal of Clinical Microbiology. 2007;45(2):564–567. doi: 10.1128/JCM.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Booton GC, Carmichael JR, Visvesvara GS, Byers TJ, Fuerst PA. Identification of Balamuthia mandrillaris by PCR assay using the mitochondrial 16S rRNA gene as a target. Journal of Clinical Microbiology. 2003;41(1):453–455. doi: 10.1128/JCM.41.1.453-455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corsaro D, Venditti D. Phylogenetic evidence for a new genotype of Acanthamoeba (Amoebozoa, Acanthamoebida) Parasitology Research. 2010;107(1):233–238. doi: 10.1007/s00436-010-1870-6. [DOI] [PubMed] [Google Scholar]
- 26.Nuprasert W, Putaporntip C, Pariyakanok L, Jongwutiwes S. Identification of a novel T17 genotype of Acanthamoeba from environmental isolates and T10 genotype causing keratitis in Thailand. Journal of Clinical Microbiology. 2010;48(12):4636–4640. doi: 10.1128/JCM.01090-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loret J-F, Greub G. Free-living amoebae: biological by-passes in water treatment. International Journal of Hygiene and Environmental Health. 2010;213(3):167–175. doi: 10.1016/j.ijheh.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Gianinazzi C, Schild M, Zumkehr B, et al. Screening of Swiss hot spring resorts for potentially pathogenic free-living amoebae. Experimental Parasitology. 2010;126(1):45–53. doi: 10.1016/j.exppara.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Kao P-M, Hsu B-M, Chen N-H, et al. Molecular detection and comparison of Acanthamoeba genotypes in different functions of watersheds in Taiwan. Environmental Monitoring and Assessment. 2012;184(7):4335–4344. doi: 10.1007/s10661-011-2267-4. [DOI] [PubMed] [Google Scholar]
- 31.Lorenzo-Morales J, Lindo JF, Martinez E, et al. Pathogenic Acanthamoeba strains from water sources in Jamaica, West Indies. Annals of Tropical Medicine and Parasitology. 2005;99(8):751–758. doi: 10.1179/136485905X65215. [DOI] [PubMed] [Google Scholar]
- 32.Aitken D, Hay J, Kinnear FB, Kirkness CM, Lee WR, Seal DV. Amebic keratitis in a wearer of disposable contact lenses due to a mixed Vahlkampfia and Hartmannella infection. Ophthalmology. 1996;103(3):485–494. doi: 10.1016/s0161-6420(96)30667-2. [DOI] [PubMed] [Google Scholar]
- 33.Kuiper MW, Valster RM, Wullings BA, Boonstra H, Smidt H, Van Der Kooij D. Quantitative detection of the free-living amoeba Hartmannella vermiformis in surface water by using real-time PCR. Applied and Environmental Microbiology. 2006;72(9):5750–5756. doi: 10.1128/AEM.00085-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan NA, Paget TA. Molecular tools for speciation and epidemiological studies of Acanthamoeba . Current Microbiology. 2002;44(6):444–449. doi: 10.1007/s00284-001-0050-4. [DOI] [PubMed] [Google Scholar]
- 35.Khan NA, Jarroll EL, Paget TA. Molecular and physiological differentiation between pathogenic and nonpathogenic Acanthamoeba. Current Microbiology. 2002;45(3):197–202. doi: 10.1007/s00284-001-0108-3. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez-Zaragoza S. Ecology of free-living amoebae. Critical Reviews in Microbiology. 1994;20(3):225–241. doi: 10.3109/10408419409114556. [DOI] [PubMed] [Google Scholar]
- 37.Thomas JM, Ashbolt NJ. Do free-living amoebae in treated drinking water systems present an emerging health risk? Environmental Science and Technology. 2011;45(3):860–869. doi: 10.1021/es102876y. [DOI] [PubMed] [Google Scholar]
- 38.Corsaro D, Feroldi V, Saucedo G, Ribas F, Loret J-F, Greub G. Novel Chlamydiales strains isolated from a water treatment plant. Environmental Microbiology. 2009;11(1):188–200. doi: 10.1111/j.1462-2920.2008.01752.x. [DOI] [PubMed] [Google Scholar]
- 39.Corsaro D, Pages GS, Catalan V, Loret J-F, Greub G. Biodiversity of amoebae and amoeba-associated bacteria in water treatment plants. International Journal of Hygiene and Environmental Health. 2010;213(3):158–166. doi: 10.1016/j.ijheh.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Valster RM, Wullings BA, Bakker G, Smidt H, Van Der Kooij D. Free-living protozoa in two unchlorinated drinking water supplies, identified by phylogenic analysis of 18S rRNA gene sequences. Applied and Environmental Microbiology. 2009;75(14):4736–4746. doi: 10.1128/AEM.02629-08. [DOI] [PMC free article] [PubMed] [Google Scholar]


