Abstract
BACKGROUND & AIMS
Biliary atresia (BA) is a progressive fibroinflammatory disorder of infants involving the extrahepatic and intrahepatic biliary tree. Its etiology is unclear but is believed to involve exposure of a genetically susceptible individual to certain environmental factors. BA occurs exclusively in the neonatal liver, so variants of genes expressed during hepatobiliary development could affect susceptibility. Genome-wide association studies previously identified a potential region of interest at 2q37. We continued these studies to narrow the region and identify BA susceptibility genes.
METHODS
We searched for copy number variants that were increased among patients with BA (n = 61) compared with healthy individuals (controls; n = 5088). After identifying a candidate gene, we investigated expression patterns of orthologues in zebrafish liver and the effects of reducing expression, with morpholino antisense oligonucleotides, on biliary development, gene expression, and signal transduction.
RESULTS
We observed a statistically significant increase in deletions at 2q37.3 in patients with BA that resulted in deletion of one copy of GPC1, which encodes glypican 1, a heparan sulfate proteoglycan that regulates Hedgehog signaling and inflammation. Knockdown of gpc1 in zebrafish led to developmental biliary defects. Exposure of the gpc1 morphants to cyclopamine, a Hedgehog antagonist, partially rescued the gpc1-knockdown phenotype. Injection of zebrafish with recombinant Sonic Hedgehog led to biliary defects similar to those of the gpc1 morphants. Liver samples from patients with BA had reduced levels of apical GPC1 in cholangiocytes compared with samples from controls.
CONCLUSIONS
Based on genetic analysis of patients with BA and zebrafish, GPC1 appears to be a BA susceptibility gene. These findings also support a role for Hedgehog signaling in the pathogenesis of BA.
Keywords: GWA, Susceptibility Loci, Animal Model, Bile Duct Growth and Development
Biliary atresia (BA) is a progressive fibroinflammatory disorder of infants involving the extrahepatic and intrahepatic biliary tree that results in obliteration of the ducts, leading to cholestasis, fibrosis, and cirrhosis. The etiology of BA is unknown but likely involves an environmental factor acting on a genetically susceptible infant, resulting in immune dysregulation leading to progressive biliary inflammation and damage. The exclusive occurrence of BA in the neonatal liver and extrahepatic biliary tree suggests that ongoing development may be key in pathogenesis, and thus genes affecting biliary development may confer susceptibility to BA.
We are interested in determining the nature of this genetic susceptibility. Others have examined genetic factors in the etiology of BA, but these studies have been hampered by the rarity of BA and a paucity of families with a clear inheritance pattern. There are reports of familial BA,1 but twin studies have been inconclusive.2–4 There is clinical heterogeneity in BA, with 20% of patients showing a distinct disorder with laterality defects,5 whereas the majority do not exhibit other anomalies.
A recent genetic study of patients with BA showed association with a region on chromosome 10,6 but most attempts to uncover a genetic cause for BA have focused on the subset of patients with laterality defects. Examination of laterality genes in patients with BA has found no mutations to date, however.7 Thus, previous studies on genetics in BA have not identified specific genes, but the existing evidence suggests that genetic susceptibility is plausible.
Multiple investigators have used genome-wide association (GWA) studies of either single nucleotide polymorphisms (SNPs) or copy number variants (CNVs) to identify susceptibility loci in complex disorders, including diabetes,8 asthma,9 and autism,10 among others. Such studies typically identify polymorphisms associated with specific disorders, but demonstration of a functional role in pathogenesis requires biological analysis, usually in an animal model. We chose to follow up GWA studies on patients with BA with functional analysis in zebrafish.
Zebrafish are a powerful animal model well suited to this application, because rapid and ex utero development of large numbers of embryos and larvae facilitates use as a screening tool for multiple genes of potential importance. There is considerable overall conservation of developmental processes and anatomic function between zebrafish and mammals, and this conservation extends to hepatobiliary development. The anatomy of the extrahepatic biliary tree is well conserved with mammals and includes a gallbladder. By 5 days postfertilization (dpf), the zebrafish liver has distinct hepatocytes and cholangiocytes, with an interconnecting ductular network.11 Conservation of hepatobiliary development at the molecular regulatory level is also evident,12–14 and several biliary disease models have been established in zebrafish, including Alagille syndrome,14 arthrogryposis–renal dysfunction–cholestasis syndrome,15 intrahepatic BA,16,17 and choledochal cysts.18
To identify genes potentially important in BA, we have performed GWA studies using SNPs and CNVs. Early findings in these studies were reported previously, in which 35 patients with BA were compared with more than 2000 controls to identify a potential region of interest at 2q37.3.19 Here we report expansion of these studies that narrow the region to include only one gene, GPC1. GPC1 encodes glypican 1, one of 6 members of the glypican family of heparan sulfate proteoglycans that are distinct from other proteoglycans in that they attach to the cell membrane by a glycosyl-phosphatidylinositol linkage.20 Glypicans mediate inflammation and modulate intercellular signaling via Hedgehog, Wnt, and fibroblast growth factor.21–23 Glypicans appear to be important in mediating duct growth and branching in developing mouse kidney21 and Drosophila trachea22 as well as in endothelial tubulogenesis.24 These processes appear similar to biliary development in zebrafish and thus may share molecular regulatory features with mammalian biliary development.
In this study, we used zebrafish to examine the role of gpc1 in biliary development and established mechanistic connections between our zebrafish studies and BA in patients, involving the importance of Hedgehog signaling. These studies thus not only identify GPC1 as a risk gene for BA but also offer mechanistic insight into the potential pathogenic role of GPC1 in BA.
Materials and Methods
Genetic Studies of BA
Sixty-one patients with BA at The Children’s Hospital of Philadelphia (CHOP) were enrolled in the study under a protocol approved by the institutional review board. DNA was prepared from peripheral blood using standard extraction procedures.19 The patients were genotyped on the Illumina Infinium II HumanHap 550 BeadChip SNP array (San Diego, CA) through the Center for Applied Genomics at CHOP. The Center for Applied Genomics also supplied genotypes of 5088 healthy controls run on the same platform. Both cohorts underwent standard quality control metrics.25 The CNVs were called using PennCNV with GC wave correction. This algorithm predicts the copy number (CN) of the genomic regions based on the probe intensity values and B allele frequencies (eg, CN = 0, CN = 1, CN = 3, CN = 4). Because the pathogenicity of deletions and duplications is inherently different, they were evaluated separately. The CNVs called from the algorithm were summarized as CNV regions, where every region is analyzed on a 2-SNP resolution. The difference in copy number between cases and controls was assessed using a 2-tailed Fisher exact test as previously described.26
Zebrafish Lines
All zebrafish experiments were performed on TL zebrafish raised in the CHOP animal facility in accordance with protocols approved by the CHOP Institutional Animal Care and Use Committee.
In Situ Hybridization Studies
In situ hybridizations were performed as described previously27 except that 0.25% acetic anhydride was added after proteinase K fixation to reduce background.28 Polymerase chain reaction (PCR) primers used to generate riboprobes to the zebrafish gpc genes are shown in Supplementary Table 1. Sequences for all genes were obtained from the Sanger Centre, Zv9 (www.sanger.ac.uk).
Morpholino Knockdown Studies and Cytokeratin Immunostaining
Morpholinos to gpc1 were obtained from Gene Tools (Philomath, OR) and are depicted in Supplementary Table 1. Morpholinos (1.5 ng) were injected at the one-cell stage, similar to previous studies, and titrated to effect. Quantitative PCR documentation of gpc1 knockdown is shown in Supplementary Figure 1. Larvae were collected and fixed at 5 dpf.
For the Hedgehog inhibition studies, treatment with cyclopamine (20 µmol/L) was initiated at 2 dpf and larvae were maintained in cyclopamine-containing E3 until killed at 5 dpf. We also examined larvae treated in 5 µmol/L and 100 µmol/L cyclopamine, as shown in Supplementary Figure 2.
For the Sonic Hedgehog (SHH) injection studies, recombinant human SHH (CYT-676; ProSpec, New Brunswick, NJ) was diluted to 100 µg/mL and injected into the yolk of 2 dpf larvae, similar to our recent studies using interferon gamma.29 Larvae were collected and fixed at 5 dpf.
Whole-mount cytokeratin staining was performed as previously described after fixation in methanol/dimethyl sulfoxide.13 Duct quantification was performed identically to previous studies.25
Quantitative PCR Studies
Samples for quantitative real-time PCR were obtained at 5 dpf. Livers from 5 dpf larvae were isolated and pooled in groups of 10 per condition in RNAlater (Qiagen, Gaithersburg, MD). RNA was isolated and reverse transcribed per standard protocols, and quantitative PCR was performed in accordance with standard protocols using StepOne Plus (Applied Biosystems, Life Technologies, Grand Island, NY). Primers for gli2a, ptch1, foxl1, znf697, and ccnd1 are noted in Supplementary Table 1. Normalization was performed using hprt. Graphs depicted are representative experiments comparing the means of quadruplicate samples per condition.
Studies on Liver Samples From Patients
Unstained liver biopsy specimens were obtained from established cases, in accordance with an approved CHOP institutional review board protocol. Immunostaining was performed by standard techniques using primary antibody against GPC1 (sc-14645; Santa Cruz Biotechnology, Santa Cruz, CA).
Results
GWA Studies of Patients With BA
To identify genes potentially important in BA, we performed a GWA study using CNVs. Previously, we investigated potentially causative large genomic alterations in 35 patients compared with more than 2000 controls and uncovered overlapping deletions in 2q37.3 in the patients with BA.19 In the current study, we extended this work to examine association with CNVs in 61 patients and 5088 healthy controls. This analysis uncovered a CNV region on chromosome 2 overlapping the previously identified region as well as additional regions (Supplementary Table 2). The region at 2q27.3 was heterozygously deleted in 6 patients (9.84%) and 4 controls (0.08%) (P = 4.4 × 10−10). The associated CNV region contained only one gene, GPC1 (Supplementary Figure 3), suggesting that GPC1 could be a BA susceptibility gene.
Expression of Glypican Family Members in Developing Zebrafish
We elected to screen promising BA candidate genes in zebrafish. Genes with a functional role in biliary development and/or inflammation in zebrafish would be plausible candidates for a similar role in the developing human liver and would thus be attractive as potential BA susceptibility genes.
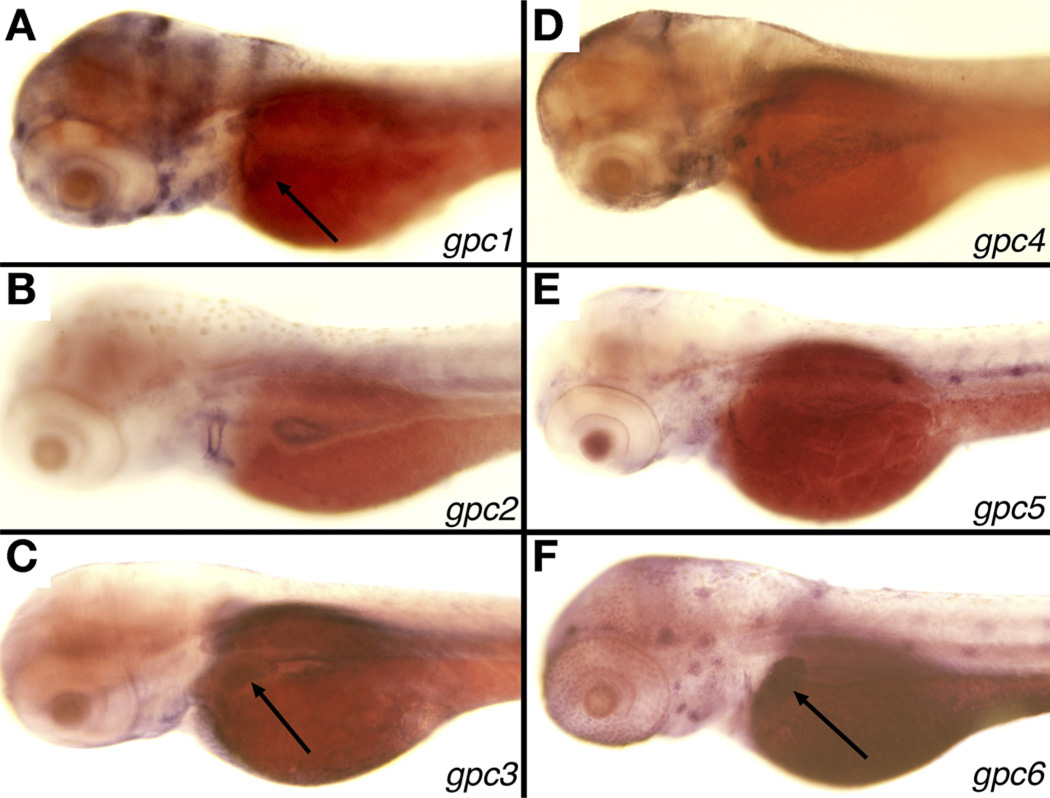
Both zebrafish and humans have 6 GPC genes. We examined tissue-specific expression of all 6, because zebrafish gpc1 may not be the functional orthologue of GPC1. We observed liver expression of gpc1, 3, and 6 at 3 dpf (Figure 1), which corresponds to the initiation of active biliary growth as evidenced by rapid proliferation of cholangiocytes and formation of nascent ducts.12,14 Genomic analysis showed synteny of these genes with their human counterparts, as did in silico analysis of amino acid sequence (not shown). Thus, we focused on gpc1, because this gene demonstrated liver-specific expression and was the likely orthologue of human GPC1.
Figure 1.
Expression of gpc genes in larval 3 dpf zebrafish. In situ hybridization studies of 3 dpf zebrafish showing liver expression (arrows) of (A) gpc1, (C) gpc3, and (F) gpc6, but not in gpc2 (B), gpc4 (D), or gpc5 (E). There is also faint intestinal expression of all 6 genes.
Morpholino-Mediated Knockdown of gpc1 Leads to Developmental Biliary Defects
To determine the importance of gpc1 in biliary development of zebrafish, we designed morpholino antisense oligonucleotides (MOs) directed against gpc1. Similar to previous studies, we designed 2 nonoverlapping MOs targeting gpc1: an MO targeting the translation start site and an MO targeting the splice acceptor site of exon 4 (see Supplementary Table 1 for sequences). Injection of gpc1 MO showed no gross defects in the appearance at 5 dpf compared with larvae injected with control MO (not shown). We confirmed knockdown of gpc1 using the splice-blocker MO and showed specificity of the MO by rescuing the phenotype with coinjection of gpc1 messenger RNA and the MO (Supplementary Figure 1).
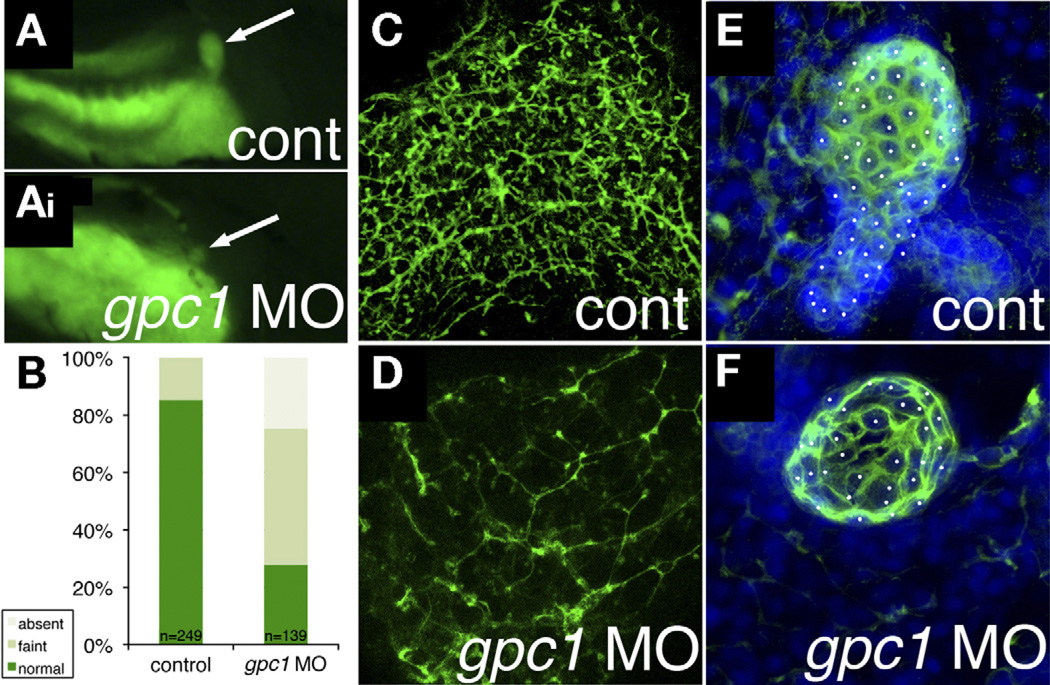
Control and MO-injected larvae were fed the fluorescent lipid reporter PED-6 for examination of biliary function. PED-6 is swallowed, absorbed by the intestine, processed through the liver, and excreted into bile, concentrating in the gallbladder. Defects in intrahepatic biliary anatomy can lead to decreased gallbladder uptake of PED-6. Gallbladders of larvae injected with gpc1 MO were significantly less intense than control larvae (Figure 2). Examination of intrahepatic biliary anatomy by cytokeratin immunostaining showed fewer and less complex-appearing ducts in gpc1 morphants (Figure 2 and Table 1, top 2 rows), similar to previous studies.12,15–17,27
Figure 2.
Knockdown of gpc1 in zebrafish leads to developmental biliary defects. (A, Ai) Right lateral views of 5 dpf zebrafish larvae after ingestion of PED-6, showing a more intense and larger gallbladder in (A) control than in (Ai) the gpc1 morphant. (B) Quantification of PED-6 uptake in control and gpc1 morphant larvae, showing a highly significant (P < .0001, χ2) decrease in gallbladder intensity in the gpc1 morphants. (C and D) Confocal projections of whole-mount cytokeratin immunostaining of livers from 5 dpf (C) control and (D) gpc1 morphant, showing decreased number and complexity of the ducts in panel D. (E and F) Confocal projections of gallbladders stained for cytokeratin and counterstained with 4’,6-diamidino-2-phenylindole (DAPI), with cells noted by white dots. Note that there are more cells in panel E.
Table 1.
Quantification of Bile Ducts in gpc1 Knockdown
| No. of total ducts | No. of interconnecting ducts |
No. of terminal ductules |
Duct length (arbitrary units) |
|
|---|---|---|---|---|
| Control | 25.5 ± 3.0 | 6.7 ± 3.3 | 54.5 ± 15.7 | 1.45 ± 0.16 |
| gpc1 MO | 19.0 ± 4.9a | 1.5 ± 1.4a | 14.7 ± 9.5b | 1.20 ± 0.39 |
| gpc1 MO + cyclopamine | 30.8 ± 10.9a | 5.3 ± 3.8a | 31.2 ± 12.2a | 1.18 ± 0.12 |
NOTE. Duct attributes from control and gpc1 MO-injected larvae. Shown are the number of total ducts, interconnecting ducts, terminal ductules, and duct length, measured in arbitrary units.
P ≤ .05,
P ≤ .005,
gpc1 MO relative to control. Also shown is quantification of duct features from gpc1 MO-injected larvae treated with cyclopamine. aP ≤ .05, relative to gpc1 MO alone.
We also examined extrahepatic biliary anatomy in 5 dpf gpc1 morphants using cytokeratin immunostaining. The gallbladders of gpc1 morphants were smaller, with fewer cells (61.3 ± 11.6 for control, 34.3 ± 6.0 for gpc1 morphants, n = 4 each, P ≤ .05), than those of control larvae (Figure 2). The biliary defects noted in the gpc1 morphants show that inhibition of gpc1 leads to both extrahepatic and intrahepatic biliary anomalies, as in BA. The extrahepatic and intrahepatic defects showed equal sensitivity to MO-mediated knockdown, because both the dose shown in Figure 2 (1.5 ng) and a lower dose (0.75 ng) affected both phenotypes equally (not shown). Our results support a potential role of GPC1 in BA.
Gene Expression Changes in gpc1 Morphants
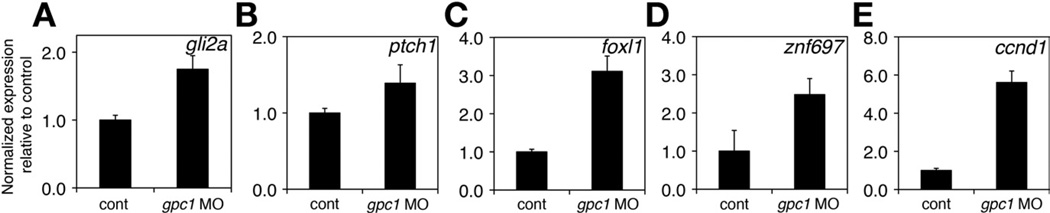
Recently, others have shown that Hedgehog activity is increased in patients with BA30 and, as mentioned previously, glypicans modulate signaling via Hedgehog. Thus, we examined the expression of Hedgehog target genes in gpc1 morphants. Expression of the target genes gli2a,ptch1, foxl1, znf697, and ccnd131 was increased at 5 dpf in livers from gpc1 morphants (Figure 3). In addition, several genes associated with fibrogenesis and epithelial-mesenchymal transition, a process linked to increased Hedgehog activity, were up-regulated in livers from gpc1 morphants (Supplementary Figure 4). This suggested that Hedgehog activity was increased in the livers of gpc1 morphants, consistent with a model in which loss of gpc1 leads to activation of Hedgehog signaling.
Figure 3.
Expression of downstream genes in gpc1 morphants. (A–E) Quantitative PCR studies of Hedgehog target genes in isolated livers from 5 dpf larvae show increased expression of (A) gli2a, (B) ptch1, (C) foxl1, (D) znf697, and (E) ccnd1. P < .05 for all.
Inhibition of Hedgehog Signaling Partially Reverses Biliary Defects in gpc1 Morphants
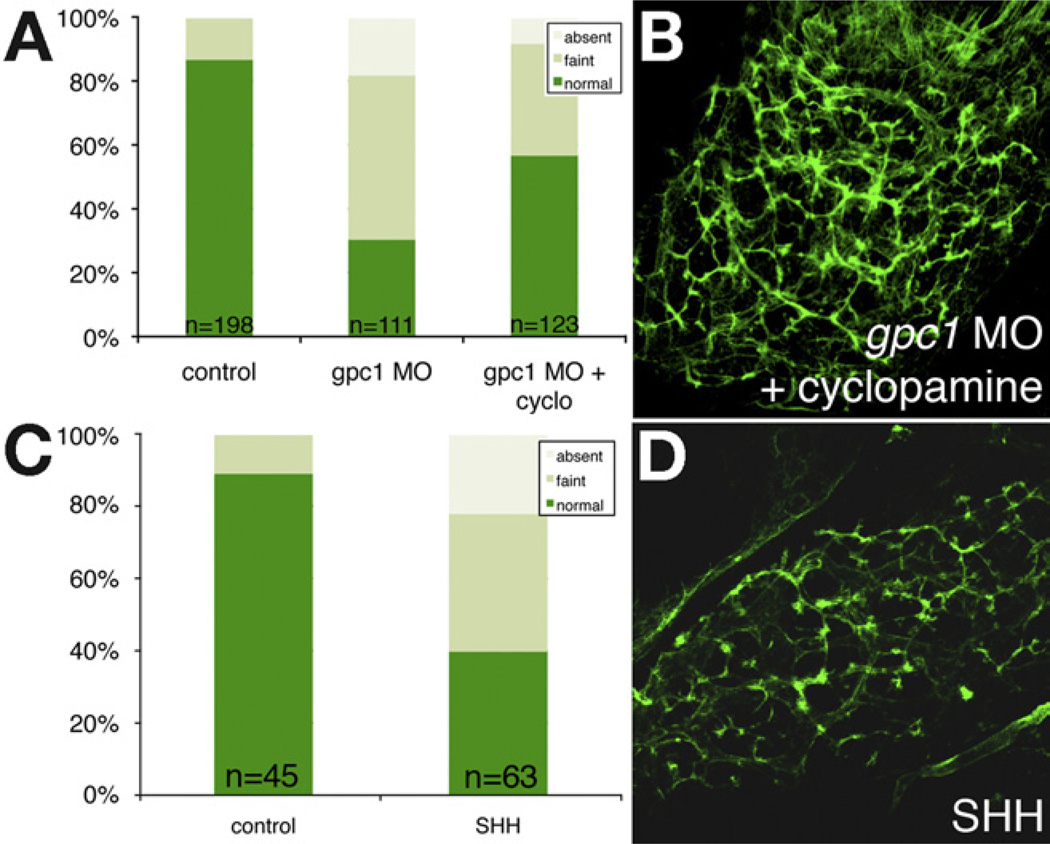
To determine the importance of increased Hedgehog activity in mediating the biliary defects in the gpc1 morphants, we treated gpc1 morphants with the Hedgehog inhibitor cyclopamine. This resulted in partial rescue of the intrahepatic biliary defects caused by gpc1 knockdown (Figure 4 and Table 1, bottom row), suggesting that increased Hedgehog activity is at least partially responsible for the biliary defects seen in gpc1 morphants. There was no effect of cyclopamine on control larvae (data not shown), but injection of SHH protein into developing larvae led to abnormalities in bile duct morphology (Figure 4), consistent with overactive Hedgehog signaling, leading to developmental biliary defects. The rescue of gpc1 knockdown by Hedgehog inhibition is consistent with a model in which glypicans modulate Hedgehog activity by acting as a “sink,” decreasing the availability of ligand; thus, absence of glypican results in increased Hedgehog signaling.
Figure 4.
Increased Hedgehog activity is associated with biliary defects. (A) Quantification of PED-6 uptake in control, gpc1 morphant larvae, and gpc1 morphant larvae treated with cyclopamine. There is a significant reduction in gallbladder intensity in the gpc1 morphants (P < .0001, χ2) that is partially rescued by treatment with cyclopamine (P < .0001). (B) Confocal projection of whole-mount cytokeratin immunostaining of liver from a 5-dpf gpc1 morphant larva treated with cyclopamine. Compare with Figure 2. (C) Similar quantification of PED-6 uptake in control and SHH-injected larvae, showing a significant (P < .0001) reduction in PED-6 gallbladder uptake in SHH-injected larvae. (D) Confocal projection of whole-mount cytokeratin immunostaining of liver from a5-dpf SHH-injected larva, showing decrease in number and complexity of intrahepatic ducts.
Patients With Cholestatic Disease Have GPC1 Abnormalities
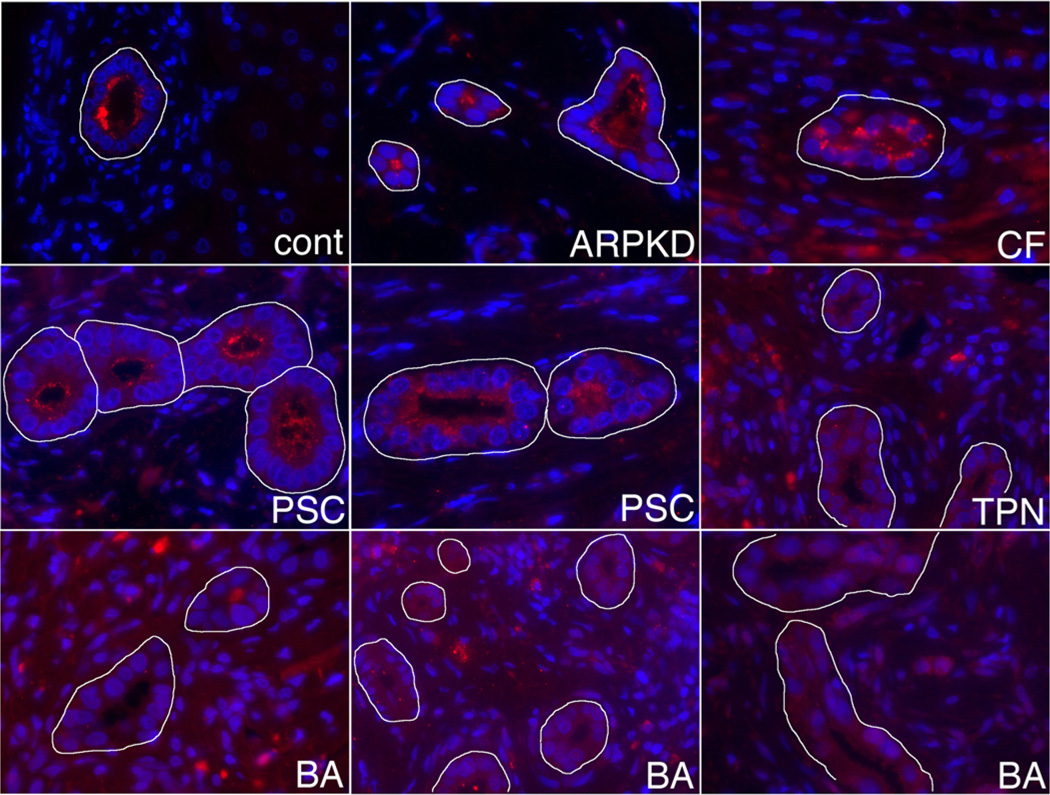
Our zebrafish studies support a role for GPC1 in mediating biliary defects in patients. To examine the potential role of GPC1 in patients with BA, we performed immunostaining on liver samples. In control patients, GPC1 staining was localized to the apical surface of cholangiocytes. GPC1 staining was also localized to the apical surface of cholangiocytes from patients with cystic fibrosis liver disease, autosomal recessive polycystic kidney disease, and primary sclerosing cholangitis (PSC). In contrast, in liver samples from patients with infantile cholestasis, including patients with BA and patients with total parenteral nutrition (TPN)-associated cholestasis, no apical GPC1 staining was visible and the amount of GPC1 appeared to be decreased globally (Figure 5). Of note, the patients with BA shown in Figure 5 were not among those with deletions in the GPC1 region. These results show localization and/or quantification defects of GPC1 in patients with infantile cholestasis, supporting the importance of GPC1 in BA that could be mediated genetically.
Figure 5.
GPC1 abnormalities in patients with cholestatic liver disease. Shown are GPC1 immunostainings from a control patient (cont), a patient with autosomal recessive polycystic kidney disease/congenital hepatic fibrosis (ARPKD), a patient with cystic fibrosis liver disease (CF), 2 patients with PSC, a patient with TPN-induced cholestasis, and 3 patients with BA. GPC1 staining is red, and samples were counterstained with DAPI. Bile ducts are outlined in white. Note that GPC1 staining is more diffusely localized and fainter in the TPN and BA samples.
Discussion
In this study, we showed that the proteoglycan gene GPC1 is a potential risk gene for BA, with evidence from genetic studies of patients with BA and from knockdown of gpc1 in zebrafish. We showed that the biliary defects elicited by gpc1 knockdown are reversed by inhibition of Hedgehog signaling and that patients with BA have abnormalities in GPC1 immunostaining. These results support the potential importance of GPC1 in BA and also show the utility of screening candidate genes uncovered in genetic association studies using zebrafish.
BA and Genetic Susceptibility
This is the first study to identify a potential BA risk gene in patients and to show functional defects in the biliary system in model organism studies. Previously, Garcia-Barcelo et al identified association of BA with a region on chromosome 10q24.2.6 This region lies between XPN-PEP1, encoding X-prolyl aminopeptidase P, and ADD3, which encodes adducin 3. X-prolyl aminopeptidase P catalyzes the breakdown of bradykinin and substance P, whereas adducin 3 is a cytoskeletal protein involved in cell motility and cell-cell contact. Both genes are expressed in biliary cells, and both proteins have functions potentially important in BA.
Other studies have adopted a more direct approach, examining the role of specific genes in the pathogenesis of BA based on overlap of clinical features with other disorders or genes and pathways believed to be important in the pathogenesis of BA. Such candidates include JAG1, the primary gene involved in Alagille syndrome, as well as laterality genes and inflammatory pathway genes. Kohsaka et al found JAG1 mutations in 9 of 102 patients with BA, and these patients appeared to have more severe disease.32 Because of the association between BA and laterality defects,5 investigators have examined patients with BA for mutations in genes involved in left/right sidedness. No mutations in INV were identified in 7 patients with BA,7 whereas CFC1 mutations were found in 5 of 10 patients with BA and laterality defects,33 although the significance of these mutations is not clear. Investigators have also examined genes important in inflammation. Polymorphisms in the gene for macrophage migration inhibitory factor (MIF) have been described in patients with BA.34 Lee et al identified polymorphisms in the vascular endothelial growth factor (VEGF) gene associated with BA,35 and this group also reported no polymorphisms in IFNG,36 IL4,37 or IL18,38 in patients with BA. Although examination of specific genes may lead to identification of some susceptibility genes, these studies are hampered by a relatively small sample size and a necessary gene selection bias that large-scale GWA studies attempt to circumvent. Additionally, because of the likely genetic heterogeneity in BA, different genes may confer susceptibility in different patients.
Specific genetic disorders have been reported to be associated with BA or a BA-like phenotype. We have reported BA in a patient with Mowat–Wilson syndrome16 and in a patient with trifunctional protein deficiency.39 Although most patients with these disorders do not have liver disease, the simultaneous occurrence of BA and a rare genetic disorder suggests a possible role for the causative gene in mediating BA.
Thus, our current study adds to a growing list of potential BA susceptibility genes (Table 2).16,18,27,40–53 In addition to the genes discussed previously, Table 2 lists genes that cause other disorders with biliary defects. Patients with North American Indian childhood cirrhosis have liver disease similar to that in patients with BA; this disorder is caused by homozygous mutation of CIRH1A.54 In addition, there are other genetic cholestatic disorders, such as ARC syndrome45 and Aagenaes syndrome.55 There are multiple conditions, including Joubert syndrome, Meckel syndrome, and Bardet–Biedl syndrome, involving genes encoding proteins important in primary cilia.40 In these disorders, biliary morphogenesis defects are associated with other findings, including laterality defects. Although the ciliopathies have not been linked to BA, genes in these disorders would be excellent BA susceptibility gene candidates. More extensive GWA studies may show that the genes in Table 2 are associated with BA and will also likely uncover unexpected genes. Continued screening of such genes using zebrafish, as reported here, will strengthen the connection to BA.
Table 2.
Genes Associated With BA, Other Cholestatic Disorders, and Biliary Development
| Gene (reference) | Function |
|---|---|
| Associated with BA | |
| GPC1 (this report) | Proteoglycan |
| XPNPEP16 | Aminopeptidase |
| ADD36 | Actin-binding |
| JAG132 | Cell fate determinant (Jagged/Notch) |
| CFC133 | Left/right determinant |
| MIF34 | Cytokine |
| VEGF35 | Growth factor |
| ZEB216 | Transcription factor |
| HADHA39 | Metabolic enzyme |
| Genetic causes of cholestasis/bile duct morphogenetic defects | |
| CIRH1A54 | Unknown |
| VIPAR44VPS33B45 | Intracellular trafficking |
| PKHD1, BBS, MKS, NPHP, others40,41 | Cilia structure/function |
| CFTR47 | Chloride channel |
| SERPINA148 | α1-antitrypsin |
| ATP8B1, ABCB11, ABCB449 | Bile component transport |
| AKR1D1, CYP7B1, HSD3B7, others50 | Bile acid synthesis |
| Animal models of cholestasis/bile duct morphogenetic defects | |
| ONECUT142 | Onecut transcription factor |
| TCF243 | Homeodomain transcription factor |
| FOXF146 | Forkhead transcription factor |
| VPS genes15,18,51 | Intracellular trafficking |
| ATP6 genes52 | Intracompartmental pH |
| ENO153 | Enolase A |
| INV7 | Left/right determinant |
| DNMT117 | DNA methylation |
| PRICKLE1, VANGL227 | Planar cell polarity |
NOTE. The genes are divided into 3 categories. The first group shows genes identified by linkage studies, direct examination of the gene in patients with BA, or reports of patients with a known mutation also with BA. The second group has genes responsible for diseases with a hepatic or biliary phenotype, and the third group has genes that lead to hepatobiliary defects in animal models.
Our studies have focused on a genetic association with BA, but we cannot rule out that genetic abnormalities in GPC1 are a risk factor for infantile cholestasis or for cholestasis in general. Our immunostaining studies showing GPC1 abnormalities in TPN cholestasis support the former. With respect to the latter, others have examined genetic influences on other cholestatic diseases such as PSC and primary biliary cirrhosis. Studies examining PSC have not shown association with SNP variants in GPC156–59 and similar studies on primary biliary cirrhosis are likewise negative,60–62 but there may be a modest association between PSC and decreased GPC1 CN (E. Ellinghaus and T. Karlsen, personal communication, December 2012). Interestingly, polymorphisms in GPC6 are associated with PSC, and lentiviral silencing of GPC6 in cholangiocytes leads to activation of proinflammatory genes.59 These latter studies, along with our studies, support the importance of glypican abnormalities in cholestasis in general and suggest that there may be genetic defects in GPC genes in multiple forms of cholestasis.
Glypicans and Morphogenesis Defects
We showed that knockdown of gpc1 leads to biliary developmental defects in zebrafish and that these defects appear at least partly due to Hedgehog signaling abnormalities. In patient samples, we observed a decrease in GPC1 at the apical surface of cholangiocytes in patients with BA. This decrease could occur via a genetic lack of GPC1 or secondary to other changes in the cholangiocyte in BA and similar disorders, such as loss of cell polarity and/or epithelial-mesenchymal transition.
Glypicans are proteoglycans anchored in the apical membrane and have been shown to modulate both Hedgehog and fibroblast growth factor signaling. There is evidence for glypicans to either augment or attenuate this signaling. Based on our data, GPC1 could act as an inhibitor for Hedgehog ligands, similar to reports previously for GPC3.63 Reduced GPC1 would then increase Hedgehog signaling.
Such a model would require the presence of Hedgehog ligand in the bile, because our evidence in patients shows GPC1 expression in the lumen. Although there is as yet no evidence for Hedgehog ligands in bile, recent studies have shown the presence of fibroblast growth factor 19 in bile,64 suggesting that other signaling proteins that interact with glypicans exist in bile. Bile salts are important signaling molecules found in bile, but the presence of protein signaling molecules in bile suggests a previously unappreciated medium for signaling potentially disrupted in cholestatic diseases such as BA.
With respect to the mechanism by which gpc1 and Hedgehog signaling disrupt biliary development in zebrafish, others have shown activation of the Hedgehog target gene Foxl1 in rodent models of cholestasis,65 and we showed increased foxl1 expression in gpc1 morphants. The Foxl1 gene is expressed in hepatocyte and cholangiocyte bipotential precursors,66 supporting a potential role for this genetic pathway in mediating biliary defects, because disruption of this pathway may negatively affect differentiation. Other potential downstream targets of Hedgehog, including transcription factors such as znf697, may also be important. Further examination of these targets may not only reveal genes important in biliary development but also may uncover additional BA susceptibility genes.
Zebrafish as a Genetic Screening Tool
Zebrafish have repeatedly shown utility as a high-throughput screening tool in forward genetic screens, small molecule screens, and other types of screens. Here we reported use of zebrafish in conjunction with GWA studies as a method to screen genes close to SNPs associated with a particular disease. To be truly useful as a screening tool for GWAS, potentially affected zebrafish require a facile functional assay, such as PED-6 screening and cytokeratin immunostaining. Given the multiple methods available to analyze larval zebrafish, numerous clinical conditions may be amenable to similar analysis.
Supplementary Material
Acknowledgments
The authors thank Michael Wittig, Eva Ellinghaus, and Tom Karlsen for copy number analysis of patients with PSC, Laura Conlin and Grace Chao for expert advice and discussion, and Jessi Erlichman for administrative assistance related to the Childhood Liver Disease Research and Education Network (ChiLDREN).
Funding
Supported by the Fred and Suzanne Biesecker Pediatric Liver Center at CHOP, other institutional support from CHOP (H.H.), and funds from the Childhood Liver Disease Research and Education Network (ChiLDREN) (pilot grant to R.P.M. and support to B.A.H. and N.B.S., National Institutes of Health grant U01 DK062481) and the National Institute of Diabetes and Digestive and Kidney Diseases (grant R01 DK090045 to M.D., B.A.H., and N.B.S.).
Abbreviations used in this paper
- BA
biliary atresia
- CHOP
The Children’s Hospital of Philadelphia
- CN
copy number
- CNV
copy number variant
- DAPI
4′,6-diamidino-2-phenylindole
- dpf
days postfertilization
- GWA
genome-wide association
- MO
morpholino antisense oligonucleotide
- PCR
polymerase chain reaction
- PSC
primary sclerosing cholangitis
- SHH
Sonic Hedgehog
- SNP
single nucleotide polymorphism
- TPN
total parenteral nutrition
Footnotes
Supplementary Materials
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://dx.doi.org/10.1053/j.gastro.2013.01.022.
Conflicts of interest
The authors disclose no conflicts.
References
Author names in bold designate shared co-first authorship.
- 1.Lachaux A, Descos B, Plauchu H, et al. Familial extrahepatic biliary atresia. J Pediatr Gastroenterol Nutr. 1988;7:280–283. doi: 10.1097/00005176-198803000-00020. [DOI] [PubMed] [Google Scholar]
- 2.Poovorawan Y, Chongsrisawat V, Tanunytthawongse C, et al. Extrahepatic biliary atresia in twins: zygosity determination by short tandem repeat loci. J Med Assoc Thai. 1996;79(Suppl 1):S119–S124. [PubMed] [Google Scholar]
- 3.Smith BM, Laberge JM, Schreiber R, et al. Familial biliary atresia in three siblings including twins. J Pediatr Surg. 1991;26:1331–1333. doi: 10.1016/0022-3468(91)90613-x. [DOI] [PubMed] [Google Scholar]
- 4.Silveira TR, Salzano FM, Howard ER, et al. Extrahepatic biliary atresia and twinning. Braz J Med Biol Res. 1991;24:67–71. [PubMed] [Google Scholar]
- 5.Davenport M, Tizzard SA, Underhill J, et al. The biliary atresia splenic malformation syndrome: a 28-year single-center retrospective study. J Pediatr. 2006;149:393–400. doi: 10.1016/j.jpeds.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Barcelo MM, Yeung MY, Miao XP, et al. Genome-wide association study identifies a susceptibility locus for biliary atresia on 10q24.2. Hum Mol Genet. 2010;19:2917–2925. doi: 10.1093/hmg/ddq196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schon P, Tsuchiya K, Lenoir D, et al. Identification, genomic organization, chromosomal mapping and mutation analysis of the human INV gene, the ortholog of a murine gene implicated in left-right axis development and biliary atresia. Hum Genet. 2002;110:157–165. doi: 10.1007/s00439-001-0655-5. [DOI] [PubMed] [Google Scholar]
- 8.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 10.Ma D, Salyakina D, Jaworski JM, et al. A genome-wide association study of autism reveals a common novel risk locus at 5p14.1. Ann Hum Genet. 2009;73:263–273. doi: 10.1111/j.1469-1809.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthews RP. Zebrafish as a model system for the study of liver development and disease. In: Arias IM, Alter HJ, Boyer JL, et al., editors. The liver: biology and pathobiology. 5th ed. Hoboken, NJ: Wiley; 2009. pp. 1067–1074. [Google Scholar]
- 12.Matthews RP, Lorent K, Russo P, et al. The zebrafish onecut gene hnf-6 functions in an evolutionarily conserved genetic pathway that regulates vertebrate biliary development. Dev Biol. 2004;274:245–259. doi: 10.1016/j.ydbio.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 13.Matthews RP, Lorent K, Pack M. Transcription factor onecut3 regulates intrahepatic biliary development in zebrafish. Dev Dyn. 2008;237:124–131. doi: 10.1002/dvdy.21407. [DOI] [PubMed] [Google Scholar]
- 14.Lorent K, Yeo SY, Oda T, et al. Inhibition of Jagged-mediated Notch signaling disrupts zebrafish biliary development and generates multi-organ defects compatible with an Alagille syndrome phenocopy. Development. 2004;131:5753–5766. doi: 10.1242/dev.01411. [DOI] [PubMed] [Google Scholar]
- 15.Matthews RP, Plumb-Rudewiez N, Lorent K, et al. Zebrafish vps33b, an ortholog of the gene responsible for human arthrogryposis-renal dysfunction-cholestasis syndrome, regulates biliary development downstream of the onecut transcription factor hnf6. Development. 2005;132:5295–5306. doi: 10.1242/dev.02140. [DOI] [PubMed] [Google Scholar]
- 16.Cui S, Erlichman J, Russo P, et al. Intrahepatic biliary anomalies in a patient with Mowat-Wilson syndrome uncover a role for the zinc finger homeobox gene zfhx1b in vertebrate biliary development. J Pediatr Gastroenterol Nutr. 2011;52:339–344. doi: 10.1097/MPG.0b013e3181ff2e5b. [DOI] [PubMed] [Google Scholar]
- 17.Matthews RP, Eauclaire SF, Mugnier M, et al. DNA hypomethylation causes bile duct defects in zebrafish and is a distinguishing feature of infantile biliary atresia. Hepatology. 2011;53:905–914. doi: 10.1002/hep.24106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadler KC, Amsterdam A, Soroka C, et al. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132:3561–3572. doi: 10.1242/dev.01918. [DOI] [PubMed] [Google Scholar]
- 19.Leyva-Vega M, Gerfen J, Thiel BD, et al. Genomic alterations in biliary atresia suggest region of potential disease susceptibility in 2q37.3. Am J Med Genet A. 2010;152A:886–895. doi: 10.1002/ajmg.a.33332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirn-Safran C, Farach-Carson MC, Carson DD. Multifunctionality of extracellular and cell surface heparan sulfate proteoglycans. Cell Mol Life Sci. 2009;66:3421–3434. doi: 10.1007/s00018-009-0096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grisaru S, Cano-Gauci D, Tee J, et al. Glypican-3 modulates BMP-and FGF-mediated effects during renal branching morphogenesis. Dev Biol. 2001;231:31–46. doi: 10.1006/dbio.2000.0127. [DOI] [PubMed] [Google Scholar]
- 22.Yan D, Lin X. Drosophila glypican Dally-like acts in FGF-receiving cells to modulate FGF signaling during tracheal morphogenesis. Dev Biol. 2007;312:203–216. doi: 10.1016/j.ydbio.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan D, Lin X. Opposing roles for glypicans in Hedgehog signalling. Nat Cell Biol. 2008;10:761–763. doi: 10.1038/ncb0708-761. [DOI] [PubMed] [Google Scholar]
- 24.Lisboa FA, Warren J, Sulkowski G, et al. Pregnancy-specific glycoprotein 1 induces endothelial tubulogenesis through interaction with cell surface proteoglycans. J Biol Chem. 2011;286:7577–7586. doi: 10.1074/jbc.M110.161810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaikh TH, Gai X, Perin JC, et al. High-resolution mapping and analysis of copy number variations in the human genome: a data resource for clinical and research applications. Genome Res. 2009;19:1682–1690. doi: 10.1101/gr.083501.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glessner JT, Wang K, Cai G, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui S, Capecci LM, Matthews RP. Disruption of planar cell polarity activity leads to developmental biliary defects. Dev Biol. 2011;351:229–241. doi: 10.1016/j.ydbio.2010.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) University of Oregon Press; 2000. [Google Scholar]
- 29.Cui S, EauClaire SF, Matthews RP. Interferon-gamma directly mediates developmental biliary defects. Zebrafish. 2013 Feb 28; doi: 10.1089/zeb.2012.0815. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omenetti A, Bass LM, Anders RA, et al. Hedgehog activity, epithelial-mesenchymal transitions, and biliary dysmorphogenesis in biliary atresia. Hepatology. 2011;53:1246–1258. doi: 10.1002/hep.24156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergeron SA, Milla LA, Villegas R, et al. Expression profiling identifies novel Hh/Gli-regulated genes in developing zebrafish embryos. Genomics. 2008;91:165–177. doi: 10.1016/j.ygeno.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kohsaka T, Yuan ZR, Guo SX, et al. The significance of human jagged 1 mutations detected in severe cases of extrahepatic biliary atresia. Hepatology. 2002;36:904–912. doi: 10.1053/jhep.2002.35820. [DOI] [PubMed] [Google Scholar]
- 33.Davit-Spraul A, Baussan C, Hermeziu B, et al. CFC1 gene involvement in biliary atresia with polysplenia syndrome. J Pediatr Gastroenterol Nutr. 2008;46:111–112. doi: 10.1097/01.mpg.0000304465.60788.f4. [DOI] [PubMed] [Google Scholar]
- 34.Arikan C, Berdeli A, Ozgenc F, et al. Positive association of macrophage migration inhibitory factor gene-173G/C polymorphism with biliary atresia. J Pediatr Gastroenterol Nutr. 2006;42:77–82. doi: 10.1097/01.mpg.0000192247.55583.fa. [DOI] [PubMed] [Google Scholar]
- 35.Lee HC, Chang TY, Yeung CY, et al. Genetic variation in the vascular endothelial growth factor gene is associated with biliary atresia. J Clin Gastroenterol. 2010;44:135–139. doi: 10.1097/MCG.0b013e3181b152c2. [DOI] [PubMed] [Google Scholar]
- 36.Lee HC, Chang TY, Yeung CY, et al. Association of interferon-gamma gene polymorphisms in Taiwanese children with biliary atresia. J Clin Immunol. 2011;30:68–73. doi: 10.1007/s10875-009-9330-8. [DOI] [PubMed] [Google Scholar]
- 37.Lee HC, Chang TY, Yeung CY, et al. Genetic variability of interleu-kin4 gene in Taiwanese children with biliary atresia. Cytokine. 2012;57:402–405. doi: 10.1016/j.cyto.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 38.Lee HC, Chang TY, Yeung CY, et al. Association of polymorphisms in the Interleukin-18 gene with susceptibility to biliary atresia. J Pediatr Gastroenterol Nutr. 2011;52:607–611. doi: 10.1097/MPG.0b013e3182111b9b. [DOI] [PubMed] [Google Scholar]
- 39.Matthews RP, Russo P, Berry GT, et al. Biliary atresia associated with a fatty acid oxidation defect. J Pediatr Gastroenterol Nutr. 2002;35:624–628. doi: 10.1097/00005176-200211000-00007. [DOI] [PubMed] [Google Scholar]
- 40.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masyuk T, Masyuk A, LaRusso N. Cholangiociliopathies: genetics, molecular mechanisms and potential therapies. Curr Opin Gastroenterol. 2009;25:265–271. doi: 10.1097/MOG.0b013e328328f4ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clotman F, Lannoy VJ, Reber M, et al. The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development. 2002;129:1819–1828. doi: 10.1242/dev.129.8.1819. [DOI] [PubMed] [Google Scholar]
- 43.Coffinier C, Gresh L, Fiette L, et al. Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1beta. Development. 2002;129:1829–1838. doi: 10.1242/dev.129.8.1829. [DOI] [PubMed] [Google Scholar]
- 44.Cullinane AR, Straatman-Iwanowska A, Zaucker A, et al. Mutations in VIPAR cause an arthrogryposis, renal dysfunction and cholestasis syndrome phenotype with defects in epithelial polarization. Nat Genet. 2010;42:303–312. doi: 10.1038/ng.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gissen P, Johnson CA, Morgan NV, et al. Mutations in VPS33B, encoding a regulator of SNARE-dependent membrane fusion, cause arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome. Nat Genet. 2004;36:400–404. doi: 10.1038/ng1325. [DOI] [PubMed] [Google Scholar]
- 46.Kalinichenko VV, Zhou Y, Bhattacharyya D, et al. Haploinsufficiency of the mouse Forkhead Box f1 gene causes defects in gall bladder development. J Biol Chem. 2002;277:12369–12374. doi: 10.1074/jbc.M112162200. [DOI] [PubMed] [Google Scholar]
- 47.Feranchak AP, Sokol RJ. Cholangiocyte biology and cystic fibrosis liver disease. Semin Liver Dis. 2001;21:471–488. doi: 10.1055/s-2001-19030. [DOI] [PubMed] [Google Scholar]
- 48.Suchy FJ. Neonatal cholestasis. Pediatr Rev. 2004;25:388–396. [PubMed] [Google Scholar]
- 49.Nicolaou M, Andress EJ, Zolnerciks JK, et al. Canalicular ABC transporters and liver disease. J Pathol. 2012;226:300–315. doi: 10.1002/path.3019. [DOI] [PubMed] [Google Scholar]
- 50.Setchell KD, Heubi JE. Defects in bile acid biosynthesis—diagnosis and treatment. J Pediatr Gastroenterol Nutr. 2006;43(Suppl 1):S17–S22. doi: 10.1097/01.mpg.0000226386.79483.7b. [DOI] [PubMed] [Google Scholar]
- 51.Schonthaler HB, Fleisch VC, Biehlmaier O, et al. The zebrafish mutant lbk/vam6 resembles human multisystemic disorders caused by aberrant trafficking of endosomal vesicles. Development. 2008;135:387–399. doi: 10.1242/dev.006098. [DOI] [PubMed] [Google Scholar]
- 52.Eauclaire SF, Cui S, Ma L, et al. Mutations in vacuolar H(+)-ATPase subunits lead to biliary developmental defects in zebrafish. Dev Biol. 2012;365:434–444. doi: 10.1016/j.ydbio.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu BR, Brindley SM, Tucker RM, et al. Alpha-enolase autoantibodies cross-reactive to viral proteins in a mouse model of biliary atresia. Gastroenterology. 2010;139:1753–1761. doi: 10.1053/j.gastro.2010.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chagnon P, Michaud J, Mitchell G, et al. A missense mutation (R565W) in cirhin (FLJ14728) in North American Indian childhood cirrhosis. Am J Hum Genet. 2002;71:1443–1449. doi: 10.1086/344580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henriksen NT, Langmark F, Sorland SJ, et al. Hereditary cholestasis combined with peripheral pulmonary stenosis and other anomalies. Acta Paediatr Scand. 1977;66:7–15. doi: 10.1111/j.1651-2227.1977.tb07801.x. [DOI] [PubMed] [Google Scholar]
- 56.Ellinghaus D, Folseraas T, Holm K, et al. Genome-wide association analysis in primary sclerosing cholangitis and ulcerative colitis identifies risk loci at GPR35 and TCF4. Hepatology. 2012 Jul 23; doi: 10.1002/hep.25977. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 57.Folseraas T, Melum E, Rausch P, et al. Extended analysis of a genome-wide association study in primary sclerosing cholangitis detects multiple novel risk loci. J Hepatol. 2012;57:366–375. doi: 10.1016/j.jhep.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melum E, Franke A, Schramm C, et al. Genome-wide association analysis in primary sclerosing cholangitis identifies two non-HLA susceptibility loci. Nat Genet. 2011;43:17–19. doi: 10.1038/ng.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karlsen TH, Franke A, Melum E, et al. Genome-wide association analysis in primary sclerosing cholangitis. Gastroenterology. 2010;138:1102–1111. doi: 10.1053/j.gastro.2009.11.046. [DOI] [PubMed] [Google Scholar]
- 60.Liu X, Invernizzi P, Lu Y, et al. Genome-wide meta-analyses identify three loci associated with primary biliary cirrhosis. Nat Genet. 2010;42:658–660. doi: 10.1038/ng.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirschfield GM, Liu X, Xu C, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med. 2009;360:2544–2555. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mells GF, Floyd JA, Morley KI, et al. Genome-wide association study identifies 12 new susceptibility loci for primary biliary cirrhosis. Nat Genet. 2011;43:329–332. doi: 10.1038/ng.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Capurro MI, Xu P, Shi W, et al. Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev Cell. 2008;14:700–711. doi: 10.1016/j.devcel.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 64.Zweers SJ, Booij KA, Komuta M, et al. The human gallbladder secretes fibroblast growth factor 19 into bile: towards defining the role of fibroblast growth factor 19 in the enterobiliary tract. Hepatology. 2012;55:575–583. doi: 10.1002/hep.24702. [DOI] [PubMed] [Google Scholar]
- 65.Sackett SD, Gao Y, Shin S, et al. Foxl1 promotes liver repair following cholestatic injury in mice. Lab Invest. 2009;89:1387–1396. doi: 10.1038/labinvest.2009.103. [DOI] [PubMed] [Google Scholar]
- 66.Sackett SD, Li Z, Hurtt R, et al. Foxl1 is a marker of bipotential hepatic progenitor cells in mice. Hepatology. 2009;49:920–929. doi: 10.1002/hep.22705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.