Abstract
Recent studies suggest that most cases of myelodysplastic syndrome (MDS) are clonally heterogeneous, with a founding clone and multiple subclones. It is not known whether specific gene mutations typically occur in founding clones or subclones. We screened a panel of 94 candidate genes in a cohort of 157 patients with MDS or secondary acute myeloid leukemia (sAML). This included 150 cases with samples obtained at MDS diagnosis and 15 cases with samples obtained at sAML transformation (8 were also analyzed at the MDS stage). We performed whole-genome sequencing (WGS) to define the clonal architecture in eight sAML genomes and identified the range of variant allele frequencies (VAFs) for founding clone mutations. At least one mutation or cytogenetic abnormality was detected in 83% of the 150 MDS patients and 17 genes were significantly mutated (false discovery rate ≤0.05). Individual genes and patient samples displayed a wide range of VAFs for recurrently mutated genes, indicating that no single gene is exclusively mutated in the founding clone. The VAFs of recurrently mutated genes did not fully recapitulate the clonal architecture defined by WGS, suggesting that comprehensive sequencing may be required to accurately assess the clonal status of recurrently mutated genes in MDS.
Keywords: myelodysplastic syndromes, MDS, acute myeloid leukemia, genomics, clonality, sequencing
INTRODUCTION
New insights into the genetic basis of myelodysplastic syndrome (MDS) have emerged in the last several years with the use of next-generation sequencing technology. Indeed, unbiased whole-genome or exome sequencing of MDS samples was instrumental in identifying novel mutations in spliceosome genes, the most commonly targeted pathway currently known in MDS, with at least one mutation in up to 50% of patients.1–4 Although the full complement of commonly mutated genes in MDS is not yet known, sequencing large numbers of candidate genes in MDS samples has identified several mutations that have independent prognostic value.5,6 These results suggest that existing MDS classification systems could be improved with the addition of mutational analysis.
The clonal architecture of MDS and acute myeloid leukemia (AML) can be defined using the variant allele frequencies (VAFs) of all somatic mutations detected by whole-genome sequencing (WGS).7–11 The vast majority of mutations present in these genomes are randomly acquired during aging (passenger mutations) and are carried forward in a cell (captured) when it is transformed.7 Although the passenger mutations are not pathogenic, they provide a genetic signature that can be used to identify clonal sub-populations. A founding clone containing hundreds of mutations is present in the MDS bone marrow, persists when patients progress to secondary AML (sAML), and gives rise to daughter subclones that contain founding clone mutations.8 Recurrently mutated genes can be detected in both founding clones and subclones.8 While recurrently mutated genes in MDS have biologic significance, it is not yet known whether specific gene mutations typically occur in the founding clone versus subclones, or whether the clonal distribution of mutations has clinical or biological importance. As targeted therapy becomes available for specific mutations, it is possible that these therapies will be more effective when the target is in the founding clone rather than in a subclone.
To begin to define the landscape of recurrently mutated genes in MDS and study their clonal distribution, we screened 150 MDS samples for mutations in 94 candidate genes using next-generation sequencing technology and calculated the VAF of each mutation. We identified recurrent mutations in known and novel genes/pathways. There was heterogeneity of VAFs for all recurrently mutated genes. These results suggest that there are many genetic paths for MDS initiation, rather than a fixed set of ‘rules’ for temporal acquisition of mutations in this disease.
MATERIALS AND METHODS
Patients
One-hundred and fifty-seven patients with de novo MDS (no antecedent chemotherapy/radiotherapy or prior hematologic disorder) or sAML arising after MDS were selected for study. All patients provided written consent on a protocol approved by the Washington University Human Studies Committee (WU #01–1014) that includes specific language authorizing WGS.
Whole-Genome Sequencing
Eight patients with de novo MDS and subsequent evolution to sAML were selected for WGS. Detailed histories for these patients are provided in the Supplementary results. Libraries were prepared using unamplified genomic DNA from unfractionated bone marrow cells (tumor) and a skin biopsy (normal) obtained at sAML diagnosis. Paired-end sequencing was performed on the Illumina platform, as previously described.8 Variant calling and validation of all mutations using liquid hybridization capture were performed, as previously described.7 In brief, custom oligonucleotide probes were designed that target >200 bp centered on all putative somatic variants. Sheared tumor and normal DNA samples were ligated to Illumina adapters with unique barcodes and hybridized in solution with targeting probes. The recovered DNA libraries were balanced by quantitative PCR, pooled and sequenced on a single Illumina HiSeq2000 (2 × 100 bp) lane. Somatic single-nucleotide variants (SNVs) were identified by SomaticSniper and VarScan2 using default parameters.12,13 Mutations were classified into tiers (as previously described),10 where Tier 1 contains all changes in the amino acid coding regions of annotated genes, consensus splice-site regions, and RNA genes (including microRNA genes).
Candidate gene sequencing
Ninety-four candidate genes were selected for recurrence screening, as described in Supplementary methods. One-hundred and fifty previously described4 de novo MDS cases were sequenced. Disease characteristics for this cohort are summarized in Table 1 (cytogenetics previously reported).4 MDS and matched normal DNA samples from all 150 MDS cases were hybridized to custom liquid capture probes covering all coding exons of the candidate genes (2227 exons; 430 777 bp of target sequence). Barcoded libraries were pooled and sequenced on the Illumina HiSeq2000 platform. Variant calling and additional validation were performed, as described in Supplementary methods.
Table 1.
Clinical characteristics of MDS patients
| No. of Patients (%) | |
|---|---|
| Total | 150 (100) |
| Gender | |
| Male | 92 (61.3) |
| Female | 58 (38.7) |
| Age | |
| <60 | 57 (38) |
| ≥60 | 93 (62) |
| Cytogenetics | |
| Very good | 3 (2.0) |
| Good | 79 (52.7) |
| Intermediate | 19 (12.7) |
| Poor | 16 (10.7) |
| Very poor | 30 (20.0) |
| NA | 3 (2.0) |
| Bone marrow blast (%) | |
| 0–2 | 53 (35.3) |
| 3–<5 | 23 (15.3) |
| 5–10 | 29 (19.3) |
| >10 | 42 (28.0) |
| NA | 3 (2.0) |
| Hemoglobin (g/dl) | |
| ≥10 | 66 (44.0) |
| 8–<10 | 76 (50.7) |
| <8 | 8 (5.3) |
| Platelets (× 109/l) | |
| ≥100 | 53 (35.3) |
| 50–<100 | 31 (20.7) |
| <50 | 66 (44.0) |
| ANC (× 109/l) | |
| ≥0.8 | 101 (67.3) |
| <0.8 | 49 (32.7) |
| IPSS-R | |
| Very low | 12 (8.0) |
| Low | 34 (22.7) |
| Intermediate | 37 (24.7) |
| High | 29 (19.3) |
| Very high | 33 (22.0) |
| NA | 5 (3.3) |
Abbreviations: ANC, absolute neutrophil count; ipss-r International Prognostic Scoring System-Revised; NA, not available.
RESULTS
Clonal architecture of sAML cases
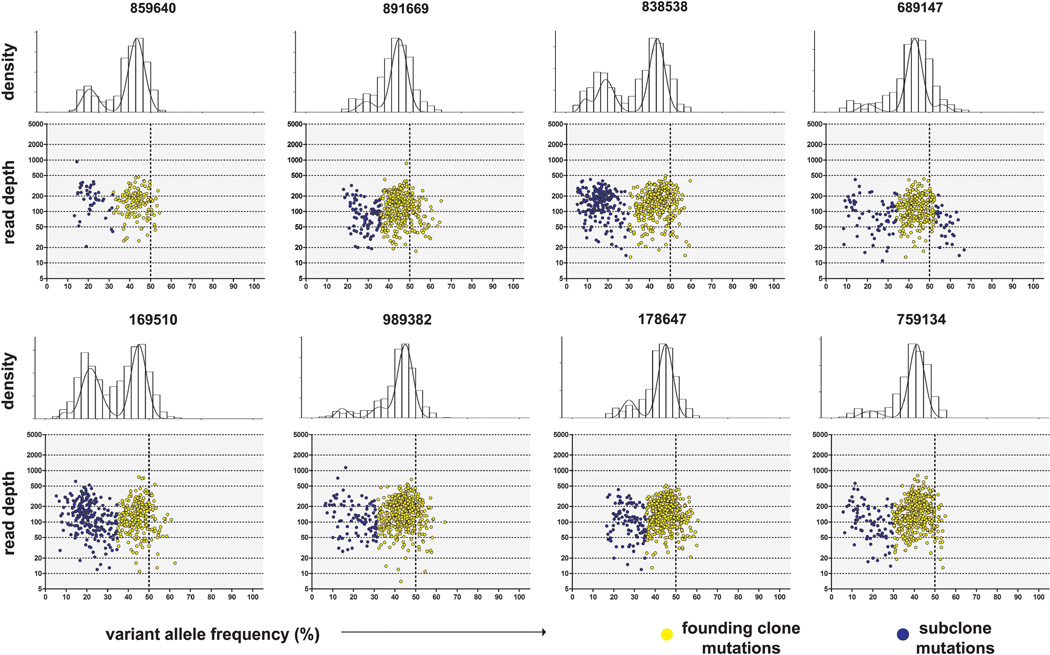
Aligned, deduplicated WGS data provided 38.34 × average haploid coverage (range 32.46–43.14 × ) for the sAML samples and 37.52 × (range 27.42–55.81 × ) for the skin samples. After orthogonal validation using hybridization capture and subsequent next-generation sequencing, an average of 602 (range 203–876) somatic SNVs or insertion/deletion events (indels) were identified in each sAML sample (Supplementary Table 1). The VAFs for all validated SNVs (restricted to mutations on autosomes without copy number alteration or uniparental disomy; Supplementary Figure 1, Supplementary Table 2) were clustered (described in supplementary methods; R package at: http://github.com/genome/sciclone) to determine the clonal composition of each tumor. All tumors contained a founding clone and at least one subclone that contained all founding clone mutations (Figure 1).
Figure 1.
Clonality of sAML samples. VAF vs read depth at the site of all validated Tier 1–3 somatic mutations in the diploid regions of eight MDS-derived sAML genomes. Above each scatterplot, the histograms depict clustered VAFs using a Bayesian approach that performs outlier detection and removal, and determines the optimal number of clusters. Founding clones (yellow) are defined by the region with the maximum density peak. Points falling outside this region are in subclones (blue). A heterozygous mutation present in every bone marrow cell would have a VAF = 50% (depicted by the vertical dashed lines in the lower plots), implying that 100% of cells are clonal in a sample.
Significantly mutated genes
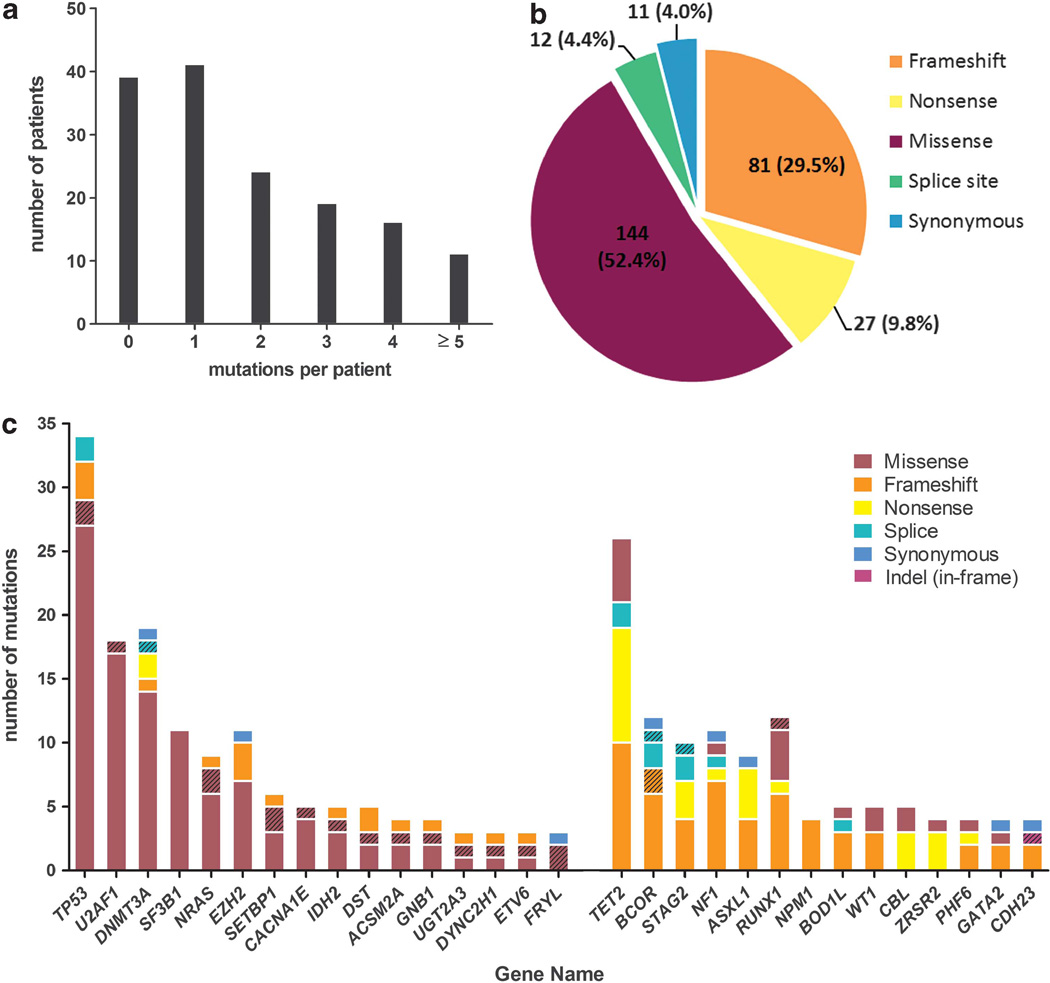
Ninety-four candidate genes (Supplementary Table 3) were screened in 150 paired MDS/normal samples. Candidate genes were selected from the list of somatic mutations identified in the 15 sAML genomes, 200 de novo AML samples (T Ley, personal communication, on behalf of The Cancer Genome Atlas AML Working Group), and the literature (additional gene selection criteria described in supplementary methods).1,3,6,8 One-hundred and fifty gigabases of sequence was produced for the MDS samples, providing an average of 70.6 × coverage (range 46.0 ×–110.3 × ) of the targeted sequence per sample. 89.9% of exons had at least 20 × coverage and 82.7% of exons had at least 30 x coverage in all MDS samples. Sixty-eight out of two-thousand two-hundred and twenty-seven (3.1%) exons in 31 genes had <20× average coverage in all samples. Two-hundred and seventy five validated somatic SNVs or indels were identified in 63 genes in 111 MDS samples (Supplementary Table 4), with an average of 1.8 genic mutations per sample (Figure 2a). The majority (96%) of mutations had predicted translational consequences (Figure 2b). With the addition of seven cases analyzed only at progression to sAML, somatic mutations were detected in 44 genes in at least 2/157 patients, 30 in ≥3 (Figure 2c), 25 in ≥4 and 18 in ≥5 patients (Table 2). Seventeen significantly mutated genes14 had a higher mutation rate than the background mutation rate of 3.39 mutations per Mbp (false discovery rate <0.05) (Table 2; also WT1, GNB1, ZRSR2, NPM1 and PHF6 with <5 mutations each).
Figure 2.
Candidate gene sequencing. (a) The number of validated somatic mutations per patient in 94 candidate genes in 150 paired MDS/ normal samples. (b) The predicted translational consequences of all mutations in 150 MDS samples. (c) Mutation spectrum in recurrently mutated genes (≥3 cases) in 150 MDS (solid fill) or seven sAML (hatched fill) samples. Left, genes with predominantly missense substitutions; Right, genes with predominantly truncating mutations (frameshift, nonsense and splice site).
Table 2.
Recurrently mutated genes in de novo MDS patientsa
| Gene Symbol |
ID | Total mutations |
Non-silent mutationsb |
MDS cases with non-silent mutations (%) (n =150) |
sAML cases with non-silent mutationsc (n = 7) |
|---|---|---|---|---|---|
| TP53 | 7157 | 34 | 34 | 27 (18) | 2 |
| TET2 | 54790 | 26 | 26 | 19 (12.7) | 0 |
| DNMT3A | 1788 | 19 | 18 | 16 (10.7) | 1 |
| U2AF1 | 7307 | 18 | 18 | 16 (10.7) | 1 |
| RUNX1 | 861 | 12 | 12 | 9 (6.0) | 1 |
| BCOR | 54880 | 12 | 11 | 8 (5.3) | 2 |
| SF3B1 | 23451 | 11 | 11 | 11 (7.3) | 0 |
| NF1 | 4763 | 11 | 10 | 10 (6.7) | 0 |
| EZH2 | 2146 | 11 | 10 | 9 (6.0) | 0 |
| STAG2 | 10735 | 10 | 10 | 9 (6.0) | 1 |
| NRAS | 4893 | 9 | 9 | 7 (4.7) | 2 |
| ASXL1 | 171023 | 9 | 8 | 8 (5.3) | 0 |
| BOD1L | 259282 | 6 | 6 | 5 (3.3) | 1 |
| SETBP1 | 26040 | 6 | 6 | 4 (2.7) | 2 |
| IDH2 | 3418 | 5 | 5 | 4 (2.7) | 1 |
| DST | 667 | 5 | 5 | 4 (2.7) | 1 |
| CACNA1E | 777 | 5 | 5 | 4 (2.7) | 1 |
| CBL | 867 | 5 | 5 | 5 (3.3) | 0 |
Abbreviations: ID, National Center for Biotechnology Information GeneID; MDS, myelodysplastic syndrome; sAML, secondary acute myeloid leukemia.
Genes mutated in ≥5 MDS or sAML cases; genes in bold are significantly mutated by significantly mutated genes test (false discovery rate ≤0.05) in MDS cases.
Includes missense, nonsense, splice site, frameshift or indel (in-frame).
Mutations in MDS plus sAML patients do not sum to total non-silent mutations in cases with compound mutations.
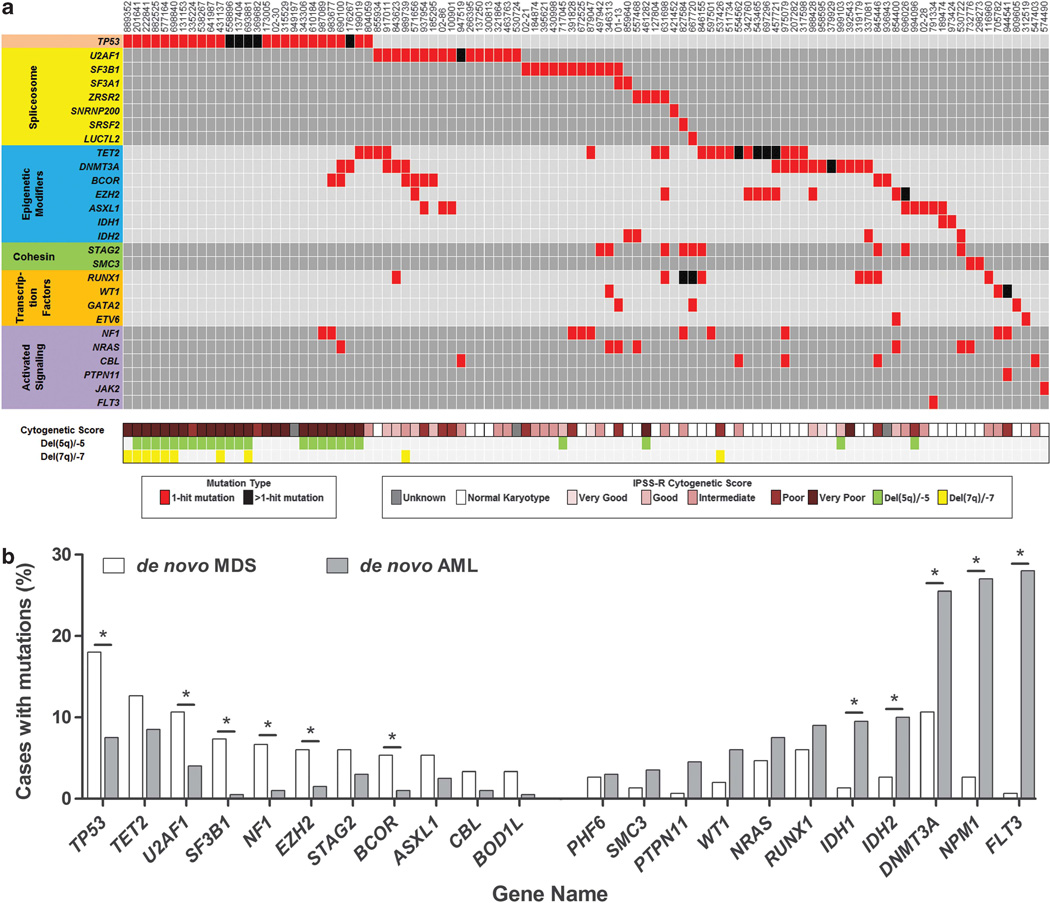
One-hundred and eleven of the 150 MDS samples (74%) had at least one mutation in the 94 genes screened and 124 samples (83%) had at least one mutation or cytogenetic abnormality. Most (15/17) of the significantly mutated genes have been implicated in MDS pathogenesis (for example, TP53, DNMT3A, U2AF1 and so on) and occurred in one of several common gene families/pathways, including spliceosome genes, epigenetic modulators, transcription factors, activated signaling/RAS pathway genes, and cohesin genes (Figure 3a). Many recurrently mutated genes in these 150 MDS samples were also mutated in 200 de novo AML samples analyzed by exome sequencing (T Ley, personal communication, on behalf of the Cancer Genome Atlas AML Working Group) (Figure 3b). However, some genes and pathways are overrepresented in MDS compared with AML, and vice versa. Mutations in TP53, NF1, U2AF1, SF3B1, EZH2 and BCOR were more common in MDS, while mutations in FLT3, NPM1, DNMT3A, IDH1 and IDH2 were more common in AML (P<0.05).
Figure 3.
Recurrently mutated genes in MDS and AML. (a) The distribution of mutations in 100 MDS samples with at least one mutation in 27 genes. Each column represents an individual patient sample and each row represents a gene with a mutation. Mutations are indicated by colored cells and gene groups/families are indicated by different colors at the left. The International Prognostic Scoring System-Revised cytogenetic score and cases with chromosome 5 and/or 7 deletions are indicated at the bottom. Normal karyotype (classified as ‘Good’ cytogenetic risk within the International Prognostic Scoring System-Revised) is indicated by a white box. (b) The frequency of genes with non-silent mutations in ≥5/150 MDS or ≥5/200 AML cases (*P<0.05).
The significantly mutated genes test corrects for background mutation rates across the exome or genome, but may be overly conservative when applied to candidate gene resequencing. Considering all the 44 genes that were mutated in at least two patients, 2 genes were recurrently mutated at the same codon (GNB1 and SETBP1). In GNB1, we identified missense mutations in 3/157 MDS/sAML samples that affected the lysine at amino-acid position 57 (K57), resulting in K57E or K57N substitutions. A fourth MDS patient had a frameshift mutation at amino-acid position 53 in GNB1. We also identified SETBP1 mutations in six MDS/sAML samples, including four mutations at amino-acid position 816 (G816S).
The landscape of somatic alterations in these MDS cases revealed several known and novel associations (Figure 3a). We performed pairwise comparisons to identify non-random patterns of mutation co-occurrence or mutual exclusivity. As previously reported for spliceosome genes, gene mutations within a group (spliceosome, transcription factor, activated signaling/RAS pathway and cohesin) were largely mutually exclusive (Supplementary Figure 2). TP53 mutations tended not to occur with other gene mutations (Figure 3a); in fact, 67% of patients with TP53 mutations had no point mutations in the other 93 candidate genes tested. However, TP53 mutations frequently co-occurred with a complex karyotype, del(5q) or del(7q), as previously reported (P<0.001) (Figure 3a), and were associated with a higher International Prognostic Scoring System-Revised risk category (P<0.001).6,15 We observed significant co-occurrence of mutations in three pairs of genes, including RUNX1 and STAG2, EZH2 and TET2, and BCOR and U2AF1 (P≤0.02) (each gene is mutated in at least 7/150 patients (~5%) and at least four samples have both mutations) (Supplementary Figure 3). Several mutated genes tended to be mutually exclusive with mutations in a separate gene family, including TP53 and spliceosome genes, and NRAS and cohesin genes (P<0.01) (each gene is mutated in at least seven patients) (Supplementary Figure 4).
Mutations in TP53 were associated with reduced hemoglobin and platelet counts, compared with patients without TP53 mutations (P<0.01). Patients with TET2 or DNMT3A mutations were significantly older at MDS diagnosis, compared with patients without these mutations (mean age of 69 vs 60 years and 68 vs 60 years, respectively; P≤0.01). Patients with SF3B1 mutations were significantly younger, compared with those without a SF3B1 mutation (mean age at diagnosis of 54 vs 62 years; P≤0.01). ASXL1 mutations were associated with poor-risk cytogenetic scores and SF3B1 mutations were associated with intermediate and poor-risk cytogenetic scores combined (P<0.01).
Clonal architecture of recurrently mutated genes
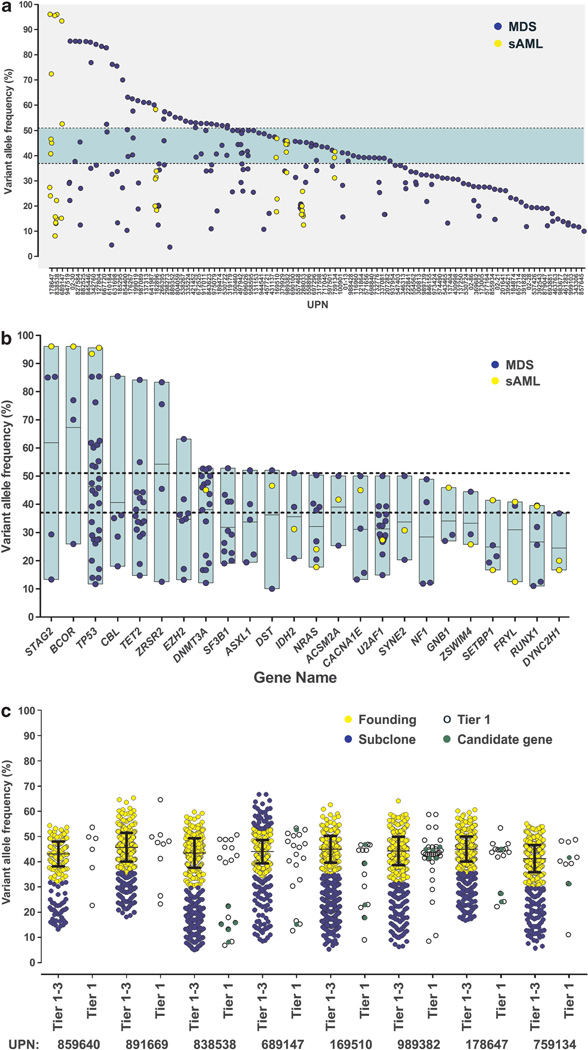
The VAFs of all validated somatic mutations (SNVs) in these MDS cases are continuously distributed (Figure 4a). WGS data from 15 MDS-derived sAML genomes presented here and elsewhere8 demonstrate that all cases have a founding clone, containing hundreds of mutations with a median VAF of 44% (±7%, 2 s.d.), that is similar to the median VAF of founding clones in eight MDS genomes of 42% (±7%, 2 s.d.) (Supplementary Figure 5). In the 94 candidate genes sequenced here, 36% of the 103 patients with at least one SNV lack even a single mutation in this range, implying that even a large panel of recurrently mutated candidate genes will not identify mutations in the founding clone in all cases using SNVs. Indels were excluded from the clonal analysis because VAFs for indels cannot be accurately quantified using this sequencing technology.
Figure 4.
Comparison of VAFs from whole-genome and candidate gene sequencing. (a) The VAFs for all validated SNVs in 157 MDS or sAML samples. Green horizontal bar, median VAF (±2 s.d.) of founding clone from WGS data (Figure 1). (b) VAFs for genes with ≥3 validated somatic SNVs. The vertical boxed regions show the range and mean (horizontal line) of VAFs for each gene. Dotted lines, boundary of founding clone (as in panel a). (c) Comparison of VAFs from capture validation of Tier 1–3 SNVs (restricted to mutations on autosomes without copy number alteration or uniparental disomy) vs only Tier 1 (open) or candidate genes (green) in eight sAML genomes. The founding clones (yellow) and subclones (blue) were identified by clustering, as described in the text. For each founding clone, the mean (±s.d.) of VAFs is shown. All data were generated simultaneously in a single liquid hybridization and deep sequencing experiment.
The range of VAFs for SNVs in recurrently mutated genes (which are more likely to be relevant for pathogenesis) is broad in all cases (Figure 4b). Several genes with VAFs >50% are located on the X chromosome (BCOR, STAG2, ZRSR2), or are in the regions prone to loss of heterozygosity, either through deletion of the wild-type allele (TP53), or partial uniparental disomy (CBL, TET2, EZH2), likely accounting for higher VAFs in some cases. A minority of these genes have consistently low VAFs, suggesting that they may be acquired later (in subclones). However, the variability of VAFs for these genes suggests that mutations in the same gene are acquired in founding clones in some patients and subclones in other patients, and that none of these genes is exclusively mutated in the MDS founding clone.
For the eight sAML samples subjected to both WGS capture validation and candidate gene sequencing in this study, direct comparisons can be made of clonal architecture defined by WGS and clonal status inferred from the VAFs of SNVs in individual genes (Figure 4c). In three of these cases (and none of seven previously published sAML genomes8; Supplementary Figure 5), sequence data from only the 94 candidate genes would fail to detect a SNV mutation in the founding clone. Six of these cases (and all other sAML genomes8; Supplementary Figure 5) have candidate gene mutations that can be assigned to the founding clone vs a subclone with little ambiguity, using the founding clone boundaries defined by their WGS data. However, several mutations have VAFs that are at the boundary between the founding clone and subclone, further highlighting the difficulty in assigning mutations accurately to a specific clone unless hundreds of mutations are available for analysis.
DISCUSSION
In this study, we defined the clonal architecture of eight sAML genomes and found them to be oligoclonal; each had a founding clone that gave rise to subclones, as we previously described for seven other sAML genomes.8 Each clone contained at least one coding gene mutation, and the majority of samples contained at least one mutation in a gene that is recurrently mutated in MDS or AML. Next, we screened 150 de novo MDS samples for mutations in 94 candidate genes and identified 264 mutations with predicted translational consequences in 59 genes. Overall, 83% of MDS samples harbored at least one mutated gene or a cytogenetic abnormality. Recurrently mutated genes typically occurred in six major gene groups/pathways, including spliceosome genes, epigenetic modifiers, transcription factors, activated signaling/RAS, cohesin factors and TP53.
Several groups have recently described mutations in candidate gene panels in de novo MDS. Bejar et al.6 screened mutational hotspots in 111 genes (excluding DNMT3A and spliceosome genes) using 439 MDS samples and reported recurrent (>1 patient) mutations in 16 genes, including TP53, EZH2, ETV6, RUNX1 and ASXL1 that were associated with reduced survival, independent of other prognostic factors. Subsequent screening of 288 low-risk MDS patients from their original cohort identified recurrent mutations in four additional genes (DNMT3A, SF3B1, SRSF2 and U2AF1).5 Our findings confirm that 17 of these 20 genes are recurrently mutated in MDS. Our sample size was insufficient to identify mutations in the other three genes (KRAS, BRAF and GNAS), which all have reported mutational frequencies of <1%.6 We found similar frequencies of mutations in most genes compared with these prior studies, including EZH2 (6% vs 6.4%), RUNX1 (6% vs 8.7%) and ETV6 (1.3% vs 2.7%), but a higher frequency of TP53 mutations (18% vs 7.5%) and a lower frequency of mutations in ASXL1 (5.3% vs 14.4%). Several other groups have profiled a variety of candidate genes (n=6–16 genes) in MDS (ranging from 88–221 samples).16–18 In these studies, at least one mutation was detected in 40–70% of MDS patients, with the frequency consistently approaching 70% when 16 or more commonly mutated genes were screened. Comparison of the frequency of specific gene mutations between studies is confounded by clinical heterogeneity. For example, the proportion of poor-risk patients (International Prognostic Scoring System-Revised >4.5) is higher in our cohort, and was associated with a higher frequency of TP53 mutations, as expected.
Similar to previous reports, these data show that patients with TP53 mutations tend to have a complex karyotype, deletions of chromosome 5 and 7, and TP53 as the only recurrently mutated gene.6,15 TP53 mutations are known to be associated with poor outcomes, and our results suggest that additional driver mutations may not be necessary to confer this poor prognosis. However, the strong association with karyotypic changes suggests that commonly deleted genes may cooperate with TP53 mutations in MDS pathogenesis.
We identified novel recurrent mutations at the same amino acid in two genes (GNB1, SETBP1), suggesting they are important for MDS pathogenesis. GNB1 was recurrently mutated at codon K57. A K57E somatic mutation was previously reported in a case of therapy-related AML3 and a K57T mutation was detected in 1/200 de novo AML samples (T Ley, personal communication, on behalf of The Cancer Genome Atlas AML Working Group). The GNB1 gene encodes the guanine nucleotide-binding protein (G protein), beta polypeptide 1. Heterotrimeric G-proteins (α and βγ subunits) transmit signals from receptors to effectors. GNB1 mRNA is robustly expressed in CD34+ hematopoietic progenitor cells harvested from normal healthy individuals and MDS patients (data not shown).19 Although the functional consequences of these mutations are not known, the mutations are located in a WD40-repeat motif that may serve as a scaffold for protein interactions. Several lines of evidence suggest that K57 mutations may have functional significance. First, the lysine at position 57 in GNB1 interacts with the guanine nucleotide-binding protein (G protein), alpha stimulating activity polypeptide 1 (GNAS).20–22 Recurrent GNAS mutations have been reported in MDS,6 resulting in substitutions of the arginine at amino-acid position 201 (R201C/H). R201 in GNAS is critical for the hydrolysis of guanosine-5’-triphosphate to guanosine diphosphate and R201 mutations produce increased levels of cyclic adenosine monophosphate due to constitutive activation of downstream effectors in the absence of receptor stimulation.23 Second, an alanine substitution at position 57 (K57A) increases the ability of GNB1 to associate with the β-adrenergic receptor kinase and decreases activation of adenylate cyclase 2 (both effectors of G-proteins).21 Finally, the K57A substitution increases GNB1-binding avidity to GSK3 (a recently identified G-protein effector kinase), which may ultimately stabilize β-catenin and activate its downstream signaling.24 Collectively, the identification of mutations in both GNB1 and GNAS in MDS samples suggests that alterations in G-couple protein receptor complexes may genetically define a novel MDS subtype. SETBP1 mutations were recently described in atypical chronic myeloid leukemia (aCML)25 and preliminary reports of recurrent SETBP1 mutations in MDS were recently presented.26,27 We observed recurrent mutations affecting codon 816 within the region homologous to the SKI protooncogene, where mutations also cluster in aCML.25 Germline mutations in SETBP1 are associated with Schinzel-Giedion syndrome (OMIM 269150), which is characterized by developmental abnormalities and predisposition to neuroepithelial tumors.28
We identified six sets of genes that are frequently mutated in MDS (Figure 3a). Mutations within a group tend not to co-occur, suggesting that a second mutation in these groups provides no additional selective advantage to a tumor, or is not tolerated. We found that mutations in RUNX1 and STAG2, or BCOR and U2AF1 co-occur more often than expected by chance. These novel observations may provide additional clues regarding the pathogenesis of MDS. Several other reported mutation co-occurrences (DNMT3A and SF3B1, SRSF2 and IDH1, SRSF2 and RUNX1, U2AF1 and DNMT3A, U2AF1 and ASXL1)16–18 were not observed in our cohort, possibly because of limited sample size, clinical heterogeneity or differences in sequencing and analytical approaches. Although many of the same genes are recurrently mutated in MDS and AML (Figure 3b), we found that some genes/ pathways are overrepresented in MDS compared with AML, and vice versa. These results highlight emerging differences in the pathophysiology of MDS and AML that are becoming evident from unbiased genomic analyses of both diseases. While the genetic landscape and clonality of MDS and AML are similar in many respects, the genetic differences between the disease groups and in individual patients likely contribute to known differences in morphology and clinical outcome.
Several groups, including our own, have reported the range of VAFs in TET2, TP53, SF3B1 and U2AF1 in MDS samples using next-generation sequencing platforms that provide digital read counts.1,4,29,30 Our results confirm and extend the observation that the clonal diversity of driver gene mutations is a common finding for the majority of recurrently mutated genes in MDS, suggesting that no single mutated gene occurs in only a founding clone or subclone. Instead, there appear to be many possible combinations of mutated genes that may arise in either the founding clone or subclones in different patients.
Accurately defining the clonal architecture of tumors based on clustering of mutations requires hundreds of data points, which can be generated using WGS in diseases with low mutational burdens, like MDS and AML. Candidate gene sequencing and exome sequencing provide fewer data points per sample, limiting statistical power for mutation clustering. Using the distribution of VAFs from mutation clusters for eight MDS genomes and 15 sAML genomes, we defined VAF ranges for SNVs that encompassed the founding clone in our samples. Using these boundaries, we observed that candidate gene sequencing results for SNVs (and exome sequencing results inferred from Tier 1 mutations) did not accurately recapitulate the clonal architecture of these genomes, and that the founding clone was missed in some samples. These results demonstrate the limitations of inferring clonal status from the VAFs of individual candidate genes (SNVs), even when applied retrospectively to tumors with clonal architecture defined by WGS. Moreover, the clonal architecture of MDS samples is probably underestimated by 30 × WGS coverage. Using standard variant calling algorithms,12,31 the minimum VAF typically achieved with 30 × WGS coverage is ~10% (that is, ~20% of cells harboring heterozygous mutations). While we identified genes with VAFs below 5% in this study using capture reagents and deep sequencing, it is possible that we are still underestimating the clonal complexity of these MDS samples. Furthermore, our analysis does not exclude the possibility that the dominant clone detected at diagnosis may be derived from an antecedent founding clone. Moving forward, a larger number of MDS genomes will need to be analyzed with WGS and deep validation before we fully understand the landscape of clonality.
Next-generation sequencing approaches are reaching the clinic. The tools required to detect driver mutations, define tumor clonality and pinpoint the location of specific mutations in the clonal architecture of tumors are available. The clinical utility of monitoring tumor clonality at specific time points (or dynamically during treatment) remains to be proven. The small sample size and variable treatments administered in this cohort (including 49/ 150 patients receiving an allogeneic transplant) limited our ability to analyze the prognostic significance of the clonal distribution of recurrent mutations in this MDS cohort. Given the genetic heterogeneity within MDS, these results suggest that a much larger cohort of MDS patients will be necessary to adequately assess whether the clonal distribution of recurrent mutations has prognostic or predictive importance.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants R01HL082973 (Graubert), RC2HL102927 (Graubert), U54HG003079 (Wilson), P01CA101937 (Ley), U01HG006517 (Ding), and a Howard Hughes Medical Institute Physician-Scientist Early Career Award (Walter). Technical assistance was provided by the Alvin J. Siteman Cancer Center Tissue Procurement Core, which is supported by an NCI Cancer Center Support Grant P30CA91842. Variant calls from WGS data have been deposited in dbGAP under accession number phs000159.v5.p2.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu
REFERENCES
- 1.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visconte V, Makishima H, Jankowska A, Szpurka H, Traina F, Jerez A, et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia. 2012;26:542–545. doi: 10.1038/leu.2011.232. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 4.Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bejar R, Stevenson KE, Caughey BA, Abdel-Wahab O, Steensma DP, Galili N, et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30:3376–3382. doi: 10.1200/JCO.2011.40.7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larson DE, Harris CC, Chen K, Koboldt DC, Abbott TE, Dooling DJ, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Research. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dees ND, Zhang Q, Kandoth C, Wendl MC, Schierding W, Koboldt DC, et al. MuSiC: identifying mutational significance in cancer genomes. Genome Res. 2012;22:1589–1598. doi: 10.1101/gr.134635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulasekararaj AG, Smith AE, Mian SA, Mohamedali AM, Krishnamurthy P, Lea NC, et al. TP53 mutations in myelodysplastic syndrome are strongly correlated with aberrations of chromosome 5, and correlate with adverse prognosis. Br J Haematol. 2013;160:660–672. doi: 10.1111/bjh.12203. [DOI] [PubMed] [Google Scholar]
- 16.Thol F, Kade S, Schlarmann C, Loffeld P, Morgan M, Krauter J, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119:3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- 17.Makishima H, Visconte V, Sakaguchi H, Jankowska AM, Abu Kar S, Jerez A, et al. Mutations in the spliceosome machinery, a novel and ubiquitous pathway in leukemogenesis. Blood. 2012;119:3203–3210. doi: 10.1182/blood-2011-12-399774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Damm F, Kosmider O, Gelsi-Boyer V, Renneville A, Carbuccia N, Hidalgo-Curtis C, et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood. 2012;119:3211–3218. doi: 10.1182/blood-2011-12-400994. [DOI] [PubMed] [Google Scholar]
- 19.Graubert TA, Payton MA, Shao J, Walgren RA, Monahan RS, Frater JL, et al. Integrated genomic analysis implicates haploinsufficiency of multiple chromosome 5q31.2 genes in de novo myelodysplastic syndromes pathogenesis. PLoS One. 2009;4:e4583. doi: 10.1371/journal.pone.0004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. The 2.0 A crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 21.Ford CE, Skiba NP, Bae H, Daaka Y, Reuveny E, Shekter LR, et al. Molecular basis for interactions of G protein betagamma subunits with effectors. Science. 1998;280:1271–1274. doi: 10.1126/science.280.5367.1271. [DOI] [PubMed] [Google Scholar]
- 22.Wall MA, Coleman DE, Lee E, Iniguez-Lluhi JA, Posner BA, Gilman AG, et al. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 23.Farfel Z, Bourne HR, Iiri T. The expanding spectrum of G protein diseases. N Engl J Med. 1999;340:1012–1020. doi: 10.1056/NEJM199904013401306. [DOI] [PubMed] [Google Scholar]
- 24.Jernigan KK, Cselenyi CS, Thorne CA, Hanson AJ, Tahinci E, Hajicek N, et al. Gbetagamma activates GSK3 to promote LRP6-mediated beta-catenin transcriptional activity. Science Signaling. 2010;3 doi: 10.1126/scisignal.2000647. ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piazza R, Valletta S, Winkelmann N, Redaelli S, Spinelli R, Pirola A, et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat Genet. 2012;45:18–24. doi: 10.1038/ng.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makishima H, Yoshida K, Nguyen N, Sanada M, Okuno Y, Ng KP, et al. Somatic mutations in Schinzel-Giedion syndrome gene SETBP1 determine progression in myeloid malignancies. ASH Annual Meeting Abstracts. 2012;120:2. [Google Scholar]
- 27.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. High throughput targeted gene sequencing in 738 myelodysplastic syndromes patients reveals novel oncogenic genes, rare driver mutations and complex molecular signatures with potential impact for patient diagnosis and prognosis in the clinic. ASH Annual Meeting Abstracts. 2012;120 LBA-5. [Google Scholar]
- 28.Hoischen A, van Bon BW, Gilissen C, Arts P, van Lier B, Steehouwer M, et al. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat Genet. 2010;42:483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 29.Jadersten M, Saft L, Smith A, Kulasekararaj A, Pomplun S, Gohring G, et al. TP53 mutations in low-risk myelodysplastic syndromes with del(5q) predict disease progression. J Clin Oncol. 2011;29:1971–1979. doi: 10.1200/JCO.2010.31.8576. [DOI] [PubMed] [Google Scholar]
- 30.Smith AE, Mohamedali AM, Kulasekararaj A, Lim Z, Gaken J, Lea NC, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood. 2010;116:3923–3932. doi: 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.