Abstract
Regulatory factor binding to genomic DNA protects the underlying sequence from cleavage by DNaseI, leaving nucleotide-resolution footprints. Using genomic DNaseI footprinting across 41 diverse cell and tissue types, we detected 45 million factor occupancy events within regulatory regions, representing differential binding to 8.4 million distinct short sequence elements. Here we show that this small genomic sequence compartment, roughly twice the size of the exome, encodes an expansive repertoire of conserved recognition sequences for DNA-binding proteins that nearly doubles the size of the human cis-regulatory lexicon. We find that genetic variants affecting allelic chromatin states are concentrated in footprints, and that these elements are preferentially sheltered from DNA methylation. High-resolution DNaseI cleavage patterns mirror nucleotide-level evolutionary conservation and track the crystallographic topography of protein-DNA interfaces, indicating that transcription factor structure has been evolutionarily imprinted on the human genome sequence. We identify a stereotyped 50 base-pair footprint that precisely defines the site of transcript origination within thousands of human promoters. Finally, we describe a large collection of novel regulatory factor recognition motifs that are highly conserved in both sequence and function, and exhibit cell-selective occupancy patterns that closely parallel major regulators of development, differentiation, and pluripotency.
Keywords: chromatin, protein occupancy, DNaseI footprinting, ENCODE, regulation
Introduction
Sequence-specific transcription factors (TFs) interpret the signals encoded within regulatory DNA. The discovery of DNaseI footprinting over 30 years ago1 revolutionized the analysis of cisregulatory sequences in diverse organisms, and directly enabled the discovery of the first human sequence-specific transcription factors2. Binding of TFs to regulatory DNA regions in place of canonical nucleosomes triggers chromatin remodeling, resulting in nuclease hypersensitivity3. Within DNaseI hypersensitive sites (DHSs), DNaseI cleavage is not uniform; rather, punctuated binding by sequence-specific regulatory factors occludes bound DNA from cleavage, leaving ‘footprints’ that demarcate TF occupancy at nucleotide resolution1,4 (Figure 1a). DNaseI footprinting has been applied widely to study the dynamics of transcription factor occupancy and cooperativity within regulatory DNA regions of individual genes5, and to identify cell- and lineage-selective transcriptional regulators6.
Figure 1. Parallel profiling of genomic regulatory factor occupancy across 41 cell types.
a, DNaseI footprinting of K562 cells identifies the individual nucleotides within the MTPN promoter that are bound by NRF1. b, Example locus harboring eight clearly defined DNaseI footprints in Th1 and SK-N-SH_RA cells, with TRANSFAC database motif instances indicated below. c, Heatmaps showing per-nucleotide DNaseI cleavage (left) and vertebrate conservation by phyloP (right) for 4,262 NRF1 motifs within K562 DHSs ranked by the local density of DNaseI cleavages. Green ticks indicate the presence of DNaseI footprints over motif instances. Blue ticks indicate the presence of ChIP-seq peaks over the motif instances. d, Lowess regression of NRF1, USF, NFE2, and NFYA K562 ChIP-seq signal intensities versus DNaseI footprinting occupancy (footprint occupancy score) at K562 DNaseI footprints containing NRF1, USF, NFE2, and NFYA motifs.
Regulatory DNA is densely populated with DNaseI footprints
To map DNaseI footprints comprehensively within regulatory DNA, we adapted digital genomic footprinting4 to human cells. The ability to resolve DNaseI footprints sensitively and precisely is critically dependent on the local density of mapped DNaseI cleavages (Supplementary Figs. 1a–d), and efficient footprinting of a large genome such as human requires substantial concentration of DNaseI cleavages within the small fraction (~1–3%) of the genome contained in DNaseI-hypersensitive regions. We selected highly enriched DNaseI cleavage libraries from 41 diverse cell types in which 53–81% of DNaseI cleavage sites localized to DNaseI-hypersensitive regions7 (Supplementary Table 1), representing nearly 10-fold higher signal-to-noise ratio vs. prior results from yeast4, and 2- to 5-fold greater enrichment than achieved using end-capture of single DNaseI cleavages8,9. We then performed deep sequencing of these libraries, and obtained 14.9 billion Illumina sequence reads, 11.2 billion of which mapped to unique locations in the human genome (Supplementary Table 1). We achieved an average sequencing depth of ~273 million DNaseI cleavages per cell type that enabled extensive and accurate discrimination of DNaseI footprints.
To detect DNaseI footprints systematically, we implemented a detection algorithm based on the original description of quantitative DNaseI footprinting1 (Supplementary Methods). We identified an average of ~1.1 million high-confidence (FDR 1%) footprints per cell type (range 434,000 to 2.3 million; Supplementary Table 1), and collectively 45,096,726 6–40 bp footprint events across all cell types. We resolved cell-selective footprint patterns to reveal 8.4 million distinct footprinted elements, each occupied in one or more cell types. At least one footprint was found in >75% of DHSs (Supplementary Figs. 1c,d and Supplementary Table 2), with detection strongly dependent on the number of mapped DNaseI cleavages within each DHS. 99.8% of DHSs with >250 mapped DNaseI cleavages contained at least one footprint, indicating that DHSs are not simply open or nucleosome-free chromatin features, but are constitutively populated with DNaseI footprints. Modeling DNaseI cleavage patterns using empirically derived intrinsic DNA cleavage propensities for DNaseI showed that only a miniscule fraction (0.24%) of discovered FDR 1% footprints from cell and tissue samples could be caused by inherent DNaseI sequence specificity (Supplementary Methods).
DNaseI footprints were distributed throughout the genome, including intergenic regions (45.7%), introns (37.7%), upstream of transcriptional start sites (8.9%), and in 5’ and 3’ UTRs (1.4% and 1.3%, respectively; Supplementary Figs. 2a,b). DNaseI footprints were enriched in promoters (3.6 fold; P < 2.2×10−16; Binomial test) and 5’ UTRs (2.4 fold; P < 2.2×10−16; Binomial test), commensurate with high DNaseI cleavage densities observed in these regions. We found that 2.0% of footprints localized within exons, raising the possibility that occupancy by DNA binding proteins could further restrict sequence diversity within coding DNA, thus superimposing an unexpected layer of constraint on codon usage.
Quantitative markers of in vivo regulatory factor occupancy
We next examined the correspondence between DNaseI footprints and known regulatory factor recognition sequences within DNaseI hypersensitive chromatin. Comprehensive scans of DNaseI hypersensitive regions for high confidence matches to all recognized TF motifs in the TRANSFAC10 and JASPAR11 databases revealed striking enrichment of motifs within footprints (P ≈ 0, Z-score = 204.22 for TRANSFAC; Z-score = 169.88 for JASPAR; Fig. 1b and Supplementary Fig. 3).
To quantify the occupancy at TF recognition sequences within DHSs genome-wide, we computed for each instance a footprint occupancy score (FOS) relating the density of DNaseI cleavages within the core recognition motif to cleavages in the immediately flanking regions (Supplementary Methods). The FOS can be used to rank motif instances by the ‘depth’ of the footprint at that position, and is expected to provide a quantitative measure of factor occupancy1. To examine this relationship for a well-studied sequence-specific regulator (NRF112), we plotted DNaseI cleavage patterns surrounding all 4,262 NRF1 motifs contained within DNaseI hypersensitive sites and ranked these by FOS. While only a subset of these motif instances (2,351) coincided with high-confidence footprints, the vast majority of NRF1 motif instances in DNaseI footprints (89%) overlapped reproducible NRF1 ChIP-seq peaks (Fig. 1c). In parallel, we analyzed nucleotide-level evolutionary conservation patterns around NRF1 binding sites, revealing that FOS closely parallels phylogenetic conservation within the core motif region, suggesting strong selection on factor occupancy (Fig. 1c). We observed a nearly monotonic relationship between FOS and ChIPseq signal intensities at NRF1 binding sites within K562 DNaseI footprints (Fig. 1d). Similarly strong correlations between footprint occupancy and either ChIP-seq signal or phylogenetic conservation were evident for diverse factors (Fig. 1d and Supplementary Figs. 4a–d). We found footprint occupancy and nucleotide-level conservation correlated for 80% of all TF motifs in the TRANSFAC database, of which 50% were statistically significant (P < 0.05; Supplementary Methods). This relationship between footprint occupancy and conservation is most readily explained by evolutionary selection on factor occupancy, with higher conservation of higher affinity binding sites. Taken together, these results indicate that footprint occupancy provides a quantitative measure of sequence-specific regulatory factor occupancy that closely parallels evolutionary constraint and ChIP-seq signal intensity.
To validate the potential for selective binding of footprints by factors predicted on the basis of motif-to-footprint matching, we developed an approach to quantify specific occupancy in the context of a complex TF milieu using targeted mass spectrometry (DNA interacting protein precipitation or DIPP; Methods). Using DIPP, we affirmed specific binding by several different classes of TFs (Supplementary Figs. 5a–e). Together with the analysis of ChIP-seq data described above, these results indicate that the localization of TF recognition motifs within DNaseI footprints can accurately illuminate the genomic protein occupancy landscape.
Footprints harbor functional variants and are sheltered from DNA methylation
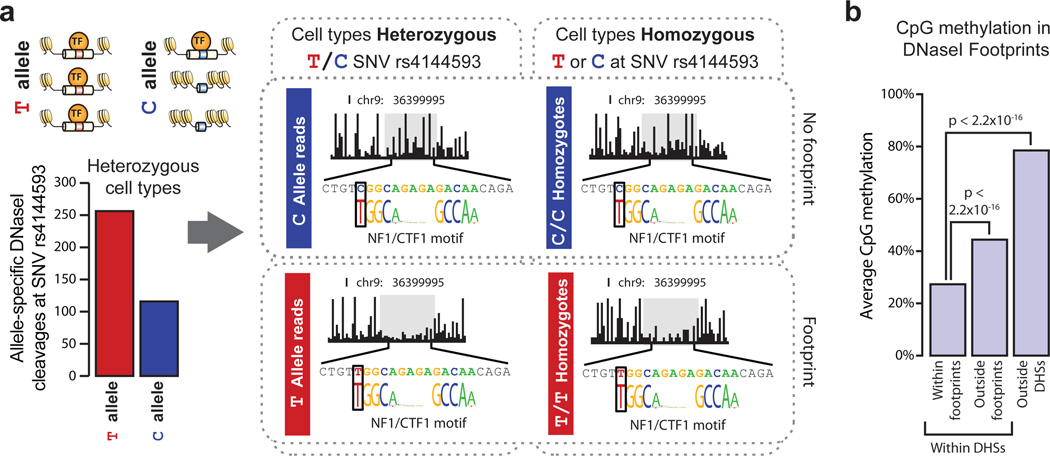
The potential for single nucleotide variants within a transcription factor recognition sequence to abrogate binding of its cognate factor is well known13. The depth of sequencing performed in the context of our footprinting experiments provided hundreds- to thousands-fold coverage of most DHSs, enabling precise quantification of allelic imbalance within DHSs harboring heterozygous variants. We scanned all DHSs for heterozygous single nucleotide variants identified by the 1000 Genomes Project14 and measured, for each DHS containing a single heterozygous variant, the proportion of reads from each allele. We identified likely functional variants conferring significant allelic imbalance in chromatin accessibility and analyzed their distribution relative to DNaseI footprints. This analysis revealed significant enrichment (P < 2.2×10−16; Fisher’s exact test) of such variants within DNaseI footprints (Supplementary Fig. 6). For example, rs4144593 is a common T/C variant that lies within a DHS on chromosome 9. This variant falls on a high-information position within an NF1/CTF1 footprint and substantially disrupts footprinting of this motif, resulting in allelic imbalance in chromatin accessibility (Fig. 2a).
Figure 2. DNaseI footprints mark sites of in vivo protein occupancy.
a, Schematic and plots showing the effect of T/C SNV rs4144593 on protein occupancy and chromatin accessibility. Bar graph y-axis is the number of DNaseI cleavage events containing either the T or C allele. Middle plots show T or C allele-specific DNaseI cleavage profiles from 10 cell lines heterozygous for the T/C alleles at rs4144593. Right plots show DNaseI cleavage profiles from 18 cell lines homozygous for the C allele at rs4144593 and 1 cell line homozygous for the T allele at rs4144593. Cleavage plots are cut off at 60% cleavage height. b, The average CpG methylation within IMR90 DNaseI footprints, IMR90 DHSs (but not in footprints) and non-hypersensitive genomic regions in IMR90 cells. CpG methylation is significantly depleted in DNaseI footprints (P < 2.2×10−16, Mann-Whitney test).
Protein-DNA interactions are also sensitive to cytosine methylation15,16. Comparing DNaseI footprints and whole genome bisulfite sequencing methylation data from pulmonary fibroblasts (IMR90), we found that CpG dinucleotides contained within DNaseI footprints were significantly less methylated than CpGs in non-footprinted regions of the same DHS (Mann-Whitney test;P < 2.2×10−16; Fig. 2b). Footprints therefore appear to be selectively sheltered from DNA methylation, suggesting a widespread connection between regulatory factor occupancy and nucleotide-level patterning of epigenetic modifications.
Transcription factor structure is imprinted on the human genome
We observed surprisingly heterogeneous base-to-base variation in DNaseI cleavage rates within the footprinted recognition sequences of different regulatory factors. And yet, the per site cleavage profiles for individual factors were highly stereotyped, with nearly identical local cleavage patterns at thousands of genomic locations (Supplementary Fig. 7). This raised the possibility that DNaseI cleavage patterns may provide information concerning the morphology of the DNA-protein interface. We obtained the available DNA-protein co-crystal structures for human transcription factors, and mapped aggregate DNaseI cleavage patterns at individual nucleotide positions onto the DNA backbone of the co-crystal model. Fig. 3a and Supplementary Fig. 8a show two examples, USF17 and SRF18. For both factors, DNaseI cleavage patterns clearly parallel the topology of the protein-DNA interface, including a marked depression in DNaseI cleavage at nucleotides involved in protein-DNA contact, and increased cleavage at exposed nucleotides such as those within the central pocket of the leucine zipper. These data show that nucleotide-level aggregate DNaseI cleavage patterns reflect fundamental features of the protein-DNA interaction interface at unprecedented resolution.
Figure 3. Footprint structure parallels TF structure and is imprinted on the human genome.
a, The co-crystal structure of Upstream Stimulatory Factor (USF) bound to its DNA ligand is juxtaposed above the average nucleotide-level DNaseI cleavage pattern (blue) at motif instances of USF in DNaseI footprints. Nucleotides that are sensitive to cleavage by DNaseI are colored as blue on the co-crystal structure. The motif logo generated from USF DNaseI footprints is displayed below the DNaseI cleavage pattern. Below is a randomly ordered heatmap showing the per-nucleotide DNaseI cleavage for each motif instance of USF in DNaseI footprints. b, The per-base DNaseI hypersensitivity (blue) and vertebrate phylogenetic conservation (red) for all DNaseI footprints in dermal fibroblasts matching three well annotated transcription factor motifs. The white box indicates width of consensus motif. The number of motif occurrences within DNaseI footprints in indicated below each graph.
We next asked how these patterns related to evolutionary conservation. Plotting nucleotide-level aggregate DNaseI cleavage in parallel with per-nucleotide vertebrate conservation calculated by phyloP19 revealed striking antiparallel patterning of cleavage vs. conservation across nearly all motifs examined (six representative examples are shown in Fig. 3b and Supplementary Fig. 8b). Surprisingly, conservation is not limited to only DNA contacting protein residues, but exhibits graded changes that mirror DNaseI accessibility across the entirety of the protein-DNA interface (Supplementary Figs. 8c,d). Taken together, these results imply that regulatory DNA sequences have evolved to fit the continuous morphology of the transcription factor-DNA binding interface.
A stereotyped 50 bp footprint localizes transcription initiation within promoters
Transcription initiation requires the binding of multi-protein complexes that position RNA polymerase II (PolII)20–23. Using a modified footprint detection algorithm designed to detect larger features (Supplementary Methods), we scanned the regions upstream from Gencode transcriptional start sites (TSSs) and identified highly stereotyped ~80bp chromatin structure comprising a prominent ~50 bp central DNaseI footprint, flanked symmetrically by ~15 bp regions of uniformly elevated DNaseI cleavage (Fig. 4a). Alignment of per-nucleotide DNaseI cleavage profiles from 5,041 prominent footprints mapped in different K562 promoters highlights the homogeneous, nearly invariant nature of the structure (Fig. 4b).
Figure 4. A highly stereotyped chromatin structural motif marks sites of transcription initiation in human promoters.
a, A 35–55 base-pair footprint is the predominant feature of many promoter DHSs and is in tight spatial coordination with the transcription start site. b, Heatmap of the per-nucleotide DNaseI cleavage pattern at 5,041 instances of this stereotypical footprint in K562 cells. c, Aggregate per-base DNaseI cleavage profile (blue line) and mean per-nucleotide conservation score (phyloP) surrounding instances of this stereotypical footprint in K562 cells (red dashed line). d, Aggregate strand corrected CAGE sequencing data (green line) and the average nearest 5’ end of a spliced EST (orange line) surrounding instances of this stereotypical footprint in K562 cells.
Plotting evolutionary conservation in parallel with DNaseI cleavage revealed two distinct peaks in evolutionary conservation within the central footprint (Fig. 4c) compatible with binding sites for paired canonical sequence-specific TFs. The density of CAGE tags (Fig. 4d; green line) and 5’ ends of expressed sequenced tags (ESTs) (Fig. 4d; orange line) relative to the central ~50 bp footprint revealed that, at the vast majority of promoters, RNA transcript initiation localized precisely within the stereotyped footprint. It is notable that the location of this footprint is often offset, typically 5’, from many Gencode-annotated TSSs. This likely derives from the incomplete nature of many of the 5’ transcript ends used to define TSSs24.
These data together define a new high-resolution chromatin structural signature of transcription initiation and the interaction of the pre-initiation complex (PIC) with the core promoter. Indeed, chromatin occupancy of TATA-binding protein (TBP), a critical component of the PIC, is maximal precisely over the center of the 50bp footprint region (Supplementary Fig. 9a). Sequence analysis of the two conservation peaks within the 50bp footprint identified motifs for GCbox- binding proteins such as SP1 and, less frequently, other general transcription factors (though with the notable absence of TATA motifs) (Supplementary Fig. 9b), suggesting that TBP (and potentially other PIC components) interact preferentially with general transcriptional factors bound to GC-box-like features in the central footprinted region. The results are therefore consistent with a model in which a limited number of sequence-specific factors function both to prime the chromatin template for recruitment of RNA polymerase II and to guide transcriptional positioning.
Differentiating DNA binding vs. indirect occupancy by TFs
Many transcriptional regulators are posited to interact indirectly with the DNA sequence of some target sites though mechanisms such as tethering25. Approaches such as ChIP-seq detect chromatin occupancy, but cannot by themselves distinguish sites of direct DNA binding from non-canonical indirect binding. We therefore asked whether DNaseI footprint data could illuminate ChIP-seq-derived occupancy profiles by differentiating directly bound factors from indirect binding events. We first partitioned ChIP-seq peaks from each of 38 ENCODE transcription factors26 mapped in K562 cells into three categories of predicted sites: ChIP-seq peaks containing a compatible footprinted motif (directly bound sites); ChIP-seq peaks lacking a compatible motif or footprint (indirectly bound sites); and ChIP-seq peaks overlying a compatible motif lacking a footprint (indeterminate sites). Predicted indirect sites showed significantly reduced ChIP-seq signal compared with predicted directly bound sites (Supplementary Fig. 10), consistent with lack of direct cross-linking to DNA (and therefore reduced ChIP efficiency). Indeterminate sites exhibited low ChIP-seq signal and were therefore excluded from further analysis (Supplementary Fig. 10).
The fraction of ChIP-seq peaks predicted to represent direct vs. indirect binding varied widely between different factors, ranging from nearly complete direct sequence-specific binding (e.g., CTCF), to nearly complete indirect binding (e.g., TBP; Supplementary Fig. 11). In many cases factors that preferentially engage in direct DNA binding at distal sites show predominantly indirect occupancy in promoter regions and vice versa (Supplementary Figs. 12a,b),
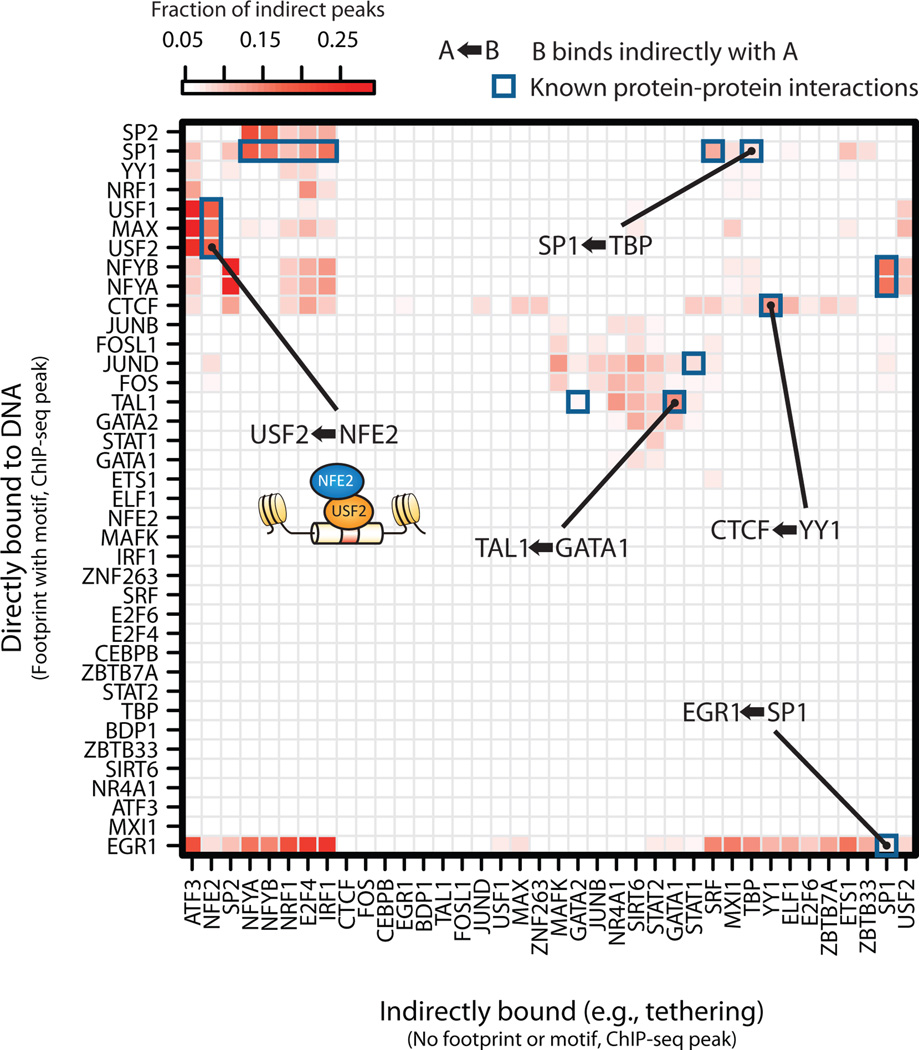
Next, we analyzed the frequency with which indirectly bound sites of one transcription factor coincided with directly bound sites of a second factor, suggestive of protein-protein interactions (e.g., tethering). This analysis recovered many known protein-protein interactions, such as CTCF/YY1 and TAL1/GATA127, as well as many novel associations (Fig. 5). We observed enrichment for NFE2 indirect interactions at promoter bound USF2 sites, compatible with their known interaction28. At distal sites, we observed the opposite, with NFE2 predominantly directly bound accompanied by USF2 indirect peaks (Supplementary Figs. 12a,b), suggesting the possibility of a reciprocal or looping mechanism. Notably, directly bound promoter-predominant transcription factors were enriched for co-localization with indirect peaks compared to distal regions (Supplementary Figs. 13a,b). These results suggest that combining DNaseI footprinting with ChIP-seq has the potential to expose a previously unappreciated landscape of complex transcription factor occupancy modes.
Figure 5. Distinguishing direct and indirect binding of transcription factors.
Heatmap of the enrichment of pairs of transcription factors in a direct-indirect association. Direct peaks are defined by ChIP occupancy accompanied by a footprint overlapping a compatible motif. Indirect peaks do not have a compatible motif. The color of each cell is determined by the fraction of indirect peaks that co-localize with the direct peaks of another factor.
Footprints encode an expansive cis-regulatory lexicon
Since the discovery of the first sequence-specific transcription factor29, considerable effort has been devoted to identifying the cognate recognition sequences of DNA-binding proteins30,31. Despite these efforts, high-quality motifs are available for only a minority of the >1,400 human transcription factors with predicted sequence-specific DNA binding domains32.
We reasoned that the genomic sequence compartment defined by DNaseI footprints in a given cell type ideally should contain much, if not all, of the factor recognition sequence information relevant for that cell type. Consequently, applying de novo motif discovery to the footprint compartments gleaned from multiple cell types should greatly expand our current knowledge of biologically active TF-binding motifs.
We performed unbiased de novo motif discovery within the footprints identified in each of the 41 cell types that yielded 683 unique motif models (Fig. 6a and Supplementary Methods). We compared these models with the universe of experimentally-grounded motif models in the TRANSFAC, JASPAR, and UniPROBE33 databases. Due to the redundancy of motif models contained within these databases, we first collapsed all duplicate models (Supplementary Methods). 394 of the 683 (58%) de novo motifs matched distinct experimentally-grounded motif models, accounting collectively for 90% of all unique entries across the three databases (Fig. 6b and Supplementary Figs. 14a–c). The wholesale de novo derivation of the vast majority of known regulatory factor recognition sequences from the small genomic compartment defined by DNaseI footprints highlights the dramatic concentration of regulatory information encoded within this sequence space.
Figure 6. De novo motif discovery expands the human regulatory lexicon.
a, Overview of de novo motif discovery using DNaseI footprints. b, Annotation of the 683 de novo-derived motif models using previously identified transcription factor motifs. 394 of these de novo-derived motifs match a motif annotated within the TRANSFAC, JASPAR or UniPROBE databases, whereas 289 are novel motifs (pie chart). The de novo consensus matching TRANSFAC, JASPAR or UniPROBE sequences cover the majority of each database (bar chart) c, Example of a DNaseI footprint found in multiple cell types that is annotated solely by one of the novel de novo-derived motifs. d, Box-andwhisker plot comparing the average nucleotide diversity at instances of the 289 novel de novoderived motif models to instances of motifs present in databases of known specificities (x-axis). The blue bar indicates the average nucleotide diversity (π) at 4-fold degenerate coding sites (width is equal to 95% confidence interval); gold bar indicates π at all coding sites (width is equal to 95% confidence interval). e, Phylogenetic conservation (red dashed) and per-base DNaseI hypersensitivity (blue) for all DNaseI footprints in dermal fibroblast cells matching two novel de novo-derived motifs. The white box indicates width of consensus motif. f, Per-nucleotide mouse liver DNaseI cleavage patterns at occurrences of the motifs in (e) at DNaseI footprints identified in mouse liver.
Strikingly, 289 of the footprint-derived motifs were absent from major databases (Fig. 6b and Supplementary Fig. 14d). These novel motifs populate millions of DNaseI footprints (Fig. 6c), and show features of in vivo occupancy and evolutionary constraint similar to motifs for known regulators, including marked anti-correlation with nucleotide-level vertebrate conservation (Figs. 6d,3).
To test whether novel motifs were functionally conserved in a distant mammal, we analyzed DNaseI cleavage patterns around human novel motifs mapped within DHSs assayed in primary mouse liver tissue (Figs. 6e,f and Supplementary Figs. 15a,b). This analysis demonstrated that many novel motifs show nearly identical DNaseI footprint patterns in both human cells and mouse liver, indicating that these novel motifs correspond to evolutionarily conserved transcriptional regulators that are functional in both mice and men.
Given the conservation of protein occupancy in a distant mammal, we assessed whether the novel motifs are under selection in human populations by analyzing nucleotide diversity across all motif instances found within accessible chromatin. Using high-quality genomic sequence data from 53 unrelated individuals34 (Supplementary Table 4), we calculated the average nucleotide diversity35 for each individual motif space (Supplementary Fig. 15c). Reduced diversity levels are indicative of functional constraint, through the elimination of deleterious alleles from the population by natural selection. We found that novel motifs are collectively under strong purifying selection in human populations. On average, the new motifs are more constrained than most motifs found in the major databases (Fig. 6d and Supplementary Fig. 15c), even following exclusion of motifs containing highly mutable CpG dinucleotides, which underlie the marked increase in nucleotide diversity seen with a subset of known motifs (Supplementary Fig. 15c, right). Collectively, these results demonstrate that DNaseI footprints encode an expansive cis-regulatory lexicon encompassing both known TF recognition sequences and novel motifs that are functionally conserved in mouse and bear strong signatures of ongoing selection in humans.
Novel motif occupancy parallels known regulators of pluripotency and cell fate
Cell-selective gene regulation is mediated by the differential occupancy of transcriptional regulatory factors at their cognate cis-acting elements. For example, the nerve growth factor gene VGF is selectively expressed only within neuronal cells (Fig. 7a), presumably due to the repressive action of the transcriptional regulator NRSF/REST at the VGF promoter in non-neuronal cell types36. Although VGF is expressed only in neuronal cells, its promoter is DNaseI-hypersensitive in most cell types (not shown). Examination of nucleotide-level cleavage patterns within the VGF promoter exposes its fundamental cis-regulatory logic, coordinated by the transcriptional regulators NRSF, SP1, USF1, and NRF1. Whereas the NRSF motif is tightly occupied in non-neuronal cells, in neuronal cells, NRSF repression is relieved, and recognition sites for the positive regulators USF1 and SP1 become highly occupied, resulting in VGF expression. These data collectively illustrate the power of genomic footprinting to resolve differential occupancy of multiple regulatory factors in parallel at nucleotide resolution.
Figure 7. Multi-lineage DNaseI footprinting reveals cell-selective gene regulators.
a, Comparative footprinting of the nerve growth factor gene (VGF) promoter in multiple cell types reveals both conserved (NRF1, USF and SP1) and cell-selective (NRSF) DNaseI footprints. b, Shown is a heatmap of footprint occupancy computed across 12 cell types (columns) for 89 motifs (rows), including well-characterized cell/tissue-selective regulators, and novel de novo-derived motifs (red text). The motif models for some of these novel de novo-derived motifs are indicated next to the heatmap. c, The proportion of motif instances in DNaseI footprints within distal regulatory regions for known (black) and novel (red) cell-type specific regulators in (b) is indicated. Also noted are these values for a small set of known promoter-proximal regulators (green).
We next extended this paradigm using genome-wide DNaseI footprints across 12 functionally distinct cell types to identify both known and novel factors showing highly cell-specific occupancy patterns. To calculate the footprint occupancy of a motif, we enumerated for each motif and cell type the number of motif instances encompassed within DNaseI footprints and normalized this by the total number of DNaseI footprints in that cell type. Fig. 7b shows a heatmap representation of cell-selective occupancy at motifs for 60 known transcriptional regulators and for 29 novel motifs. This approach appropriately identified a number of known cell-selective transcriptional regulators including; (1) the pluripotency factors OCT4, SOX2, KLF4, and NANOG in human embryonic stem cells37; (2) the myogenic factors MEF2A and MYF6 in skeletal myocytes38; and (3) the erythrogenic regulators GATA1, STAT1, and STAT5A in erythroid cells39–41 (Fig. 7b).
Many of the footprint-derived novel motifs displayed markedly cell-selective occupancy patterns highly similar with the aforementioned well-established regulators. This suggests that many novel motifs correspond to recognition sequences for important but uncharacterized regulators of fundamental biological processes. Notably, both known and novel motifs with high cell-selective occupancy predominantly localized to distal regulatory regions (Fig. 7c), further highlighting the role of distal regulation in developmental and cell-selective processes42,43.
Perspective
We describe an expansive map of regulatory factor occupancy at millions of precisely demarcated sequence elements across the human genome revealed by genomic DNaseI footprinting applied to a wide spectrum of cell types. These elements collectively define a highly information rich genomic sequence compartment which encodes the recognition landscape of hundreds of DNA binding proteins. This compartment has been extensively shaped by evolutionary forces to match closely the physical properties of its cognate interacting proteins. Mining footprint sequences for recognition motifs has nearly doubled the human cis-regulatory lexicon, exposing a previously hidden trove of elements with evolutionary, structural, and functional profiles that parallel the collections of experimentally-derived genomic regulators brought to light during the past 30 years. Because the ability to resolve footprints is dependent on sequencing depth, and the sequencing level of DNaseI cleavage events in most DHSs is not saturating (even in cell types with >500 million mapped unique DNaseI cleavages), the present study, while extensive in many respects, represents only an initial foray into this biologically rich space. Identification of the cognate DNA binding proteins for novel recognition sequences presents a significant challenge, though one which can be addressed with confidence using emerging technologies and our extensive experimental data demonstrating both occupancy in vivo and strong evolutionary signatures of function. On a broader level, the approach we describe here can, in principle, be applied to derive the cis-regulatory lexicon of any organism. We anticipate that the extensive new resources we describe, particularly in combination with other ENCODE data, will help to advance many aspects of human gene regulation research.
Methods Summary
DNaseI digestion and high-throughput sequencing were performed on intact human nuclei from various cell types, following published methods4,44. Briefly, roughly 10 million cells were grown in appropriate culture media and nuclei were extracted using NP-40 in an isotonic buffer. The NP-40 detergent was removed and the nuclei were incubated for 3 minutes at 37°C with limiting concentrations of the DNA endonuclease, deoxyribonuclease I (DNaseI) (Sigma) supplemented with Ca2+ and Mg2+. The digestion was stopped with EDTA and the samples were treated with proteinase K. The small ‘double-hit’ fragments (<500 bp) were recovered by sucrose ultra-centrifugation, endrepaired and ligated with adapters compatible with the Illumina sequencing platform. High quality libraries from each cell type were sequenced on the Illumina platform to an average depth of 273 million uniquely mapping single-end tags. The sequencing tags were aligned to the human reference genome and per-nucleotide cleavage counts were generated by summing the 5’ ends of the aligned sequencing tags at each position in the genome. FDR 1% DNaseI footprints were identified using an iterative search method based upon optimization of the footprint occupancy score. De novo motif discovery was performed using a full enumeration algorithm.
Supplementary Material
Acknowledgements
This work was supported by NIH grants HG004592 (J.A.S.) and RC2HG005654 (J.A.S. and M.G.). J.V. is supported by a National Science Foundation Graduate Research Fellowship. This work was supported in part by the University of Washington Proteomics Resource (UWPR95794). We thank Sam John and Fyodor Urnov for critical readings of the manuscript and many helpful discussions, and Sean Thomas for many helpful insights.
Footnotes
Author Contributions
J.A.S., A.B.S., S.N., M.T.M., B.V., and J.V. designed the experiments. S.N., J.V., A.B.S., A.P.R., B.V., M.T.M., R.E.T., E.H. and R.S. carried out the analysis, J.A.S., J.V., A.B.S., S.N., and A.P.R. wrote the paper, and all others carried out various aspects of experimental data collection. The authors declare no competing interests.
Data Availability
All genomic DNaseI footprinting data are are available through the NCBI Gene Expression Omnibus (GEO) data repository (accessions GSE26328 and GSE18927), and also through the UCSC browser under the Digital Genomic Footprinting (DGF) table designation. De novo motif models are available through the ENCODE Consortium data release website.
References
- 1.Galas DJ, Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dynan WS, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- 3.Gross DS, Garrard WT. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 4.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 6.Tsai SF, et al. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 7.Thurman RE, et al. The accessibe chromatin landscape of the human genome. Nature. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabo PJ, et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyle AP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matys V, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryne JC, et al. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan JY, Han XL, Kan YW. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. U.S.A. 1993;90:11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rockman MV, Wray GA. Abundant raw material for cis-regulatory evolution in humans. Mol. Biol. Evol. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- 14.1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 16.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferré-D'Amaré AR, Pognonec P, Roeder RG, Burley SK. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellegrini L, Tan S, Richmond TJ. Structure of serum responsefactor core bound to DNA. Nature. 1995;376:490–498. doi: 10.1038/376490a0. [DOI] [PubMed] [Google Scholar]
- 19.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pugh BF, Tjian R. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev. 1991;5:1935–1945. doi: 10.1101/gad.5.11.1935. [DOI] [PubMed] [Google Scholar]
- 21.Kim TH, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buratowski S, Hahn S, Guarente L, Sharp PA. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- 23.Kim TK, et al. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc. Natl. Acad. Sci. U.S.A. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Project ACSHET. Post-transcriptional processing generates a diversity of 5'-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biddie SC, et al. Transcription factor AP1potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wadman IA, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, et al. USF and NF-E2 cooperate to regulate the recruitment and activity of RNA polymerase II in the beta-globin gene locus. 2010;285:15894–15905. doi: 10.1074/jbc.M109.098376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert W, Müller-Hill B. Isolation of the lac repressor. Proc. Natl. Acad. Sci. U.S.A. 1966;56:1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukherjee S, et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 33.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drmanac R, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 35.Nei M, Li WH. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. U.S.A. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 38.Yun K, Wold B. Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr. Opin. Cell Biol. 1996;8:877–889. doi: 10.1016/s0955-0674(96)80091-3. [DOI] [PubMed] [Google Scholar]
- 39.Pevny L, et al. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 40.Socolovsky M, et al. Ineffective erythropoiesis in Stat5a(-/-)5b(-/-) mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 41.Halupa A, et al. A novel role for STAT1 in regulating murine erythropoiesis: deletion of STAT1 results in overall reduction of erythroid progenitors and alters their distribution. Blood. 2005;105:552–561. doi: 10.1182/blood-2003-09-3237. [DOI] [PubMed] [Google Scholar]
- 42.Treisman R, Maniatis T. Simian virus 40 enhancer increases number of RNA polymerase II molecules on linked DNA. 1985;315:73–75. doi: 10.1038/315072a0. [DOI] [PubMed] [Google Scholar]
- 43.Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 44.Sabo PJ, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.