Abstract
Common genetic variation frequently accounts for only a modest amount of inter-individual variation in quantitative traits and complex disease susceptibility. Circulating adiponectin, an adipocytokine implicated in metabolic disease, is a model for assessing the contribution of genetic and clinical factors to quantitative trait variation. The adiponectin locus, ADIPOQ, is the primary source of genetically-mediated variation in plasma adiponectin levels. This study sought to define the genetic architecture of ADIPOQ in the comprehensively phenotyped Hispanic (n=1151) and African American (n=574) participants from the Insulin Resistance Atherosclerosis Family Study (IRASFS). Through resequencing and bioinformatic analysis, rare/low frequency (<5% MAF) and common variants (>5% MAF) in ADIPOQ were identified. Genetic variants and clinical variables were assessed for association with adiponectin levels and contribution to adiponectin variance in the Hispanic and African American cohorts. Clinical traits accounted for the greatest proportion of variance (POV) at 31% (p=1.16×10−47) and 47% (p=5.82×10−20), respectively. Rare/low frequency variants contributed more than common variants to variance in Hispanics: POV=18% (p= 6.40×10−15) and POV=5% (p=0.19), respectively. In African Americans, rare/low frequency and common variants both contributed approximately equally to variance: POV=6% (p=5.44×10−12) and POV=9% (P=1.44×10−10), respectively. Importantly, single low frequency alleles in each ethnic group were as important as, or more important than, common variants in explaining variation in adiponectin. Cumulatively, these clinical and ethnicity-specific genetic contributors explained half or more of the variance in Hispanic and African Americans and provide new insight into the sources of variation for this important adipocytokine.
Keywords: adiponectin, proportion of variation, rare variants, common variants, clinical traits
Introduction
Genome Wide Association Studies (GWAS), based on evaluation of common genetic variation, have made remarkable contributions to the understanding of the genetic basis for many complex traits and common diseases. With these successes, it is now recognized that much of the source of inter-individual variation in quantitative traits or susceptibility to disease remains to be explained, the so-called “missing heritability” [Manolio, et al. 2009]. Numerous contributors to “missing heritability” beyond common variations have been proposed: copy number variations, epigenetic mechanisms, and microRNAs, to name a few. Low frequency (LF) or rare variants, especially coding mutations, as a source of variance in traits have recently become of great interest with the advent of next generation DNA sequencing methods. In this report we assessed the relative contributions of common genetic variants (>5% minor allele frequency; MAF), LF and rare genetic variants (<5% MAF), and clinical contributors to variation in a model trait, plasma adiponectin.
Adiponectin is an adipocytokine that is secreted primarily by the adipose tissue and has been implicated in glucose homeostasis and fatty acid oxidation [Fruebis, et al. 2001; Hotta, et al. 2000]. Adiponectin is the most abundant adipocytokine found in the plasma, accounting for 0.01% of total plasma protein. Low circulating adiponectin levels are observed in a wide variety of conditions including insulin resistance [Berg, et al. 2001], type 2 diabetes (T2D) [Hotta, et al. 2000; Scherer 2006], obesity [Arita, et al. 1999; Yamamoto, et al. 2002], hypertension [Adamczak, et al. 2003; Ohashi, et al. 2006], dyslipidemia [Matsubara, et al. 2002], atherosclerosis [Okamoto, et al. 2002], and metabolic syndrome [Comuzzie, et al. 2001].
Family studies have shown plasma adiponectin levels to be highly heritable with values ranging from 40-70% [Chuang, et al. 2004; Comuzzie, et al. 2001; Lindsay, et al. 2003]. Variation in circulating adiponectin levels has been associated with multiple genetic loci including at the adiponectin coding gene ADIPOQ on chromosome 3 [Guo, et al. 2006], and chromosomes 5, 14 [Comuzzie, et al. 2001; Richards, et al. 2009], 9 [Lindsay, et al. 2003], and 16 [Ling, et al. 2009] in various populations. Importantly, efforts to date strongly suggest that the ADIPOQ gene itself is the major genetic determinant of variance in circulating adiponectin [Dastani, et al. 2012; Heid, et al. 2010]. Common variations, i.e. single nucleotide polymorphisms (SNPs), in the ADIPOQ gene have been genotyped in multiple studies. Heid et al [Heid, et al. 2010] reported that common variants in ADIPOQ cumulatively accounted for 6.7% of the variance in adiponectin levels in a study of European-derived individuals. Recently, we described a low frequency coding variant (G45R) in Hispanic Americans which alone accounts for approximately 17% of the variance in plasma adiponectin levels in the Insulin Resistance Atherosclerosis Family Study (IRASFS) Hispanic American population [Bowden, et al. 2010].
While the association of ADIPOQ polymorphisms with circulating adiponectin levels is now compellingly demonstrated [Heid, et al. 2010], the case for association of these variants with biomedical traits such as diabetes, obesity, and insulin sensitivity is less compelling. Fundamental to a clearer understanding of the influence of adiponectin on biomedical traits is a more comprehensive understanding of the contributors, i.e. genetic, clinical, and biometric, to variation in adiponectin.
There have been few studies in which both genetic and non-genetic contributions to variance of a trait have been documented in detail. In light of previously documented contributions of both genetic and clinical variables, adiponectin is an ideal trait to assess the magnitude of different influences on variation in levels of the circulating protein. The IRASFS provides a unique opportunity to study this question with its comprehensive phenotyping in a large number of Hispanic and African Americans which include detailed assessments of insulin sensitivity, adipose tissue distribution as measured by computed tomography, and serum biomarkers.
Results
The goal of this study was to measure the individual and cumulative contributions of common genetic variants, LF and rare genetic variants, and clinical measures to variation in circulating adiponectin in two different ethnicities: Hispanic and African Americans.
Clinical measures
The mean (SD) circulating adiponectin level was 13.5 (7.0) μg/ml and 9.06 (5.23) μg/ml in the Hispanic (n=1151) and African American (n=574) IRASFS samples, respectively. It was associated with a wide range of clinical traits in univariate analyses (Table 1 and Table 2). The most strongly associated clinical trait was high density lipoproteins (HDL) with a p-value of 9.29×10−77 and 2.03×10−29 in Hispanic (n=1148) and African Americans (n=574), respectively. Lipids, adiposity measures, and glucose homeostasis variables were highly correlated with plasma adiponectin levels in both cohorts. These associations with clinical traits are consistent with prior studies [Hanley, et al. 2007; Hanley, et al. 2011] documenting the negative correlation of adiponectin levels and metabolic derangement.
Table 1. Summary statistics for IRASFS Hispanic American participants.
For the descriptive demographic features gender and T2D status the total number of samples evaluated (n) with the number of samples for the trait (all samples) and corresponding percentage are listed. For quantitative demographic feature adiponectin levels and age as well as traits by trait class, the total number of samples evaluated (n), the mean trait value (mean) and standard deviation are listed. For each trait, the results of univariate association with adiponectin are listed as the beta ± standard error (β±SE) with the corresponding p-value.
| Trait Class | Trait | n | all samples | percentage | β±SE | p-value (assoc with adp) |
|---|---|---|---|---|---|---|
| Gender (n, female, %) | 1151 | 672 | 58.4 | 0.28 ± 0.025 | 2.32E-27 | |
| T2D (n, affected, percent total) | 1149 | 126 | 11 | −0.0036 ± 0.044 | 0.934 | |
| n | mean | standard deviation | ||||
| Adiponectin (ug/mL) | 1151 | 13.5 | 7 | NA | ||
| Age (years) | 1151 | 41.1 | 13.9 | 0.0039 ± 0.0010 | 0.0002 | |
| Insulin Sensitivity (SI; ×10−5 min−1 pmol/L) | 964 | 2.21 | 1.88 | 0.24 ± 0.027 | 1.60E-18 | |
| Acute Insulin Response (AIR; pmol/L) | 964 | 775 | 659 | −0.0059 ± −0.0013 | 1.34E-05 | |
| Fasting Glucose (mg/dL) | 1023 | 93.2 | 9.4 | −0.012 ± 0.0014 | 7.44E-18 | |
| Glucose Homeostasis | Fasting Insulin (uU/mL) | 1021 | 14.7 | 10.6 | −0.21 ± 0.019 | 3.09E-26 |
| Body Mass Index (kg/m2) | 1144 | 28.8 | 6.2 | −0.021 ± 0.0023 | 6.57E-19 | |
| Visceral Adipose Tissue (cm2) | 1101 | 110 | 59 | −0.048 ± 0.0048 | 2.66E-22 | |
| Subcutaneous Adipose Tissue (cm2) | 1101 | 338 | 155 | −0.0088 ± 0.0034 | 0.0092 | |
| Adiposity | Visceral Subcutaneous Ratio | 1061 | 0.36 | 0.2 | −0.23 ± 0.026 | 1.19E-17 |
| Triglycerides (mg/dL) | 1146 | 121 | 85 | −0.24 ± 0.022 | 1.38E-27 | |
| Lipids | HDL (mg/dL) | 1148 | 43.6 | 12.9 | 0.85 ± 0.042 | 9.29E-77 |
| Systolic Blood Pressure (mmHg) | 1151 | 118 | 17 | −0.19 ± 0.10 | 0.0598 | |
| Hypertension | Diastolic Blood Pressure (mmHg) | 1151 | 76.1 | 9.8 | −0.64 ± 0.11 | 1.30E-09 |
| C-reactive protein (mg/dL) | 1071 | 3.5 | 4.28 | −0.056 ± 0.012 | 2.58E-06 | |
| PAI-1 (ng/mL) | 1150 | 42.7 | 37.7 | −0.16 ± 0.015 | 5.20E-25 | |
| Inflammation | Fibrinogen (mg/dL) | 1149 | 264 | 62 | −0.023 ± 0.062 | 0.708 |
Footnote: sample sizes range from 964 to 1151 due to incomplete data for some measures. In particular, the FSIGT was not performed in those with diabetes, reducing the sample size for SI and AIR
Table 2. Summary statistics for IRASFS African American participants.
For the descriptive demographic features gender and T2D status the total number of samples evaluated (n) with the number of samples for the trait (all samples) and corresponding percentage are listed. For quantitative demographic feature adiponectin levels and age as well as traits by trait class, the total number of samples evaluated (n), the mean trait value (mean) and standard deviation are listed. For each trait, the results of univariate association with adiponectin are listed as the beta ± standard error (β±SE) with the corresponding p-value.
| Trait Class | Trait | n | all samples | percentage | β±SE | p-value (assoc with adp) |
|---|---|---|---|---|---|---|
| Gender (n, female, %) | 574 | 339 | 59.1 | 0.34 ± 0.036 | 2.23E-19 | |
| T2D (n, affected, percent total) | 574 | 62 | 10.8 | −0.013 ± 0.063 | 0.834 | |
| n | mean | standard deviation | ||||
| Adiponectin (ug/mL) | 574 | 9.06 | 5.23 | |||
| Age (years) | 572 | 42.5 | 13.9 | −0.0026 ± 0.0015 | 0.0839 | |
| Insulin Sensitivity (SI; ×10−5 min−1 pmol/L) | 498 | 1.63 | 1.17 | 0.47 ± 0.044 | 9.98E-25 | |
| Glucose Homeostasis | Acute Insulin Response (AIR; pmol/L) | 497 | 1010 | 828 | −0.0032 ± 0.0018 | 0.0739 |
| Fasting Glucose (mg/dL) | 512 | 94.6 | 9.7 | −0.017 ± 0.0020 | 3.85E-17 | |
| Fasting Insulin (uU/mL) | 512 | 14.3 | 11.3 | −0.20 ± 0.027 | 3.00E-12 | |
| Body Mass Index (kg/m2) | 570 | 30 | 6.8 | −0.018 ± 0.0029 | 2.00E-09 | |
| Adiposity | Visceral Adipose Tissue (cm2) | 522 | 93.3 | 59.5 | −0.061 ± 0.0066 | 7.16E-19 |
| Subcutaneous Adipose Tissue (cm2) | 522 | 355 | 192 | −0.011 ± 0.0040 | 4.79E-03 | |
| Visceral Subcutaneous Ratio | 504 | 0.3 | 0.19 | −0.27 ± 0.035 | 1.26E-13 | |
| Lipids | Triglycerides (mg/dL) | 574 | 81.5 | 70.5 | −0.27 ± 0.033 | 8.88E-16 |
| HDL (mg/dL) | 574 | 47.2 | 12.7 | 0.86 ± 0.072 | 2.03E-29 | |
| Hypertension | Systolic Blood Pressure (mmHg) | 574 | 119 | 18.6 | −0.086 ± 0.14 | 0.539 |
| Diastolic Blood Pressure (mmHg) | 574 | 74.9 | 10.5 | −0.53 ± 0.15 | 2.55E-04 | |
| Inflammation | C-reactive protein (mg/dL) | 563 | 3.93 | 4.93 | −0.084 ± 0.015 | 5.16E-08 |
| PAI-1 (ng/mL) | 574 | 28.4 | 27.6 | −0.16 ± 0.020 | 4.44E-15 | |
| Fibrinogen (mg/dL) | 574 | 279 | 65.1 | −0.21 ± 0.089 | 0.0207 | |
Footnote: sample sizes range from 497 to 574 due to incomplete data for some measures. In particular, the FSIGT was not performed in those with diabetes, reducing the sample size for SI and AIR
ADIPOQ variant identification in Hispanic Americans by direct sequencing
Previously we identified a G45R ADIPOQ mutation using a linkage based strategy [Bowden, et al. 2010]. The G45R allele had a large effect on adiponectin levels suggesting a detailed survey of the ADIPOQ gene in an effort to find additional variants that influence adiponectin levels. ADIPOQ was sequenced in DNAs from subjects in the bottom decile of plasma adiponectin levels in both the Hispanic (n=115) and African American (n=60) IRASFS sample following the logic that variations in the coding sequence were most likely to result in mutations that lower the amount of circulating adiponectin. The individuals in the top decile of adiponectin levels in the Hispanic American cohort (n=115) were sequenced for completeness, but no additional variants were identified. The results of this survey in the Hispanic American cohort are summarized in Supplementary Table 1. In the Hispanics, this sequencing identified a promoter variant, 3 variants in exon 2, and 1 variant in exon 3. The promoter variant (C-186T) was not previously identified. Three of the four coding variants were previously identified. Of the three previously identified coding variants, rs2241766 (G15G, MAF 0.14) and rs17366743 (Y111H, MAF 0.022), are well documented and in the dbSNP database. The other previously identified variant is a novel SNP, G133C (G45R, MAF 0.011), reported by Bowden et al in 2010 [Bowden, et al. 2010]. The coding SNP C75T (MAF 0.0087), which results in a synonymous mutation at amino acid 25, had not previously been identified in the IRASFS. In a parallel sequencing effort in African Americans (An et al submitted), five coding variants were identified, rs2241766, G113A, C163T, rs17366743 including two well documented variants also identified in the Hispanic cohort, rs2241766 and rs17366743. The remaining coding variant was a low frequency variant, G113A (G38D, MAF 0.014).
Genotyping and analysis of common and LF/rare variants
Genotype data from the SNPs were analyzed for association with plasma adiponectin levels using a variance component model implemented in SOLAR (Table 3 and Table 4) for both the Hispanic (n=1151) and African American (n=574) cohorts.
Table 3. Association of SNPs with plasma adiponectin levels in the IRASFS Hispanic American cohort.
SNPs are divided into two categories based on frequency, rare/low frequency (LF) variants and common variants. For each SNP the position (hg18), location relative to the ADIPOQ gene, and corresponding amino acid change (if applicable) is listed. The alleles for each SNP (major/minor) are listed with the corresponding minor allele frequency (MAF). Genotypic means ± standard deviation are listed by genotype where 1 and 2 are the major and minor alleles, respectively. The unadjusted association p-value is listed with the beta value indicating direction of effect.
| Amino Acid Change | Genotypic mean +/− SD |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Position | Location | Alleles1 | MAF2 | 1 / 1 | 1 / 2 | 2 / 2 | P-value3 | beta | ||
| rare/LF variants | rs17300539 | 188042154 | 5′ | G / A | 0.047 | 13.3 ± 6.9 | 16.4 ± 10.9 | 24.5 ± 11.0 | 0.0079 | 0.13 | |
| rs9877202 | 188052301 | intron 1 | A / G | 0.0066 | 13.6 ± 7.4 | 11.8 ± 4.2 | - | 0.81 | −0.038 | ||
| rs2036373 | 188052885 | intron 1 | T / G | 0.044 | 13.7 ± 7.5 | 13.2 ± 5.9 | 19.2 | 0.67 | −0.020 | ||
| G133C | 188053674 | exon 2 | G45R | G / C | 0.011 | 13.9 ± 7.3 | 2.6 ± 2.2 | - | 5.03E-40 | −1.17 | |
| rs62625753 | 188054720 | exon 3 | G90S | G / A | 0.0044 | 13.7 ± 7.4 | 10.3 ± 4.1 | - | 0.025 | −0.38 | |
| rs17366743 | 188054783 | exon 3 | Y111H | T / C | 0.016 | 13.7 ± 7.4 | 12.2 ± 4.4 | - | 0.77 | −0.029 | |
| rs6444174 | 188055883 | 3′ UTR | T / C | 0.029 | 13.7 ± 7.5 | 12.9 ± 5.9 | - | 0.97 | 0.0027 | ||
| rs9842733 | 188058176 | 3′ UTR | A / T | 0.0044 | 13.6 ± 7.4 | 10.2 ± 5.6 | - | 0.77 | 0.074 | ||
| rs1403697 | 188059387 | 3′ | A / G | 0.0045 | 13.6 ± 7.4 | 10.2 ± 5.6 | - | 0.77 | 0.074 | ||
| rs1403696 | 188062560 | 3′ | C / T | 0.024 | 13.6 ± 7.4 | 14.5 ± 6.8 | - | 0.17 | 0.096 | ||
| common variants | rs7648121 | 68183280 | 5′ | C / T | 0.060 | 13.7 ± 7.5 | 13.0 ± 6.6 | - | 0.25 | 0.048 | |
| rs6810075 | 188031259 | 5′ | T / C | 0.44 | 13.8 ± 7.3 | 13.7 ± 7.5 | 13.1 ± 7.4 | 0.74 | −0.0070 | ||
| rs10937273 | 188032389 | 5′ | G / A | 0.33 | 14.0 ± 7.6 | 13.3 ± 7.2 | 13.6 ± 7.1 | 0.062 | −0.042 | ||
| rs1648707 | 188034405 | 5′ | A / C | 0.42 | 13.8 ± 7.4 | 13.7 ± 7.5 | 13.1 ± 7.2 | 0.66 | −0.0093 | ||
| rs864265 | 188036986 | 5′ | G / T | 0.19 | 13.6 ± 7.8 | 13.4 ± 6.6 | 15.3 ± 6.7 | 0.25 | 0.031 | ||
| rs822387 | 188038731 | 5′ | T / C | 0.060 | 13.4 ± 6.9 | 15.8 ± 10.7 | 24.5 ± 11.0 | 0.021 | 0.11 | ||
| rs16861194 | 188042119 | 5′ | A / G | 0.14 | 13.6 ± 6.8 | 13.9 ± 9.2 | 11.6 ± 4.0 | 0.67 | −0.012 | ||
| rs266729 | 188042168 | 5′ | C / G | 0.28 | 13.7 ± 8.1 | 13.5 ± 6.8 | 14.0 ± 6.5 | 0.72 | 0.0082 | ||
| rs710445 | 188044212 | intron 1 | A / G | 0.44 | 13.5 ± 7.4 | 13.6 ± 7.4 | 13.9 ± 7.6 | 0.67 | 0.0090 | ||
| rs16861205 | 188044328 | intron 1 | G / A | 0.13 | 13.4 ± 6.7 | 14.4 ± 9.7 | 13.0 ± 3.9 | 0.64 | 0.014 | ||
| rs16861209 | 188045808 | intron 1 | C / A | 0.064 | 13.3 ± 6.9 | 16.1± 10.6 | 18.3 ± 13.2 | 0.068 | 0.083 | ||
| rs822391 | 188046497 | intron 1 | T / C | 0.19 | 13.6 ± 7.8 | 13.5 ± 6.7 | 15.6 ± 7.1 | 0.0072 | 0.070 | ||
| rs16861210 | 188049192 | intron 1 | G / A | 0.942 | 13.4 ± 7.0 | 15.3 ± 10.1 | 15.0 ± 12.6 | 0.32 | 0.01344 | ||
| rs822394 | 188049422 | intron 1 | C / A | 0.19 | 13.5 ± 7.8 | 13.4 ± 5.6 | 15.6 ± 7.1 | 0.013 | 0.064 | ||
| rs822396 | 188049571 | intron 1 | A / G | 0.20 | 13.7 ± 7.9 | 13.4 ± 6.6 | 15.5 ± 7.0 | 0.025 | 0.058 | ||
| rs12495941 | 188050874 | intron 1 | G / T | 0.32 | 14.0 ± 7.8 | 13.4 ± 7.0 | 13.1 ± 6.9 | 0.090 | −0.039 | ||
| rs7649121 | 188051479 | intron 1 | A / T | 0.76 | 13.7 ± 7.8 | 13.6 ± 7.0 | 12.6 ± 6.4 | 0.31 | 0.00123 | ||
| rs17366568 | 188053147 | intron 1 | G / A | 0.066 | 13.7 ± 7.5 | 12.9 ± 6.2 | 3.2 | 0.24 | −0.0562 | ||
| rs2241766 | 188053586 | exon 2 | G15G | T / G | 0.18 | 13.6 ± 7.4 | 13.6 ± 7.4 | 14.1 ± 8.2 | 0.32 | −0.0257 | |
| rs2241767 | 188053890 | intron 2 | A / G | 0.18 | 13.6 ± 7.4 | 13.6 ± 7.4 | 14.1 ± 8.2 | 0.30 | −0.027 | ||
| rs3774261 | 188054253 | intron 2 | G / A | 0.43 | 13.5 ± 6.8 | 13.5 ± 7.8 | 14.3 ± 7.4 | 0.85 | −0.0039 | ||
| rs3821799 | 188054430 | intron 2 | C / T | 0.51 | 13.6 ± 7.1 | 13.5 ± 7.6 | 13.7 ± 7.5 | 0.30 | 0.0086 | ||
| rs3774262 | 188054508 | intron 2 | G / A | 0.18 | 13.6 ± 7.4 | 13.6 ± 7.4 | 14.4 ± 8.1 | 0.48 | −0.0185 | ||
| rs6773957 | 188056399 | 3′ UTR | G / A | 0.43 | 13.5 ± 6.8 | 13.5 ± 7.7 | 14.3 ± 7.5 | 0.90 | −0.0026 | ||
| rs2082940 | 188056858 | 3′ UTR | C / T | 0.19 | 13.6 ± 7.4 | 13.5 ± 7.4 | 13.95 ± 8.13 | 0.35 | −0.024 | ||
| rs7639352 | 188061168 | 3′ | C / T | 0.24 | 13.2 ± 7.0 | 14.3 ± 8.2 | 13.3 ± 5.8 | 0.44 | 0.019 | ||
| rs7641507 | 188061267 | 3′ | C / T | 0.094 | 13.6 ± 7.4 | 12.3 ± 2.9 | - | 0.53 | 0.16 | ||
| rs6444175 | 188062438 | 3′ | G / A | 0.25 | 13.3 ± 7.0 | 14.2 ± 8.2 | 13.3 ± 5.9 | 0.54 | 0.015 | ||
| rs13085499 | 188063534 | 3′ | G / A | 0.49 | 14.2 ± 7.5 | 13.5 ± 7.6 | 13.4 ± 7.0 | 0.78 | 0.0057 | ||
| rs7628649 | 188068075 | 3′ | C / T | 0.21 | 13.6 ± 7.4 | 13.6 ± 7.3 | 14.3 ± 8.3 | 0.36 | −0.023 | ||
| rs17373414 | 188068221 | 3′ | C / T | 0.051 | 13.5 ± 6.9 | 15.5 ± 13.1 | 18.4 ± 10.5 | 0.052 | 0.11 | ||
Alleles listed as major allele / minor allele
Assuming the Bonferroni correction for 41 SNPs we have power of 0.63, 0.87, 0.96, and 0.99 in Hispanic Americans for variants explaining 0.02, 0.03, 0.04 and 0.05 of the variation in adiponectin.
A strict Bonferroni correction for 41 tests (SNPs) would result in a significant threshold P<0.0012.
Table 4. Association of SNPs with plasma adiponectin levels in the IRASFS African American cohort.
SNPs are divided into two categories based on frequency, rare/low frequency (LF) variants and common variants. For each SNP the position (hg18), location relative to the ADIPOQ gene, and corresponding amino acid change (if applicable) is listed. The alleles for each SNP (major/minor) are listed with the corresponding minor allele frequency (MAF). Genotypic means ± standard deviation are listed by genotype where 1 and 2 are the major and minor alleles, respectively. The unadjusted association p-value is listed with the beta value indicating direction of effect.
| Amino Acid Change | Genotypic mean +/− SD |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Position | Location | Alleles1 | MAF2 | 1 / 1 | 1 / 2 | 2 / 2 | P-value3 | beta | ||
| rare/LF variants | rs17300539 | 188042154 | 5′ | G / A | 0.028 | 8.98 ± 5.25 | 11.16 ± 5.21 | - | 0.0018 | 0.32 | |
| rs822391 | 188046497 | intron 1 | T / C | 0.031 | 8.97 ± 5.11 | 9.49 ± 4.34 | 8.13 ± 2.94 | 0.67 | −0.031 | ||
| rs822394 | 188049422 | intron 1 | C / A | 0.029 | 8.93 ± 5.06 | 9.92 ± 4.62 | 5.87 ± 3.14 | 0.88 | −0.010 | ||
| rs17366568 | 188053147 | intron 1 | G / A | 0.037 | 9.11 ± 5.34 | 8.36 ± 3.77 | 13.13 | 0.56 | 0.052 | ||
| rs2241766 | 188053586 | exon 2 | G15G | T / G | 0.031 | 8.97 ± 5.04 | 9.29 ± 5.01 | - | 0.62 | 0.043 | |
| C113T | 188053654 | exon 2 | G38D | G / A | 0.019 | 9.04 ± 5.28 | 9.81 ± 4.33 | - | 0.76 | −0.041 | |
| C163T | 188053704 | exon 2 | R55C | C / T | 0.010 | 9.06 ± 5.01 | 1.20 ± 0.37 | - | 0.00030 | −1.00 | |
| rs2241767 | 188053890 | intron 2 | A / G | 0.028 | 9.06 ± 5.27 | 9.52 ± 5.26 | - | 0.31 | 0.087 | ||
| rs3774262 | 188054508 | intron 2 | G / A | 0.028 | 9.05 ± 5.26 | 9.52 ± 5.26 | - | 0.30 | 0.088 | ||
| rs17366743 | 188054783 | exon 3 | Y111H | T / C | 0.010 | 9.02 ± 5.04 | 7.28 ± 4.04 | - | 0.35 | −0.20 | |
| T491C | 188054943 | exon 3 | I164T | T / C | 0 | 9.07 ± 5.27 | 10.45 | - | 1.00 | 0.00 | |
| rs1865762 | 188062094 | 3′ | A / G | 0.010 | 8.95 ± 5.00 | 11.50 ± 5.69 | - | 0.17 | 0.19 | ||
| rs17373414 | 188068221 | 3′ | C / T | 0.020 | 8.96 ± 4.99 | 11.05 ± 6.10 | - | 0.47 | 0.10 | ||
| common variants | rs7648121 | 68183280 | 5′ | C / T | 0.061 | 8.63 ± 4.85 | 10.68 ± 5.53 | - | 0.0052 | 0.15 | |
| rs6810075 | 188031259 | 5′ | T / C | 0.43 | 8.78 ± 4.83 | 9.19 ± 5.36 | 9.72 ± 62021 | 0.72 | 0.011 | ||
| rs10937273 | 188032389 | 5′ | G / A | 0.13 | 8.66 ± 4.94 | 10.07 ± 5.19 | 6.74 ± 3.54 | 0.15 | 0.063 | ||
| rs1648707 | 188034405 | 5′ | A / C | 0.39 | 9.25 ± 4.73 | 9.19 ± 5.09 | 8.48 ± 5.10 | 0.017 | −0.072 | ||
| rs864265 | 188036986 | 5′ | G /T | 0.065 | 9.09 ± 5.39 | 9.00 ± 4.91 | 10.31 ± 2.02 | 0.89 | −0.0060 | ||
| rs822387 | 188038731 | 5′ | T / C | 0.33 | 9.43 ± 5.10 | 8.88 ± 4.96 | 7.68 ± 4.95 | 0.23 | −0.042 | ||
| rs16861194 | 188042119 | 5′ | A / G | 0.32 | 8.98 ± 5.13 | 8.93 ± 4.93 | 9.51 ± 4.83 | 0.79 | 0.010 | ||
| rs266729 | 188042168 | 5′ | C / G | 0.11 | 8.92 ± 4.83 | 9.38 ± 5.73 | 14.01 | 0.44 | −0.040 | ||
| rs710445 | 188044212 | intron 1 | A / G | 0.44 | 8.63 ± 4.74 | 9.30 ± 5.44 | 9.11 ± 4.50 | 0.86 | 0.0054 | ||
| rs16861205 | 188044328 | intron 1 | G / A | 0.26 | 8.86 ± 5.16 | 9.36 ± 4.85 | 8.88 ± 5.04 | 0.76 | 0.011 | ||
| rs16861209 | 188045808 | intron 1 | C / A | 0.14 | 8.90 ± 5.20 | 9.26 ± 4.51 | 9.16 ± 5.61 | 0.034 | 0.086 | ||
| rs822396 | 188049571 | intron 1 | A / G | 0.19 | 9.34 ± 5.18 | 8.75 ± 5.44 | 7.44 ± 5.00 | 0.060 | −0.071 | ||
| rs12495941 | 188050874 | intron 1 | G / T | 0.39 | 7.51 ± 4.43 | 9.57 ± 5.61 | 9.08 ± 4.94 | 0.77 | −0.0091 | ||
| rs9877202 | 188052301 | intron 1 | A / G | 0.13 | 9.26 ± 5.19 | 8.06 ± 4.31 | 6.94 ± 3.94 | 0.21 | −0.059 | ||
| rs2036373 | 188052885 | intron 1 | T / G | 0.083 | 8.92 ± 5.07 | 10.43 ± 6.46 | 6.39 ± 4.09 | 0.88 | −0.0092 | ||
| rs3774261 | 188054253 | intron 2 | G / A | 0.40 | 8.44 ± 4.68 | 9.22 ± 5.56 | 9.20 ± 5.10 | 0.16 | 0.041 | ||
| rs6444174 | 188055883 | 3′ UTR | T / C | 0.21 | 9.15 ± 5.12 | 8.73 ± 4.88 | 8.57 ± 4.30 | 0.16 | −0.056 | ||
| rs6773957 | 188056399 | 3′ UTR | G / A | 0.40 | 8.44 ± 4.68 | 9.22 ± 5.56 | 9.20 ± 5.10 | 0.16 | 0.041 | ||
| rs2082940 | 188056858 | 3′ UTR | C / T | 0.23 | 9.31 ± 5.47 | 8.52 ± 4.79 | 9.39 ± 4.91 | 0.20 | −0.047 | ||
| rs9842733 | 188058176 | 3′ UTR | A / T | 0.13 | 9.11 ± 5.03 | 8.73 ± 5.13 | 6.11 ± 2.65 | 0.78 | 0.015 | ||
| rs1403697 | 188059387 | 3′ | A / G | 0.18 | 9.04 ± 5.07 | 8.81 ± 4.82 | 9.77 ± 5.97 | 0.90 | 0.0055 | ||
| rs7639352 | 188061168 | 3′ | C / T | 0.33 | 8.62 ± 4.67 | 9.18 ± 5.76 | 10.20 ± 4.94 | 0.026 | 0.072 | ||
| rs7641507 | 188061267 | 3′ | C / T | 0.094 | 9.03 ± 5.09 | 9.11 ± 4.77 | 7.89 ± 0.43 | 0.61 | −0.030 | ||
| rs6444175 | 188062438 | 3′ | G / A | 0.33 | 8.74 ± 4.64 | 8.97 ± 5.33 | 10.21 ± 4.98 | 0.024 | 0.076 | ||
| rs1403696 | 188062560 | 3′ | C / T | 0.26 | 9.19 ± 5.48 | 8.71 ± 4.67 | 9.58 ± 5.52 | 0.39 | −0.032 | ||
| rs13085499 | 188063534 | 3′ | G / A | 0.34 | 9.41 ± 5.10 | 9.18 ± 5.56 | 8.06 ± 4.64 | 0.072 | −0.054 | ||
| rs7628649 | 188068075 | 3′ | C / T | 0.33 | 9.17 ± 5.64 | 8.87 ± 4.70 | 9.60 ± 5.46 | 0.48 | −0.023 | ||
Alleles listed as major allele / minor allele
Assuming the Bonferroni correction for 40 SNPs, we have power of 0.29, 0.53, 0.72 and 0.86 in African Americans for variants explaining 0.02, 0.03, 0.04 and 0.05 of the variation in adiponectin.
A strict Bonferroni correction for 40 tests (SNPs) would result in a significant threshold P<0.0013.
In the Hispanic American cohort (Table 3), there was nominal evidence of association (p<0.05) with plasma adiponectin levels in four common SNPs including rs822387, rs822391, rs822394, and rs822396 with p-values ranging from 0.0072 to 0.025 and the beta values ranging from 0.058 to 0.11. There were a total of 10 polymorphic LF/rare variants (MAF<5%) genotyped in the sample. The frequency of these variants ranged from 0.0044 to 0.047 in Hispanic Americans. The majority of the LF/rare variants selected for genotyping from the literature were not found in this Hispanic sample. The most strongly associated LF variant was the previously reported G45R variant [Bowden, et al. 2010] with a p-value of 5.03 × 10−40 for association with plasma adiponectin. Two additional rare variants, rs17300539 (–363 bp 5’ of the ADIPOQ promoter) and rs62625753 (a G90S coding mutation) showed association with plasma adiponectin levels with p-values of 0.0079 and 0.025, respectively. Individuals carrying the minor allele (A) of rs17300539 had higher levels of adiponectin in a dose dependent manner, with mean (SD) adiponectin levels of 13.3 (6.9) μg/mL, 16.4 (10.9) μg/mL, and 24.5 (11.0) μg/mL for 0, 1, 2 copies of the minor allele, respectively. In contrast, individuals with the minor allele (A) of rs62625753 had lower adiponectin levels with mean (SD) adiponectin levels of 13.7 (7.4) μg/mL and 10.3 (4.1) μg/mL for individuals with 0 and 1 copy of the minor allele, respectively.
Of the common SNPs found to be associated with adiponectin levels in Hispanic Americans, rs822391 (located in intron 1) had the strongest evidence of association with a p-value of 0.0072. This SNP showed some differences in mean adiponectin between individuals with 2 copies of the minor allele (15.6 ± 7.1 μg/mL) and those with no copies of the minor allele (13.6 ± 7.8 μg/mL), however these differences were not as dramatic as those seen with the LF variants. The only other common variant with similar differences in mean plasma adiponectin levels was rs822387 (–3786 bp 5’ of the ADIPOQ promoter), in which the minor allele led to higher levels of adiponectin in a dose-dependent manner. Average (SD) adiponectin levels were 13.4(6.9) μg/mL, 15.8 (10.7) μg/mL, and 24.5 (11.0) μg/mL for individuals with 0, 1, 2 copies of the minor allele, respectively.
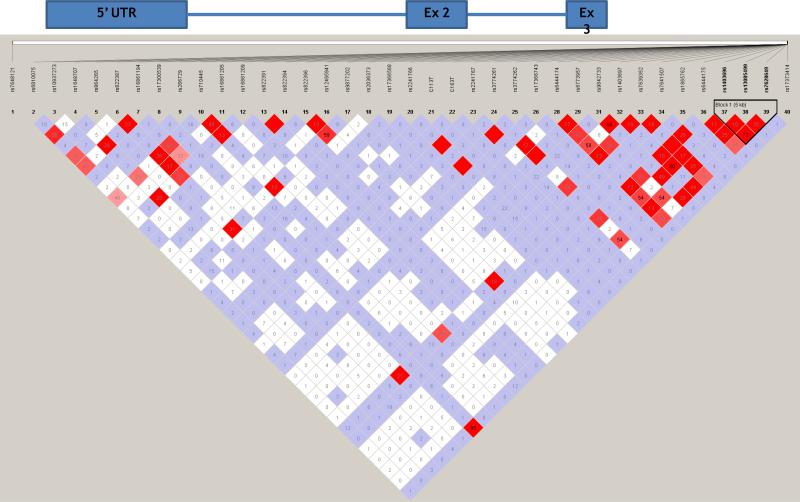
Figure 1 shows a gene map of the ADIPOQ locus with a linkage disequilibrium (LD) plot of all genotyped SNPs showing five major LD blocks within the Hispanic American sample. There were three SNPs (rs822387, rs7649121, and rs3821799) associated with adiponectin that did not fall within any of the LD blocks. The remaining seven associated variants were found within the LD blocks, with four of the variants (rs822391, rs16861210, rs822394, and rs822396) within block 3.
Figure 1.
Gene map of ADIPOQ matched to the haploview-generated LD matrix for ADIPOQ +/−50kb in the Hispanic IRASFS sample. Figures generated with Haploview 4.0. Regions of high LD (D’ = 1 and logarithm of odds [LOD]>2) are shown in the darkest shade. Markers with lower LD (0.45<D’<1 and LOD>2) are shown in dark through light shades, with the color intensity decreasing with decreasing D’ value. Regions of low LD and low LOD scores (LOD<2) are shown in white. The number within each box indicates the r2 statistic value between the corresponding two SNPs. Black triangles represent LD blocks based on the confidence intervals [Gabriel, et al. 2002].
In a similar analysis of the IRASFS African Americans (Table 4), seven SNPs were associated with adiponectin (P<0.05), two of which were low frequency. Of all the SNPs associated with plasma adiponectin levels, the low frequency SNPs, rs17300539 and the novel SNP, R55C, were most strongly associated, with p-values of 0.0018 and 0.00030, respectively. rs17300539 is a low frequency variant (MAF 0.028) found −363 bp 5’ of the ADIPOQ promoter. Individuals with the minor allele (A) for rs17300539 had higher plasma adiponectin levels with mean (SD) adiponectin levels of 8.98 (5.25) μg/mL and 11.16 (5.21) μg/mL with 0 and 1 copy of the minor allele, respectively. The most strongly associated variant was the R55C coding variant (p=0.00030), initially identified through sequencing analysis in the IRASFS. It is located in exon 2 of the ADIPOQ gene in the collagen-like domain of the adiponectin protein. Individuals with the minor allele (T) had substantially lower levels of adiponectin with mean (SD) adiponectin levels of 9.06 (5.01) μg/mL and 1.20 (0.37) μg/mL with 0 and 1 copy of the minor allele, respectively. For both these low frequency variants, there were no individuals homozygous for the minor allele.
Of the common variants associated with plasma adiponectin levels in the African American cohort, rs7648121 was the most strongly associated variant with a p-value of 0.0052. This SNP is located in the intron of FAM19A1, a gene upstream of ADIPOQ. The differences in the genotypic means for the variant were not as dramatic in individuals with 0 (8.63 ± 4.85 μg/mL) and 1 (10.68 ± 5.53 μg/mL) copy of the minor allele (T), compared to the genotypic means seen in LF variants.
Figure 2 shows a gene map of the ADIPOQ gene with a LD plot of all the genotyped SNPs within the African American cohort. There is only one LD block within the ADIPOQ gene. None of the seven associated variants were located within the LD block.
Figure 2.
Gene map of ADIPOQ matched to the haploview-generated LD matrix for ADIPOQ +/−50kb in the African American IRASFS sample. Figures generated with Haploview 4.0. Regions of high LD (D’ = 1 and logarithm of odds [LOD]>2) are shown in the darkest shade. Markers with lower LD (0.45<D’<1 and LOD>2) are shown in dark through light shades, with the color intensity decreasing with decreasing D’ value. Regions of low LD and low LOD scores (LOD<2) are shown in white. The number within each box indicates the r2 statistic value between the corresponding two SNPs. Black triangles represent LD blocks based on the confidence intervals [Gabriel, et al. 2002].
Proportion of variance explained by clinical factors, LF/rare variants, and common variants
The proportion of the variance (POV) in plasma adiponectin levels was determined using a variance component analysis implemented in SOLAR for each trait category (clinical traits, LF / rare SNPs, and common SNPs) in both the Hispanic and African American cohorts (Table 5). In the Hispanic American cohort, clinical traits explained 31% of the POV (n=820), the ten LF/rare variants included in the analysis explained 18% of the POV (n=1104), and finally, the remaining 31 common variants explained 5% of the POV (n=890). When the clinical traits and genetic variants were combined, they explained 49% of the POV in the IRASFS Hispanic American sample (n=628). The POV estimates were highly consistent and the p-values for association of the clinical traits with adiponectin level are highly significant (p-value of 1.16×10−47). The second most significant contributor to adiponectin variation was LF/rare SNPs, with a p-value of 6.40×10−15. Common SNPs did not have a statistically significant association with adiponectin levels with a p-value of 0.19. Clinical traits had the strongest association, even conditional on both common and rare SNPs with a p-value of 1.21×10−54 (data not shown). Rare / LF variants were also significantly associated conditional on common variants and clinical traits with a p-value of 5.53×10−19 and common SNPs are not statistically significant with adiponectin levels conditional on rare SNPs and clinical traits, with a p-value of 0.301 (data not shown).
Table 5. Proportion of variance explained by clinical characteristics, common variants, and LF/rare variants in IRASFS Hispanic and African American cohorts.
For each population, the contributors to the variance delineated with the number of observations, proportion of variance explained for the full cohort or limited to individuals with complete data for all contributors and the corresponding p-value.
| Contributors to variance | N | Proportion of Variation Explained | N | Proportion of Variation Explained | p-value | |
|---|---|---|---|---|---|---|
| Hispanics | None | 1151 | -- | 628 | -- | -- |
| Clinical Characteristics | 820 | 0.31 | 628 | 0.32 | 1.16E-47 | |
| Rare SNPs | 1104 | 0.18 | 628 | 0.16 | 6.40E-15 | |
| Common SNPs | 890 | 0.050 | 628 | 0.069 | 0.194 | |
| All SNPs | 880 | 0.23 | 628 | 0.21 | 1.16E-09 | |
| Clinical + Rare SNPs | 785 | 0.44 | 628 | 0.44 | 5.39E-66 | |
| Clinical + Common SNPs | 637 | 0.38 | 628 | 0.38 | 3.13E-40 | |
| Clinical + All SNPs | 628 | 0.49 | 628 | 0.49 | 1.33E-56 | |
| African Americans | None | 574 | -- | 360 | -- | -- |
| Clinical Characteristics | 437 | 0.44 | 360 | 0.47 | 5.82E-20 | |
| Rare SNPs | 494 | 0.057 | 360 | 0.063 | 5.44E-12 | |
| Common SNPs | 479 | 0.085 | 360 | 0.081 | 1.44E-10 | |
| All SNPs | 477 | 0.14 | 360 | 0.15 | 4.68E-09 | |
| Clinical + Rare SNPs | 374 | 0.50 | 360 | 0.51 | 7.77E-18 | |
| Clinical + Common SNPs | 362 | 0.54 | 360 | 0.54 | 6.87E-16 | |
| Clinical + All SNPs | 360 | 0.59 | 360 | 0.59 | 4.48E-13 | |
The proportion of variance analysis was also performed in the African American cohort of the IRASFS for comparison (Table 5). It is noteworthy that 11 SNPs that were classified as LF/rare (MAF<5%) in the Hispanic sample were classified as common variants (MAF>5%) in the African American sample. Clinical traits explained 44% (n=437) of the variation with LF/rare and common variants explained 5.7% (n=494) and 8.5% (n=479), respectively. Thus, in African Americans, the LF/rare variants and the common variants explained comparable proportions of the variance. Cumulatively, the clinical traits and genetic variants accounted for 59% of the variance in adiponectin levels (n=360). The most significant contributor to adiponectin variation was clinical traits with a p-value of 5.82×10−20. The second most significant contributor to adiponectin variation is LF/rare variants with a p-value of 5.44×10−12, with common variants contributing similarly to the POV (1.44×10−10).
Discussion
In this study we assessed the relative contributions of common, LF and rare genetic variants, and clinical factors to the variance of a model quantitative trait, plasma adiponectin in Hispanic and African Americans. Plasma adiponectin as a model trait has several advantages. The quantitative assay measures mass rather than activity, which could vary with protein variants. Although common variation in other genes, including LYZL1, CDH13 [Ling, et al. 2009] and ARL15 [Richards, et al. 2009] have been shown to be associated with adiponectin levels, prior studies have documented the ADIPOQ locus itself is the primary GWAS signal for plasma adiponectin levels [Dastani, et al. 2012; Heid, et al. 2010] and thus likely the primary source of variation. The IRASFS is an ideal sample in which to assess contributions to variance due to the extensive and detailed phenotyping of participants with an emphasis on metabolic measures. In addition, subjects in the two ethnic groups were ascertained and examined using a common protocol. As a result, this study design enabled us to assess the contributions of both clinical and genetic phenotypes for their effects on adiponectin variance in a uniform analysis.
We found that clinical traits and LF/rare variants contributed the largest POV in plasma adiponectin levels in the Hispanic American cohort. Clinical traits accounted for the largest component of variance in adiponectin levels at 31% with the previously identified LF variant, G45R, having the largest genetic contribution (16.7%) [Bowden, et al. 2010]. There was only one other LF/rare variant (rs62625753), found to be associated with lower plasma adiponectin levels in a Caucasian population (p=0.016) [Vasseur, et al. 2002], that was associated with adiponectin (p=0.025). It is noteworthy that (1) other LF/rare SNPs added little additional influence on adiponectin variance beyond the G45R, and (2) common variants contributed only modestly to the variance. Thus, if G45R was absent from the Hispanic American population, the contributions to variance by LF/rare variants would be dramatically changed. The G45R mutation has only been observed in Hispanic Americans [Bowden, et al. 2010].
In contrast and in the absence of a strong variant like the G45R, LF/rare variants explained a modest but significant amount of the variation of adiponectin in African Americans (6.3%). When comparing the two groups, it is interesting to note that there were only two SNPs that remained LF/rare variants in both cohorts when classified based on MAF<5%. Eight variants, classified as LF/rare in Hispanic Americans, were reclassified as common variants in the African American cohort. Similar to Hispanic Americans, a single rare variant, the R55C, made the largest single contribution to the variance. Common variants were able to explain a nominally greater percentage of the variation (8.1%). However, LF/rare variants were more strongly associated than common variation with plasma adiponectin levels with p-values of 5.44 × 10−12 and 1.44 × 10−10, respectively, in spite of the smaller amount of variation explained by rare variants. This highlights the importance of LF/rare SNPs in adiponectin variation.
Cumulatively, the clinical traits and genetic variants included in this analysis accounted for a substantial POV (Hispanic American: 49%; African American: 59%) in adiponectin levels. While these estimates are unlikely to be precise measures, this suggests that approximately half of the variance is still unexplained. We have estimated the heritability of adiponectin in the IRASFS sample to be 71% and 64% in Hispanic and African Americans, respectively [Guo, et al. 2006]. We estimate that the common, LF, and rare variants tested in this study account for approximately 50–60% of the heritability leaving 40–50% of unexplained residual heritability. This means that there are most likely other genetic and non-genetic factors yet to be identified. The analysis of of the clinical and genetic components shows that clinical traits are the most significant contributors to adiponectin variation (p=1.16×10−47 and 5.82×10−20 in Hispanic and African Americans, respectively). Additional non-genetic contributors to adiponectin variation that were not included in these analyses could be traits such as vitamin D levels [Vaidya, et al. 2012] and dairy intake [Stancliffe, et al. 2011]. In addition, our results show that genetic variation is also an important contributor to adiponectin variation (p=1.16×10−9 and 4.68×10−9 in Hispanic and African Americans, respectively). Rare/LF variants were the most strongly associated genetic variants (p=6.40×10−15 and 5.44×10−12 in the Hispanic and African American cohorts, respectively). Common SNPs explained similar variation in African Americans (p=1.44×10−10), and more interestingly, did not contribute significantly in Hispanic Americans (p=0.19).
LF/rare variants explained the greatest proportion of genetic risk (16%) in the Hispanic Americans and were more strongly associated with adiponectin variation (p=6.40×10−15). In comparison, LF/rare variants explained less of the variation than common SNPs in the African Americans, but were more strongly associated with adiponectin variation than common variants with p=5.44×10−12 vs. 1.44×10−10. It is interesting to note that we have also resequenced ADIPOQ in a large number of European Americans and found no mutations with the G45R/R55C-like phenotype, i.e. mutations resulting in a significant reduction in circulating adiponectin levels. Thus, we predict that LF/rare coding variants in ADIPOQ will have a modest contribution to variance in European-derived populations. Thus, there is no consistent pattern between ethnicities and the impact of LF/rare variants.
The source(s) of the remaining contributors to adiponectin variation remain to be identified. Although there are other genes, e.g. ARL15 [Richards, et al. 2009], CDH13, LYZL1 [Ling, et al. 2009], FER [Qi, et al. 2011], and chromosomal intervals, e.g. 16q23.2, 19q13.11, 12q24.31, 8q24.13, 6p21.1 and 1q41 [Dastani, et al. 2012], identified from GWAS that have been associated with adiponectin levels and may contribute additional common variants as a source of variance. Common variants in the ADIPOQ locus contribute only a small proportion of variance and extending this analysis to common variations in other, weaker contributors seems unlikely to capture additional sources of variation. There is a counter to this argument. Yang et al [Yang, et al. 2010] recently proposed that common variants account for a greater proportion of variance than explained by variants identified solely by highly stringent statistical criteria. Another possible source of variation, epigenetic influences, seems unlikely to have a large impact on adiponectin levels as there are no CpG islands within the ADIPOQ locus with the closest island approximately 15 kb upstream. Copy number variants have not been identified in the ADIPOQ locus, and thus not a likely source of variation. Studies have shown that microRNAs, such as miR-369-5p and miR-37I, down-regulate and up-regulate ADIPOQ expression respectively [Bork, et al. 2010] and are thus a plausible source of variance. Finally LF/rare coding variants in other genes that have the potential to modulate plasma adiponectin levels, such as ADIPOQR1, ADIPOR2, CDH13, ARL15, LYZL1, and FER. While a single LF/rare variant in the Hispanic American cohort explained a significant proportion of the variance, there could be multiple LF/rare SNP in and/or outside of ADIPOQ that could be important in African Americans. Resources from exome sequencing in Hispanic Americans are not yet available to systematically test this possibility and there are limited resources for African American exomes. These are possibilities given we have an example in hand, i.e. the high impact G45R and R55C variants. It is widely recognized that such coding variants are frequently found in genes in which common variants have been shown to influence a trait [Johansen, et al. 2010]. Another consideration for future studies is the use of high molecular weight (HMW) adiponectin, which may have some effect on the results. The G45R mutation, for example, results in low levels of total adiponectin, but even lower levels of HMW adiponectin [Bowden, et al. 2010].
Most prior studies have looked solely at common SNPs for their contribution to adiponectin variation [Menzaghi, et al. 2004] or LF SNPs for their functional impact [Morandi, et al. 2010]. These studies have primarily been conducted in European-derived or Asian populations. Among these studies, the amount of adiponectin variation explained has not been comparable with Heid et al. explaining 6.7% in a sample of 4,659 Europeans [Heid, et al. 2010] and Warren et al explaining 6% in a sample of 14,002 individuals (including Europeans, African Americans, and Indian Asians) [Warren, et al. 2012]. This study is novel in that it was conducted in well-phenotyped samples of large Hispanic and African American families in which we have examined a variety of contributions to adiponectin variation.
While not the focus of this study, the implications of the observations summarized here are provocative in relation to understanding the role of adiponectin variation in human health. We have estimated that half or more of variation in adiponectin is explained with clinical relationships being more important than genetic influences. The relatively minor influences of common variations make it unlikely that they directly influence clinically relevant phenotypes. It is striking that Hispanic Americans with the G45R mutation were clinically little different from subjects without the G45R variant [Bowden, et al. 2010]. We have made similar observations with R55C in the African American population (An et al., submitted). What is striking is that both variants reduce adiponectin levels >80% without obvious metabolic impact. Given the well documented action of adiponectin in cell and animal models, this suggests that individuals with these mutations somehow compensate for the loss of adiponectin-mediated metabolic effects.
In conclusion, we have assessed the contributions to variation of a model trait, plasma adiponectin, in a comprehensively phenotyped bi-ethnic sample. Simple genetic variations in the gene itself, either common or low frequency and rare variation, in combination with clinical measures account for a substantial proportion of the variation in adiponectin. These results highlight the importance of rare variants in adiponectin variation. With this observation though, it is clear that other sources of variation remain to be identified.
Research Design and Methods
Insulin Resistance Atherosclerosis Family Study (IRASFS)
Characteristics of the study sample are summarized in Tables 1 and 2. The study design, recruitment, and phenotyping for IRASFS have been described in detail [Henkin, et al. 2003]. Briefly, the IRASFS was designed to identify the genetic and environmental basis of insulin resistance and adiposity. Subjects included in this report were recruited from clinical centers in San Luis Valley, Colorado (a rural Hispanic population), San Antonio, Texas (an urban Hispanic population) (Table 1), and Los Angeles, California (an urban African American population) (Table 2). Family members were recruited to obtain an average of 12-13 family members. The exam included a fasting blood draw and medical history interview. The clinical examination included an insulin-modified frequently sampled intravenous glucose tolerance test (FSIGT) using the reduced sampling protocol [Steil, et al. 1993]. Glucose homeostasis parameters were computed with the MINMOD analysis program [Bergman, et al. 1985]. Height, weight, and waist and hip circumferences were measured and computed tomography (CT) was used to estimate visceral and subcutaneous fat areas and liver density [Wagenknecht, et al. 2009]. Glucose, insulin, triglycerides, HDL, CRP, PAI-1, and fibrinogen levels were assayed using standard laboratory methods.
Laboratory Methods
Biomarkers
Total plasma adiponectin levels were measured by radioimmunoassay (RIA; Linco Research, St. Charles, MO). This RIA uses a polyclonal anti-adiponectin antibody which recognizes trimers and higher multimers of adiponectin and includes recognition of the globular domain. In addition, a subset of 200 samples was measured with a monoclonal antibody-based ELISA, with good correlation with the RIA [Bowden, et al. 2010].
DNA Isolation
Genomic DNA was purified using PUREGENE DNA isolation kits (Gentra Inc., Minneapolis, MN, USA). Total genomic DNA was quantified using a fluorometric assay by Hoefer DyNA Quant 200 fluorometer (Hoefer Pharmacia Biotech Inc., San Francisco, CA, USA).
Sequencing and Genotyping
Targeted promoter and exonic sequences of ADIPOQ were PCR amplified and directly sequenced using Big Dye Ready Reaction Mix on an ABI3730xl sequencer (Applied Biosystems, Foster City, CA). Sequence data was visualized using Sequencher Software version 4.9 (GeneCodes Corporation, Ann Arbor, MI). The coding regions of ADIPOQ were sequenced, as well as the promoter region as defined by Kita et al. [Kita, et al. 2005]. SNPs identified in prior association studies [Heid, et al. 2010; Hivert, et al. 2008; Vasseur, et al. 2002], functional studies [Waki, et al. 2003], and LF and rare variants identified from DNA sequencing described above and studies in diverse populations as summarized in Waki et al [Waki, et al. 2003] were also identified for genotyping. In addition, common SNPs across the ADIPOQ gene and the surrounding +/−50kb region were selected using HapMap YRI and CEU populations (www.hapmap.org). Thirty-eight tag-SNPs were selected with an r2 threshold of 0.8 and MAF>5%. A total set of 41 SNPs was genotyped in the IRASFS Hispanic American sample (n=1240). SNP genotyping was performed on a Sequenom MassARRAY Genotyping System (Sequenom, San Diego, CA) using methods previously described [Palmer, et al.]. The genotyping efficiency was >97% and 48 blind duplicate samples included to evaluate genotyping accuracy were 100% concordant. These data were combined with data from six SNPs previously genotyped in IRASFS [Bowden, et al. 2010; Guo, et al. 2006; Sutton, et al. 2005] resulting in a total of 47 SNPs. Of the 47 SNPs included in the analysis, six of them were monomorphic (T31I, C36S, G38D, R55C, I164T, and rs1865762) and thus excluded from further study. In parallel, the same tag SNPs and rare variants identified in the literature as described above were also genotyped in the African American cohort. Similarly, this genotype data was combined with previously genotyped three SNPs previously in the IRASFS [Bowden, et al. 2010; Guo, et al. 2006; Sutton, et al. 2005] resulting in a total of 44 SNPs. In addition, there were four monomorphic SNPs (C36S, G45R, G84R, and rs62625753) that were excluded from further analysis in the African American sample.
Statistical Analysis
All variants were examined for Mendelian inconsistencies using PEDCHECK [O'Connell and Weeks 1998], resulting in <0.05% discrepancies which were converted to missing. Plasma adiponectin levels were log transformed to best approximate the distributional assumptions of the test and to minimize heterogeneity of the variance. Tests of association between the variants and plasma adiponectin were computed using a variance component model as implemented in Sequential Oligogenic Linkage Analysis Routines (SOLAR) [Almasy and Blangero 1998] with primary inference based on the additive genetic model adjusting for age, gender, recruitment center, BMI, and admixture proportions. The covariates for admixture were estimated using principal components analysis on 80 ancestry informative markers (AIMs) in the Hispanic American population [Palmer, et al. 2010] and 36 AIMs selected in African Americans population [Palmer, et al. 2010].
SOLAR was also used to determine the contribution of genetic factors to adiponectin levels. A variance components analysis of family data decomposes the total variance of the phenotype into components that are due to genetic (polygenic) effects (additive genetic variance), measured covariates, and random environmental effects. Clinical and phenotypic traits were selected for inclusion in the models based on prior reports of association from these data with adiponectin levels [Hanley, et al. 2007; Hanley, et al. 2011] or at least weak suggestive evidence of association in univariate analysis (P<0.20). Additionally, fibrinogen was included as a clinical trait in the analysis despite lack of association due to published association of fibrinogen with diabetes, cardiovascular disease, and subclinical inflammation [Rooney, et al. 2011; Tosetto, et al. 2011; Zhao, et al. 2011], conditions that are also associated with adiponectin levels. The glucose homeostasis measures included adjustment for insulin sensitivity (SI) and acute insulin response (AIR). The adiposity measures included in analysis were body mass index (BMI), visceral adipose tissue (VAT), subcutaneous adipose tissue (SAT), and the ratio of VAT to SAT. Additional factors included fasting insulin, fasting glucose, HDL, triglycerides, PAI-1, fibrinogen, CRP, and systolic and diastolic blood pressure. Analysis was completed on all individuals with complete phenotypic and genotypic data. Additionally, the proportion of variance explained was calculated in SOLAR following adjustment for clinical traits, genetic variants (both LF/rare and common), and the two in combination. To test for the statistical significance of the sets of clinical characteristics, rare, or common SNPs, the corresponding likelihood ratio tests were computed based on the individuals with full clinical and genotype data. For example, the test of the rare SNPs conditional on the clinical characteristics used the difference in −2 log (likelihood) from the models that only contained the clinical characteristics versus those that contained both the clinical characteristics and the rare SNPs.
Supplementary Material
Acknowledgements
This research was supported in part by NIH grants HL060894, HL060931, HL060944, HL061019, HL061210, DK066358, DK085175 and DK91076. As the corresponding author and guarantor of this manuscript I, Dr. Donald Bowden, take full responsibility for the work as a whole, including the study design, access to data, and the decision to submit and publish the manuscript.
Footnotes
Conflict of Interest
There are no conflicts of interest to report.
References
- Adamczak M, Wiecek A, Funahashi T, Chudek J, Kokot F, Matsuzawa Y. Decreased plasma adiponectin concentration in patients with essential hypertension. Am J Hypertens. 2003;16(1):72–5. doi: 10.1016/s0895-7061(02)03197-7. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita Y, Kihara S, Ouchi N, Takahashi M, Maeda K, Miyagawa J, Hotta K, Shimomura I, Nakamura T, Miyaoka K. Paradoxical decrease of an adipose-specific protein, adiponectin, in obesity. Biochem Biophys Res Commun. 1999;257(1):79–83. doi: 10.1006/bbrc.1999.0255. others. [DOI] [PubMed] [Google Scholar]
- Berg AH, Combs TP, Du X, Brownlee M, Scherer PE. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat Med. 2001;7(8):947–53. doi: 10.1038/90992. [DOI] [PubMed] [Google Scholar]
- Bergman RN, Finegood DT, Ader M. Assessment of insulin sensitivity in vivo. Endocr Rev. 1985;6(1):45–86. doi: 10.1210/edrv-6-1-45. [DOI] [PubMed] [Google Scholar]
- Bork S, Horn P, Castoldi M, Hellwig I, Ho AD, Wagner W. Adipogenic differentiation of human mesenchymal stromal cells is down-regulated by microRNA-369-5p and up-regulated by microRNA-371. J Cell Physiol. 2010 doi: 10.1002/jcp.22557. [DOI] [PubMed] [Google Scholar]
- Bowden DW, An SS, Palmer ND, Brown WM, Norris JM, Haffner SM, Hawkins GA, Guo X, Rotter JI, Chen YD. Molecular basis of a linkage peak: exome sequencing and family-based analysis identify a rare genetic variant in the ADIPOQ gene in the IRAS Family Study. Hum Mol Genet. 2010;19(20):4112–20. doi: 10.1093/hmg/ddq327. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LM, Chiu YF, Sheu WH, Hung YJ, Ho LT, Grove J, Rodriguez B, Quertermous T, Chen YD, Hsiung CA. Biethnic comparisons of autosomal genomic scan for loci linked to plasma adiponectin in populations of Chinese and Japanese origin. J Clin Endocrinol Metab. 2004;89(11):5772–8. doi: 10.1210/jc.2004-0640. others. [DOI] [PubMed] [Google Scholar]
- Comuzzie AG, Funahashi T, Sonnenberg G, Martin LJ, Jacob HJ, Black AE, Maas D, Takahashi M, Kihara S, Tanaka S. The genetic basis of plasma variation in adiponectin, a global endophenotype for obesity and the metabolic syndrome. J Clin Endocrinol Metab. 2001;86(9):4321–5. doi: 10.1210/jcem.86.9.7878. others. [DOI] [PubMed] [Google Scholar]
- Dastani Z, Hivert MF, Timpson N, Perry JR, Yuan X, Scott RA, Henneman P, Heid IM, Kizer JR, Lyytikainen LP. Novel Loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 2012;8(3):e1002607. doi: 10.1371/journal.pgen.1002607. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruebis J, Tsao TS, Javorschi S, Ebbets-Reed D, Erickson MR, Yen FT, Bihain BE, Lodish HF. Proteolytic cleavage product of 30-kDa adipocyte complement-related protein increases fatty acid oxidation in muscle and causes weight loss in mice. Proc Natl Acad Sci U S A. 2001;98(4):2005–10. doi: 10.1073/pnas.041591798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–9. doi: 10.1126/science.1069424. others. [DOI] [PubMed] [Google Scholar]
- Guo X, Saad MF, Langefeld CD, Williams AH, Cui J, Taylor KD, Norris JM, Jinagouda S, Darwin CH, Mitchell BD. Genome-wide linkage of plasma adiponectin reveals a major locus on chromosome 3q distinct from the adiponectin structural gene: the IRAS family study. Diabetes. 2006;55(6):1723–30. doi: 10.2337/db05-0428. others. [DOI] [PubMed] [Google Scholar]
- Hanley AJ, Bowden D, Wagenknecht LE, Balasubramanyam A, Langfeld C, Saad MF, Rotter JI, Guo X, Chen YD, Bryer-Ash M. Associations of adiponectin with body fat distribution and insulin sensitivity in nondiabetic Hispanics and African-Americans. J Clin Endocrinol Metab. 2007;92(7):2665–71. doi: 10.1210/jc.2006-2614. others. [DOI] [PubMed] [Google Scholar]
- Hanley AJ, Wagenknecht LE, Norris JM, Bergman R, Anderson A, Chen YI, Lorenzo C, Haffner SM. Adiponectin and the incidence of type 2 diabetes in Hispanics and African Americans: the IRAS Family Study. Diabetes Care. 2011;34(10):2231–6. doi: 10.2337/dc11-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid IM, Henneman P, Hicks A, Coassin S, Winkler T, Aulchenko YS, Fuchsberger C, Song K, Hivert MF, Waterworth DM. Clear detection of ADIPOQ locus as the major gene for plasma adiponectin: results of genome-wide association analyses including 4659 European individuals. Atherosclerosis. 2010;208(2):412–20. doi: 10.1016/j.atherosclerosis.2009.11.035. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkin L, Bergman RN, Bowden DW, Ellsworth DL, Haffner SM, Langefeld CD, Mitchell BD, Norris JM, Rewers M, Saad MF. Genetic epidemiology of insulin resistance and visceral adiposity. The IRAS Family Study design and methods. Ann Epidemiol. 2003;13(4):211–7. doi: 10.1016/s1047-2797(02)00412-x. others. [DOI] [PubMed] [Google Scholar]
- Hivert MF, Manning AK, McAteer JB, Florez JC, Dupuis J, Fox CS, O'Donnell CJ, Cupples LA, Meigs JB. Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring Study. Diabetes. 2008;57(12):3353–9. doi: 10.2337/db08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K, Funahashi T, Arita Y, Takahashi M, Matsuda M, Okamoto Y, Iwahashi H, Kuriyama H, Ouchi N, Maeda K. Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler Thromb Vasc Biol. 2000;20(6):1595–9. doi: 10.1161/01.atv.20.6.1595. others. [DOI] [PubMed] [Google Scholar]
- Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, Martins RA, Kennedy BA, Hassell RG, Visser ME. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42(8):684–7. doi: 10.1038/ng.628. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita A, Yamasaki H, Kuwahara H, Moriuchi A, Fukushima K, Kobayashi M, Fukushima T, Takahashi R, Abiru N, Uotani S. Identification of the promoter region required for human adiponectin gene transcription: Association with CCAAT/enhancer binding protein-beta and tumor necrosis factor-alpha. Biochem Biophys Res Commun. 2005;331(2):484–90. doi: 10.1016/j.bbrc.2005.03.205. others. [DOI] [PubMed] [Google Scholar]
- Lindsay RS, Funahashi T, Krakoff J, Matsuzawa Y, Tanaka S, Kobes S, Bennett PH, Tataranni PA, Knowler WC, Hanson RL. Genome-wide linkage analysis of serum adiponectin in the Pima Indian population. Diabetes. 2003;52(9):2419–25. doi: 10.2337/diabetes.52.9.2419. [DOI] [PubMed] [Google Scholar]
- Ling H, Waterworth DM, Stirnadel HA, Pollin TI, Barter PJ, Kesaniemi YA, Mahley RW, McPherson R, Waeber G, Bersot TP. Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: the GEMS Study. Obesity (Silver Spring) 2009;17(4):737–44. doi: 10.1038/oby.2008.625. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara M, Maruoka S, Katayose S. Decreased plasma adiponectin concentrations in women with dyslipidemia. J Clin Endocrinol Metab. 2002;87(6):2764–9. doi: 10.1210/jcem.87.6.8550. [DOI] [PubMed] [Google Scholar]
- Menzaghi C, Ercolino T, Salvemini L, Coco A, Kim SH, Fini G, Doria A, Trischitta V. Multigenic control of serum adiponectin levels: evidence for a role of the APM1 gene and a locus on 14q13. Physiol Genomics. 2004;19(2):170–4. doi: 10.1152/physiolgenomics.00122.2004. [DOI] [PubMed] [Google Scholar]
- Morandi A, Maffeis C, Lobbens S, Bouatia-Naji N, Heude B, Pinelli L, Meyre D, Froguel P. Early detrimental metabolic outcomes of rs17300539-A allele of ADIPOQ gene despite higher adiponectinemia. Obesity (Silver Spring) 2010;18(7):1469–73. doi: 10.1038/oby.2009.403. [DOI] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–66. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, Kihara S, Ouchi N, Kumada M, Fujita K, Hiuge A, Hibuse T, Ryo M, Nishizawa H, Maeda N. Adiponectin replenishment ameliorates obesity-related hypertension. Hypertension. 2006;47(6):1108–16. doi: 10.1161/01.HYP.0000222368.43759.a1. others. [DOI] [PubMed] [Google Scholar]
- Okamoto Y, Kihara S, Ouchi N, Nishida M, Arita Y, Kumada M, Ohashi K, Sakai N, Shimomura I, Kobayashi H. Adiponectin reduces atherosclerosis in apolipoprotein E-deficient mice. Circulation. 2002;106(22):2767–70. doi: 10.1161/01.cir.0000042707.50032.19. others. [DOI] [PubMed] [Google Scholar]
- Palmer ND, Langefeld CD, Ziegler JT, Hsu F, Haffner SM, Fingerlin T, Norris JM, Chen YI, Rich SS, Haritunians T. Candidate loci for insulin sensitivity and disposition index from a genome-wide association analysis of Hispanic participants in the Insulin Resistance Atherosclerosis (IRAS) Family Study. Diabetologia. 2010;53(2):281–9. doi: 10.1007/s00125-009-1586-2. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Menzaghi C, Salvemini L, De Bonis C, Trischitta V, Hu FB. Novel locus FER is associated with serum HMW adiponectin levels. Diabetes. 2011;60(8):2197–201. doi: 10.2337/db10-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JB, Waterworth D, O'Rahilly S, Hivert MF, Loos RJ, Perry JR, Tanaka T, Timpson NJ, Semple RK, Soranzo N. A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet. 2009;5(12):e1000768. doi: 10.1371/journal.pgen.1000768. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney T, Scherzer R, Shigenaga JK, Graf J, Imboden JB, Grunfeld C. Levels of plasma fibrinogen are elevated in well-controlled rheumatoid arthritis. Rheumatology (Oxford) 2011;50(8):1458–65. doi: 10.1093/rheumatology/ker011. [DOI] [PubMed] [Google Scholar]
- Scherer PE. Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes. 2006;55(6):1537–45. doi: 10.2337/db06-0263. [DOI] [PubMed] [Google Scholar]
- Stancliffe RA, Thorpe T, Zemel MB. Dairy attentuates oxidative and inflammatory stress in metabolic syndrome. Am J Clin Nutr. 2011;94(2):422–30. doi: 10.3945/ajcn.111.013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steil GM, Volund A, Kahn SE, Bergman RN. Reduced sample number for calculation of insulin sensitivity and glucose effectiveness from the minimal model. Suitability for use in population studies. Diabetes. 1993;42(2):250–6. doi: 10.2337/diab.42.2.250. [DOI] [PubMed] [Google Scholar]
- Sutton BS, Weinert S, Langefeld CD, Williams AH, Campbell JK, Saad MF, Haffner SM, Norris JM, Bowden DW. Genetic analysis of adiponectin and obesity in Hispanic families: the IRAS Family Study. Hum Genet. 2005;117(2-3):107–18. doi: 10.1007/s00439-005-1260-9. [DOI] [PubMed] [Google Scholar]
- Tosetto A, Prati P, Baracchini C, Manara R, Rodeghiero F. Association of plasma fibrinogen, C-reactive protein and G-455>A polymorphism with early atherosclerosis in the VITA Project cohort. Thromb Haemost. 2011;105(2):329–35. doi: 10.1160/TH10-08-0522. [DOI] [PubMed] [Google Scholar]
- Vaidya A, Williams JS, Forman JP. The Independent Association Between 25-Hydroxyvitamin D and Adiponectin and Its Relation With BMI in Two Large Cohorts: The NHS and the HPFS. Obesity (Silver Spring) 2012;20(1):186–91. doi: 10.1038/oby.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur F, Helbecque N, Dina C, Lobbens S, Delannoy V, Gaget S, Boutin P, Vaxillaire M, Lepretre F, Dupont S. Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet. 2002;11(21):2607–14. doi: 10.1093/hmg/11.21.2607. others. [DOI] [PubMed] [Google Scholar]
- Wagenknecht LE, Scherzinger AL, Stamm ER, Hanley AJ, Norris JM, Chen YD, Bryer-Ash M, Haffner SM, Rotter JI. Correlates and heritability of nonalcoholic fatty liver disease in a minority cohort. Obesity (Silver Spring) 2009;17(6):1240–6. doi: 10.1038/oby.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waki H, Yamauchi T, Kamon J, Ito Y, Uchida S, Kita S, Hara K, Hada Y, Vasseur F, Froguel P. Impaired multimerization of human adiponectin mutants associated with diabetes. Molecular structure and multimer formation of adiponectin. J Biol Chem. 2003;278(41):40352–63. doi: 10.1074/jbc.M300365200. others. [DOI] [PubMed] [Google Scholar]
- Warren LL, Li L, Nelson MR, Ehm MG, Shen J, Fraser DJ, Aponte JL, Nangle KL, Slater AJ, Woollard PM. Deep Resequencing Unveils Genetic Architecture of ADIPOQ and Identifies a Novel Low-Frequency Variant Strongly Associated With Adiponectin Variation. Diabetes. 2012 doi: 10.2337/db11-0985. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Hirose H, Saito I, Tomita M, Taniyama M, Matsubara K, Okazaki Y, Ishii T, Nishikai K, Saruta T. Correlation of the adipocyte-derived protein adiponectin with insulin resistance index and serum high-density lipoprotein-cholesterol, independent of body mass index, in the Japanese population. Clin Sci (Lond) 2002;103(2):137–42. doi: 10.1042/cs1030137. [DOI] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–9. doi: 10.1038/ng.608. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang J, Wu J. Diabetes mellitus is associated with shortened activated partial thromboplastin time and increased fibrinogen values. PLoS One. 2011;6(1):e16470. doi: 10.1371/journal.pone.0016470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.