Abstract
G proteins orchestrate critical cellular functions by transducing extracellular signals into internal signals and controlling cellular responses to environmental cues. G proteins typically function as switches that are activated by G protein-coupled receptors (GPCRs) and negatively controlled by regulator of G protein signaling (RGS) proteins. In the human fungal pathogen Cryptococcus neoformans, three G protein α subunits (Gpa1, Gpa2, and Gpa3) have been identified. In a previous study, we identified the RGS protein Crg2 involved in regulating the pheromone response pathway through Gpa2 and Gpa3. In this study, a role for Crg2 was established in the Gpa1-cAMP signaling pathway that governs mating and virulence. We show that Crg2 physically interacts with Gpa1 and crg2 mutations increase cAMP production. crg2 mutations also enhance mating filament hyphae production, but reduce cell-cell fusion and sporulation efficiency during mating. Although crg2 mutations and the Gpa1 dominant active allele GPA1Q284L enhanced melanin production under normally repressive conditions, virulence was attenuated in a murine model. We conclude that Crg2 participates in controlling both Gpa1-cAMP-virulence and pheromone-mating signaling cascades and hypothesize it may serve as a molecular interface between these two central signaling conduits.
Introduction
Cells respond to environmental cues through a complex network involving a variety of cell surface receptors and signal transduction pathways. Signal transduction by seven transmembrane (7-TM) domain G protein-coupled receptors (GPCRs) controls a vast array of physiological processes in eukaryotic organisms. GPCR mediated signal activation involves a complicated intracellular network of signaling molecules, including G proteins and their regulators such as regulator of G protein signaling (RGS) proteins.
Heterotrimeric guanine-nucleotide binding proteins (G proteins) play a central role in transducing extracellular cues into intrinsic signals effecting appropriate biochemical and physiological responses. Heterotrimeric G proteins are activated via GPCRs. The conformational change of a GPCR following ligand binding enhances binding to the corresponding Gα to promote exchange of GDP to GTP, leading to the dissociation of Ga from Gβγ. Freed Gα and Gβγ can each activate downstream signaling pathways. RGS proteins are GTPase-activating proteins for Gα, and they function primarily as GTPase accelerating proteins (GAPs) to increase the hydrolysis rate of GTP bound to Gα subunits, thereby inactivating Gα (Dohlman and Thorner, 2001; Siderovski and Willard, 2005). RGS proteins are therefore physiologically and pathophysiologically important negative regulators of GPCR signaling.
Since their discovery in the 1990s, RGS proteins have emerged as crucial regulators of GPCR signaling. In humans, over 20 RGS proteins have been identified (Jean-Baptiste et al., 2006; Wieland et al., 2007). In Saccharomyces cerevisiae, the RGS protein Sst2 was first identified as a negative regulator of the pheromone response pathway, which is controlled by the pheromone receptors Ste2 and Ste3, and its downstream Gα subunit Gpa1 (Apanovitch et al., 1998; Dohlman et al., 1996). New features of this regulator have been revealed recently, indicating that Sst2 can directly bind via its DEP domain to the carboxyl terminal tail of the pheromone receptor Ste2, and thereby functions as a principal regulator of mating pheromone signaling (Ballon et al., 2006; Chasse et al., 2006). Similar to S. cerevisiae, the Sst2 RGS homolog in the human fungal pathogen Candida albicans also controls mating responses (Dignard and Whiteway, 2006). Rgs2 is the second RGS domain protein in S. cerevisiae and negatively regulates glucose signaling via the GPCR Gpr1 and its coupled Gα subunit Gpa2, which control cAMP-PKA signaling (Versele et al., 1999).
While only two RGS proteins are present in the model yeast S. cerevisiae, additional RGS proteins exist in most other fungal systems that have been studied (Li et al., 2007b). Functional studies in filamentous fungi revealed RGS proteins regulate signals that control vegetative growth, sporulation, stress responses and pathogenicity, in organisms as diverse as Aspergillus nidulans (Han et al., 2004; Lafon et al., 2005; Lafon et al., 2006; Yu, 2006), the rice blast fungus Magnaporthe grisea (Liu et al., 2007), and the Chestnut blight fungus Cryphonectria parasitica (Segers et al., 2004). Interestingly, in Arabidopsis thaliana a hybrid protein has been identified (AtRGS1) containing both 7-TM and RGS domains, and shown to play an important role in plant cell proliferation (Chen et al., 2003).
Cryptococcus neoformans is a major human fungal pathogen that can infect the central nervous system to cause meningitis, leading to significant morbidity and mortality (Casadevall and Perfect, 1998). This fungus has emerged as a model system to study signal transduction and several major signaling cascades have been identified and well characterized. In the mating pathway, the receptors Ste3α/a sense pheromones from cells of the opposite mating type and activate a G protein complex that includes the G protein a subunits Gpa2 and Gpa3, the Gβ subunit Gpb1, and the Gγ subunits Gpg1 and Gpg2 (Hsueh et al, 2007; Li et al., 2007a; Wang and Heitman, 1999). Following the activation of Gpa2 and Gpa3, the Gβ subunit Gpb1 is released to activate the downstream MAP kinase cascade to trigger mating responses (Hsueh et al., 2007; Li et al., 2007a). One RGS protein, Crg1, has been identified as a negative regulator of the pheromone response pathway (Nielsen et al., 2003; Wang et al., 2004). Cells lacking Crg1 are hypersensitive to mating pheromones and produce abundant conjugation tubes in confrontation assays (Wang et al., 2004). Recently, we also identified a second RGS protein, Crg2, that plays an important role in modulating mating (Hsueh et al., 2007).
In addition to the pheromone response pathway, the Gpa1-cAMP signaling pathway has also been discovered to play a central role in virulence of C. neoformans (Alspaugh et al., 1997; D'Souza et al., 2001; Lengeler et al., 2000; Pukkila-Worley and Alspaugh, 2004; Wang and Heitman, 1999). In this pathway, multiple receptors may be involved in activating the downstream Gα protein Gpa1. In turn Gpa1 governs the production of the secondary messenger cAMP and activation of protein kinase A (PKA) to promote melanin and capsule production and thereby control virulence. The Gpa1-cAMP cascade also influences mating. Gpr4 has been identified as one receptor for the cAMP pathway, and shown to promote capsule production and mating, but not to be important for virulence in a murine inhalation model (Xue et al., 2006). Recently, Gib2 was identified as a Gβ subunit that interacts with both Gpa1 and the Gγ subunits Gpg1 and Gpg2 to form a G protein complex (Palmer et al., 2006). But how the Gpa1 G protein signal is modulated in this pathway was unclear.
In this study, we show that Crg2 physically interacts with Gpa1. Unlike Crg1, mutation of CRG2 enhances cAMP production in response to glucose. Our results provide additional evidence showing that the RGS protein Crg2 is not only a negative regulator of the pheromone response pathway, but also of the Gpa1-cAMP signaling pathway controlling virulence of C. neoformans. Thus, we hypothesize that Crg2 could serve as an interface regulating the activities of these two parallel signaling pathways that coordinately evoke key developmental transitions leading to mating or pathogenicity.
Material and methods
Strains, media and growth conditions
C. neoformans strains used in this study are listed in Table 1. crg2 gpa1 and crg2 gpr4 double deletion mutants were generated by crossing individual mutants and screening the progeny basidiospores isolated by micromanipulation. Strains were grown at 30°C on yeast extract-peptone-dextrose (YPD) agar medium and synthetic (SD) medium. V8 medium (pH=5.0) was used for mating assays. MS medium was used for mating and sporulation assays and prepared as previous described with modification (Xue et al., 2007). In a one-liter volume, 100 ml 10× basal salt mixture (Sigma M5524) was mixed with 40 grams of agar and the pH was adjusted to 5, and 1 ml of 1000× vitamin mixture (Sigma M7150) was added after autoclaving. Niger-seed medium was used to test for melanin production. Dulbecco modified Eagles (DME) medium for assessing capsule production was prepared as previously described (Bahn et al., 2004). All other media were prepared as described previously (Alspaugh et al., 1997; Bahn et al., 2005b; Granger et al., 1985)
Table 1.
C. neoformans strains used in this study
| Strain | Genotype | Source/reference |
|---|---|---|
| H99 | MATα | (Perfect et al., 1993) |
| KN99a | MATa | (Nielsen et al., 2003) |
| YPH276 | MATα crg1:NAT | This study |
| PPW196 | MATa crg1∷URA5 | (Wang et al., 2004) |
| YSB83 | MATα gpa1∷NAT | (Bahn et al., 2004) |
| YSB85 | MATa gpa1∷NEO | (Bahn et al., 2004) |
| JKH7 | MATα pka1∷URA5 | (Hicks et al., 2004) |
| JKH74 | MATa pka∷URA5 | (Hicks et al., 2004) |
| JKH63 | MATα pde1∷NAT | (Hicks et al., 2005) |
| CDX40 | MATa gpa1∷URA5 PGPD1-GPA1Q284L | (Xue et al., 2006) |
| CDX50 | MATα crg2∷NAT | This study |
| CDX51 | MATa crg2∷NAT | This study |
| CDX111 | MATa crg2∷NEO | This study |
| CDX112 | MATα crg2∷NAT CRG2∷NEO | This study |
| CDX114 | MATa crg1∷URA5 crg2∷NAT | This study |
| CDX115 | MATα crg1∷NAT crg2∷NEO | This study |
| CDX118 | MATα crg2∷NEO gpa1∷NAT | This study |
| CDX119 | MATa crg2∷NAT gpa1∷NEO | This study |
| CDX122 | MATα gpr4∷NAT crg2∷NEO | This study |
| CDX123 | MATa gpr4∷NAT crg2∷NEO | This study |
| CDX153 | MATα gpa1∷URA5 PGPD1-GPA1Q284L | This study |
| CDX154 | MATα PGPD1-CRG2∷NEO | This study |
| CDX155 | MATa PGPD1-CRG2∷NEO | This study |
| CDX156-2 | MATα PGPD1-GPA1Q284L | This study |
| CDX156-4 | MATα PGPD1-GPA1Q284L | This study |
| CDX159 | MATa crg2∷NAT CRG2∷NEO | This study |
| CDX173 | MATα crg1∷NAT crg2∷NEO | This study |
| CDX174 | MATa crg1∷NAT crg2∷NEO | This study |
| CDX180 | MATα crg2∷NAT CRG2∷NEO | This study |
| CDX181 | MATa crg2∷NAT CRG2∷NEO | This study |
| CDX182 | MATα crg2∷NAT CRG2-TM∷NEO | This study |
| CDX183 | MATa crg2∷NAT CRG2-TM∷NEO | This study |
| CDX184 | MATα crg2∷NAT CRG2-TM+CAAX∷NEO | This study |
| CDX185 | MATa crg2∷NAT CRG2-TM+CAAX∷NEO | This study |
| CDX186 | MATα pka1∷URA5 PGPD1-CRG2∷NEO | This study |
| CDX187 | MATa pka1∷URA5 PGPD1-CRG2∷NEO | This study |
| CDX188 | MATα gpa1∷NAT PGPD1-CRG2∷NEO | This study |
| CDX189 | MATa pde1∷NAT PGPD1-CRG2∷NEO | This study |
| CDX190 | MATα crg2∷NAT PGPD1-GPA1Q284L | This study |
| CDX191 | MATa crg2∷NAT PGPD1-GPA1Q284L | This study |
| CDX192 | MATα crg2∷NAT crg3∷NEO | This study |
| CDX193 | MATa crg2∷NAT crg3∷NEO | This study |
Disruption of the CRG2 gene and construction of a crg2 + CRG2 complemented strain
The crg2 null mutant was generated in the congenic C. neoformans serotype A MATα (H99) and MATa (KN99a) strains by overlap PCR as previously described (Davidson et al., 2002). The 5’ and 3’ regions of the CRG2 gene were amplified with primers JH14775/JH14776 (see Supplemental Table 1 for primer sequences) and JH14777/JH14778 from H99 or KN99 genomic DNA, and the dominant selectable markers (Natr or Neor) were amplified with the M13 primers (M13F/M13R) from plasmid pNATSTM#209 or pJAF1 (Fraser et al., 2003), respectively. The CRG2 gene replacement cassette was generated by overlap PCR with primers JH14775/JH14778, precipitated onto 600-µg gold microcarrier beads (0.8 µm; Bioworld Inc, Dublin OH), and strains H99 or KN99a were biolistically transformed as described previously (Davidson et al., 2000). The same crg2 gene replacement cassette was also used to transform the crg1 mutant to generate crg1 crg2 double mutants. Stable transformants were selected on YPD medium containing nourseothricin (100 mg/L) or G418 (200 mg/L). To screen for crg2 mutants, diagnostic PCR was performed by analyzing the 5’ junction of the disrupted crg2 alleles with primers JH14774/JH8994. Positive transformants identified by PCR screening were further confirmed by Southern blot analysis.
To construct the crg2 + CRG2 complemented strains, H99 genomic DNA containing the entire CRG2 gene and 1.5-kb 5’ promoter region and 0.6-kb 3’ untranslated region (UTR) was amplified with primers JH17832/JH17833 and sequence verified. A crg2 mutant was transformed with an overlap PCR product containing the 3.7-kb CRG2 PCR fragment and the NEO gene via biolistic transformation. The crg2 + CRG2 complemented strain was selected from transformants containing the full-length CRG2 gene.
A similar strategy was employed to generate CRG2 expression strains expressing either a CRG2 allele lacking the TM domains or in which the TM domains are replaced by the Ras1 CAAX motif. The genomic fragment containing CRG2 promoter and a CRG2 allele with the TM domains truncated was amplified with primers JH17832/JH20163. The 3’ UTR of CRG2 was amplified with primers JH20164/JH17833. To generate the CRG2 allele containing the CAAX box, the CRG2 allele and 3’ UTR were PCR amplified with JH17832/JH20165, and JH20166/JH17833, respectively. The expression cassettes were amplified with primers JH17832/JH12954 in an overlap PCR to combine the CRG2 allele, 3’ UTR, and NEO marker. All PCR products were sequence verified, and used to transform crg2 gene deletion mutants. Stable transformants were selected and single colony purified. The expression of CRG2 transcripts was verified in a Northern blot assay.
To generate crg2 crg3 double mutants, spores were dissected from an a crg2 × α crg3 genetic cross. Progeny were screened for double mutants via PCR with JH14779/JH14780 (crg2) and JH14786/JH14787 (crg3). Positive double mutants were confirmed by Southern blot.
Assays for melanin and capsule production
Melanin production was assayed by inoculating C. neoformans strains into 2 ml YPD liquid medium, and incubating overnight at 30°C. 5 µl of each overnight culture was placed on Niger seed agar medium. The agar plates were incubated at 30°C or 37°C for two days, and pigmentation of fungal colonies was assessed.
To examine capsule production, 5 µl of overnight cultures were inoculated onto DME agar medium or with 10 mM cAMP or 4–10% CO2 and incubated at 30°C or 37°C for 3 days. The high CO2 condition was generated in an anaerobic container (Scientific Devices, IL) with a CO2 gas pack (BD Bioscience, MD). Capsule was visualized with India ink staining and observed with a 100× Zeiss Axioskop 2 equipped with an AxioCam MRM digital camera (Carl Zeiss, Thornwood, NY). Quantitative measurement of capsule size was performed as described previously (Zaragoza et al., 2003) by measuring the diameters of the capsule and the cell using Axio Vision 4.2 software (Carl Zeiss, Thornwood, NY). The relative capsule diameter (100(Dw – Dc)/Dw, where Dw indicates the diameter of the cell plus capsule and Dc indicates the diameter of the cell only) was statistically compared between each mutant and wild-type strains by the student’s t-test (P values of <0.05 were considered as significant).
Assays for mating and cell fusion
In a mating assay, C. neoformans cells of opposite mating type were homogenized and co-cultured on V8 or MS agar medium at 25°C in the dark for several days and filamentation was examined by light microscopy. For confrontation assays, cells of two strains were grown in parallel lines on filamentation agar (FA) plates or V8 plates at a distance of 2 to 5 mm, and incubated at room temperature for 10 days in the dark.
Cell fusion efficiency was measured by mixing 2 × 106 cells of two strains. 5 µl culture suspension was inoculated onto V8 agar medium (pH=5.0) and incubated for 24 h at room temperature in the dark. Four replicate plates were prepared for each of the three cell mixtures. Cells were then harvested and resuspended in 1 ml dH2O and 200 µl of the suspension (~105 total cells) was plated onto YPD medium containing nourseothricin (NAT) and G418. The number of colonies formed on YPD containing both NAT and G418 were counted on each plate. The cell fusion efficiency between H99 and KN99a was scored as 100%, and the cell fusion efficiency of crg2 mutants was calculated relative to wild type.
Overexpression of the CRG2 gene
The CRG2 gene genomic ORF was amplified using primers JH18786/JH18787, and cloned into vector pXL1 between the FseI and PacI sites to generate plasmid pDX112, in which CRG2 is under the control of the constitutively expressed promoter from the GPD1 gene. All constructs generated are confirmed by sequencing. Wild type strains H99 and KN99a, and gpa1, pka1, and pde1 phosphodiesterase mutant strain were transformed with plasmid pDX112, and overexpression of the CRG2 gene was confirmed by RNA expression based on Northern blot analysis.
Generation of the GPA1 dominant active allele
The GPA1 dominant active allele (GPA1Q284L), corresponding to GPA2Q300L in S. cerevisiae, was introduced by overlap PCR using primers JH12497/JH12500 and JH12498/JH12499. The overlap PCR product was amplified with primers JH12497/JH12498 and cloned into the integrating vector pRCD83 (Davidson et al., 2002), generating pDX10. This expression construct was sequence verified, linearized with the enzyme NotI, and then introduced into the wild-type strain H99 through biolistic transformation to generate dominant active strains. This construct was also used to transform the crg2 mutant to evaluate the epistatic relationship between Gpa1 and Crg2. Strains generated were confirmed by Southern blot.
Assays for cAMP production
cAMP assays were conducted as described previously (Xue et al., 2006). Briefly, a single colony of each C. neoformans strain was inoculated into 10 ml of YPD liquid medium and incubated for 24 h at 30°C with shaking. Cells were collected and washed twice with dH2O, once with MES buffer (10 mM MES, 0.5 mM EDTA), and resuspended in 20 ml MES buffer. 15 µl of diluted cells (OD600 = 2.0) were incubated for 2 h at 30°C for glucose starvation. 1 ml of cells was filtered through a wet Millipore filter on a vacuum manifold (pore size 0.45 micron, HVLP02500) for the 0 time point. 1.5 ml of 20% glucose was added to the remaining 14 ml of cell suspension. 1 ml was removed and filtered at 30 sec, 1 min, and 3 min. At each time point, filters were immediately removed, placed into Petri-dishes containing 1 ml of formic acid solution (9.2 ml of 100% formic acid, 190.8 ml dH2O, and 50 ml butanol), and agitated for 1 h to lyse cells on a table-top orbital shaker. Cell suspensions were spun down and the supernatants were transferred to fresh tubes and lyophilized. Pellets were resuspended in 400 µl of assay buffer and 100 µl was used for each sample. cAMP concentrations were determined using the cAMP Biotrak Enzyme immunoassay (EIA) system (Amersham, Piscataway NJ) and normalized to the wet weight of the cells.
Protein-protein interaction assays using the yeast two-hybrid system and split-ubiquitin system
Yeast two-hybrid interaction assays were performed as described (Osman, 2004). The CRG2 partial cDNA lacking the transmembrane regions was cloned into the bait vector pGADT7 and the prey vector pGBKT7, respectively. The full length GPA1 cDNA was cloned into plasmid pGBKT7. All inserted cDNA sequences were confirmed by sequencing. Both bait constructs and prey constructs were co-transformed into yeast strain PJ69-4A. Transformants growing on medium lacking histidine and adenine were considered positive interactions and were further confirmed and quantified by β-galactosidase enzyme activity assays using Chlorophenolred-β–D-galactopyranoside (CRPG; Calbiochem, San Diego, CA) as substrate as described previously (Idnurm and Heitman, 2005).
The split-ubiquitin system was also utilized to investigate the interaction between Gpr4 and Crg2. Vectors and yeast strains were those included in the DUALmembrane Kit 2 (Dualsystem Biotech, Switzerland). GPR4 full-length cDNA was cloned into yeast expression vector pCCW (the N-terminal half of the ubiquitin Cub protein was fused to the C-terminus of Gpr4), or pNCW (Cub was fused to the N-terminus of Gpr4), respectively. CRG2 full-length cDNA was cloned into the pDSL-XN vector (the mutated C-terminal half of ubiquitin NubG protein was fused to the Crg2 N-terminus). All cDNA sequences were confirmed by DNA sequencing. Cub and NubG fusion constructs were co-transformed into host yeast strain NMY32. Two constructs, pAI-Alg5 and pDL2-Alg5, express a fusion of the endogenous ER protein Alg5 to the Nub portion of yeast ubiquitin. pAI-Alg5 contains a wild type Nub that interacts with the Cub portion of the ubiquitin from the bait vectors, and serves as a positive control. pDL2-Alg5 contains a Nub portion bearing an isoleucine to glycine mutation that prevents nonspecific interaction with the Cub portion from bait vectors, and serves as a negative control. Interaction was determined by the growth of yeast transformants on medium lacking histidine or adenine, and also by measuring β-galactosidase activity.
In vitro binding assay between Crg2 and Gpa1
The Crg2-polyHistidine fusion construct was generated by cloning the CRG2 partial cDNA (1–909 nt) lacking the transmembrane regions into vector pRSET-C (Invitrogen, Carlsbad, CA), between the BamHI and EcoRI sites. The Gpa1-GST fusion construct was generated by cloning the GPA1 cDNA into vector pGEX-KG (provided by Dr. Jin-Rong Xu), between the BamHI and EcoRI sites.
For protein expression, E. coli BL21 (DE3) cells expressing Crg2–6× His or GST-Gpa1 or GST alone (pGEX-KG) were grown to an OD600= 0.5 and induced with 1 mM IPTG. After 4 hr, cells were collected and washed once with TBS and once with lysis buffer (50 mM HEPES [pH 7.6], 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 1 mM EGTA, protease inhibitor cocktail (Calbiochem), and 0.5 mM PMSF). Cells were resuspended in lysis buffer and incubated on ice for 30 min. Resuspensions were spun down and supernatants were used as extracts.
In vitro binding between Crg2 and Gpa1 was performed using the ProFound™ Pull-down GST protein kit (Pierce, Rockford, IL). Briefly, 60 µg protein extracts containing GST-Gpa1 or GST alone extracts were incubated with GDP (50 µM) alone, GTPγS (50 µM) alone, or GDP (50 µM), AlCl3 (30 µM), and NaF (10 mM) for 30 min at 25°C. 50 µl glutathione-sepharose beads were equilibrated with 400 µl washing buffer for 5 times. Beads were span 1250 g × 30 sec each time. Pretreated protein extracts were incubated with glutathione-sepharose beads at 4°C. After 1 hr, beads were washed 2 times with 400 µl washing buffer. 60 µg of His-Crg2 crude extract was added and incubated with GDP (50 µM) alone, GTPγS (50 µM) alone, or GDP (50 µM), AlCl3 (30 µM), and NaF (10 mM) for 2 hr at 4°C with gentle shaking. After precipitation, beads were washed 5 times with washing buffer. Bound proteins were boiled and separated by SDS-PAGE and Western blotted with anti-His (Sigma, St. Louis), or anti-GST antibodies (Sigma, St Louis).
Virulence study
Yeast strains were grown at 30°C overnight and cultures were washed twice with 1× phosphate-buffered saline (PBS), and resuspended at a final concentration of 2 × 106 CFU/ml. Groups of 10 female A/Jcr mice (NCI/Charles River Laboratories) were intranasally infected with 105 yeast cells of each strain as previously described (Cox et al., 2000). Animals that appeared moribund or in pain were sacrificed by CO2 inhalation. Survival data from the murine experiments were statistically analyzed between paired groups using the long-rank test using the PRISM program 4.0 (GraphPad Software, San Diego, CA) (P values of <0.001 were considered significant).
Histopathology and organ fungal burden
Infected animals were sacrificed at 7, 14, and 21 days post-infection, respectively. Infected lungs were isolated and fixed in 10% formalin solution, and sent to the AML laboratory for section preparation (AML Labs, Inc. Rosedale, MD). Tissue slides were stained with hematoxylin and eosin (H & E), and examined by light microscopy. Infected lungs were also isolated and homogenized using a homogenizer in 1× PBS buffer. Resuspensions were diluted and 50 µl of each dilution was spread on Niger seed medium and colonies were determined after 3 days incubation at 30°C.
Scanning Electron Microscopy
Mating between crg2 mutants were performed on MS medium and incubated at room temperature in the dark for 2 weeks. The mating plate containing mating filaments was fixed with 0.1 M Na cacodylate buffer (pH 6.8) containing 3% glutaraldehyde by flooding the entire dish for 24 hrs at 4°C. The plate was washed with 0.1 M Na cacodylate buffer (pH 6.8), and areas of interest were excised into 1 mm3 blocks from the cultures, and incubated in the fixation buffer for several weeks at 4°C. Samples were then rinsed in three 30-minute changes of cold 0.1 M Na cacodylate buffer, and post-fixed in 2% osmium tetroxide in 0.1 M Na cacodylate buffer for 2.5 hrs at 4°C before being viewed by SEM.
Results
Three proteins contain RGS domains in C. neoformans
Three genes encoding proteins containing RGS domains were identified in the Cryptococcus genome (Loftus et al., 2005). One, Crg1, is a functional homolog of Sst2 in S. cerevisiae, has both a DEP and an RGS domain, and functions as a negative regulator of Gpa2 and Gpa3 to regulate the pheromone response pathway (Li et al., 2007a; Nielsen et al., 2003; Wang et al., 2004).
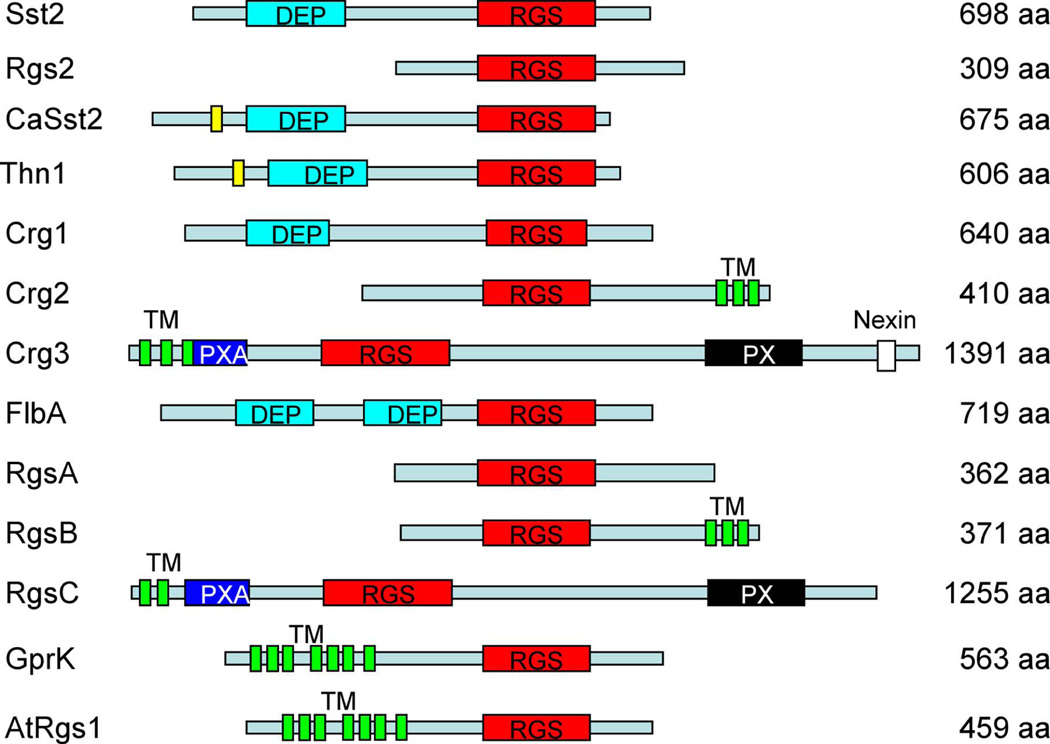
Two other putative RGS proteins, Crg2 and Crg3, contain additional functional domains besides the RGS domain. Crg2 has three transmembrane (TM) domains at its C-terminus, indicating that it may be integrally membrane associated. Crg3 also contains TM domains at the N-terminus, as well as a PXA domain and a PX domain, similar to RgsC, an RGS protein from A. nidulans (Figure 1). The PXA domain is known to associate with PX domains, a novel PI (phosphoinositide)-binding domain (Zheng et al., 2001). This study focuses on the functions of the Crg2 protein.
Figure 1. RGS proteins in fungi.
A schematic of RGS domain proteins in S. cerevisiae (Sst2 and Rgs2), C. albicans (CaSst2), S. commune (Thn1), C. neoformans (Crg1, Crg2, and Crg3), A. nidulans (FlbA, RgsA, RgsB, RgsC, and GprK), and Arabidopsis thaliana (AtRGS1). Names of different domains are indicated. DEP, domain found in Dishevelled, Egl-10, and Pleckstrin; RGS, regulator of G protein signaling domain; TM, transmembrane domain; PXA, PX association domain; PX, Phox homology domain, a novel PI (phosphoinositide)-binding domain.
Crg2 is important for sexual spore formation and involved in Gpa1-cAMP signaling
A previous study revealed that both Crg1 and Crg2 are negative regulators of the pheromone response pathway. They play in part a redundant role in mating regulation, yet crg1 and crg2 mutant phenotypes differ (Hsueh et al., 2007). Although pheromone gene expression is highly induced in both crg1 and crg2 mutants, Crg1 and Crg2 have distinct expression patterns under different induction conditions. While CRG1 expression is only induced during mating or in a gpa3 mutant when grown on V8 mating medium, and is not expressed in wild type or gpa1 or gpa2 mutant backgrounds, the CRG2 gene is constitutively expressed under the same growth conditions based on previous study, indicating Crg2 could have a broader role than Crg1 (Hsueh et al., 2007).
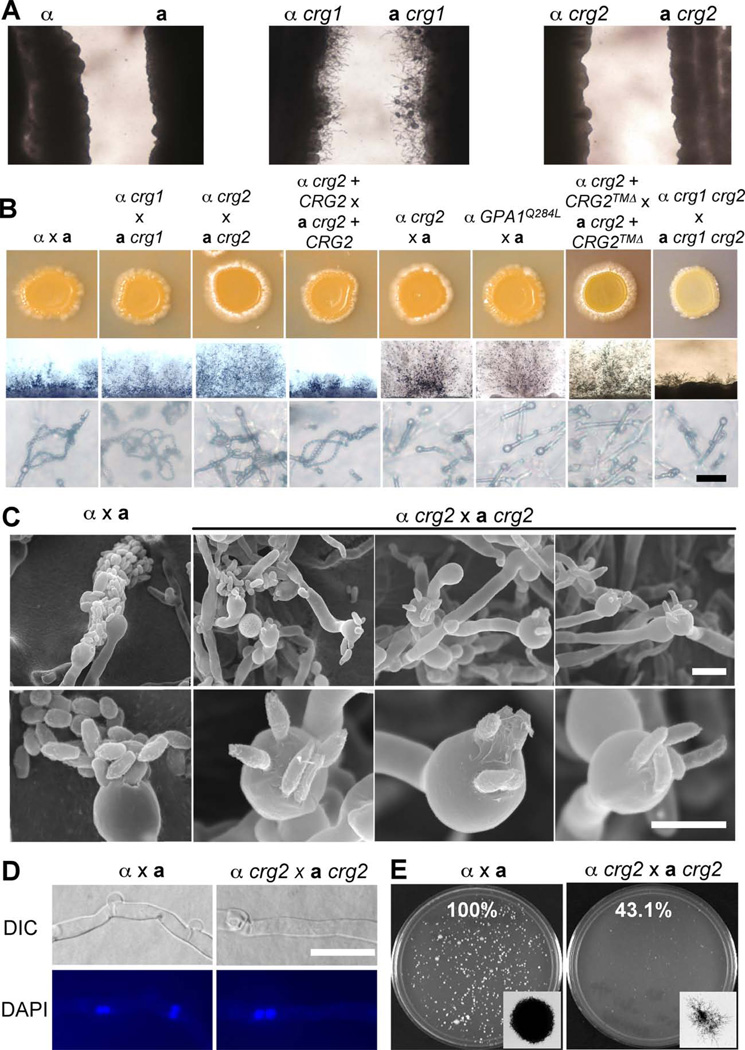
Although crg2 mutations increase dikaryotic mating hyphae production, similar to crg1 mutations, they confer a different response in a confrontation assay. The confrontation assay is used to detect the response and sensitivity of a Cryptococcus strain to pheromone. When yeast cells sense pheromone from cells of the opposite mating type, conjugation tubes are produced and grow towards the source of pheromone. Cryptococcus neoformans var. neoformans wild type strains produce robust conjugation tubes in this assay, but var. grubii wild type strains typically do not. While crg1 mutants in the var. grubii background are hypersensitive to pheromone and produce abundant conjugation tubes after 10 days of incubation in a confrontation assay, no conjugation tubes were produced by confronting crg2 mutants, similar to wild type strains, indicating that crg1 mutants are more sensitive than crg2 mutants or wild type cells to pheromone (Figure 2A).
Figure 2. crg2 mutations enhance filamentation but reduce sporulation in mating.
A) Confrontation assay of wild type (H99 and KN99a), crg1 (YPH276 and PPW196), and crg2 (CDX50 and CDX51) mutant strains on V8 medium. Two opposite mating type strains were inoculated on V8 medium in parallel at a distance of 2–5 mm. Conjugation tubes were only observed with crg1 × crg1 after 7 days incubation in the dark at 25°C. B) Mating filament production and sporulation. Matings were performed on V8 medium or MS medium. Mating results on V8 medium (upper panel), mating structures at 40× magnification (middle panel), and 400× magnification (lower panel) were photographed after 2 weeks of incubation in the dark at 25°C. Bar, 20 µM. C) Matings were performed on MS medium and incubated in the dark at 25°C for 10 days. The edge of mating cultures from both wild type (H99 × KN99a) and crg2 mutants (CDX50 × CDX51) were fixed and viewed by SEM at two different magnifications, 2500× (upper panel) and 75000× (lower panell). Bar, 5 µM. D) Nuclei were stained with 4’-6-Diamidino-2- phenylindole (DAPI). Two nuclei were observed in each cell in the mating filaments of both wild type α × a and crg2 × crg2 mutant crosses. Bar, 5 µM. D) Cell fusion assays were performed for both wild type (H99 × KN99a) and crg2 mutants (CDX50 and CDX51) at 30°C. In each experiment, the percentage of cell fusion products relative to the wild type cell fusion (100%) was calculated by averaging results from duplicated plates from three independent experiments.
The mating efficiency of crg2 mutants was quantified by scoring colony forming units (CFU) in a cell fusion assay. At 30°C, α crg2 × a crg2 cell fusion efficiency was reduced to ~43% of the efficiency of wild type strains. Colonies formed by dikaryon or diploid fusion products in α crg2 × a crg2 mutant mating were smaller and formed extensive filaments at early time points, while wild type cells produced large colonies with shorter peripheral filaments (Figure 2E). We also analyzed the mating of the crg1 crg2 double mutant; very interestingly, instead of showing any additive or synergistic enhancement of mating filament production, two independently generated crg1 crg2 double mutants (CDX114 and CDX115) exhibited a mating defect and only produced sporadic short dikaryotic mating hyphae in a bilateral cross (Figure 2B), whereas both produced more abundant mating filaments in unilateral crosses with wild type (data not shown). crg3 mutants have also been generated, but no obvious phenotype in mating, capsule or melanin production has been observed (data not shown). We also generated crg2 crg3 double mutants and observed no significant additional phenotype compared to crg2 mutants (Figure 6D and data not shown). Thus, the functions of the Crg3 protein remain to be established.
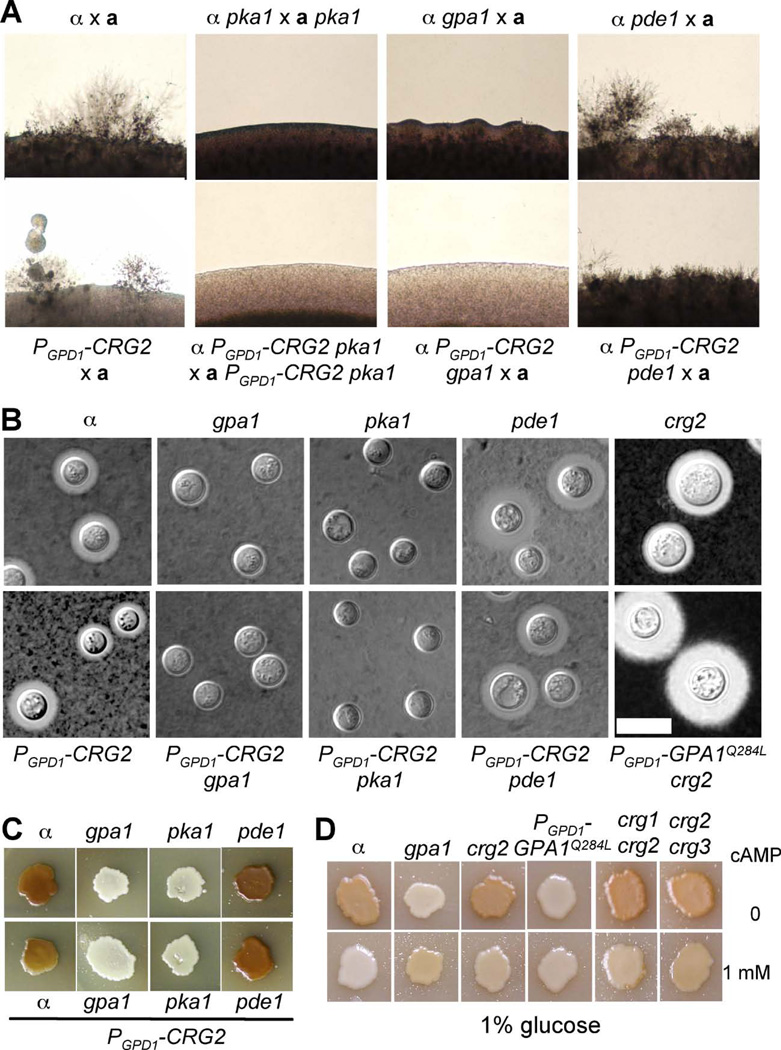
Figure 6. Crg2 functions upstream of Gpa1 in cAMP signaling regulation.
A) Mating assays for the following strains were cocultured on MS medium for 7 days in the dark at 25°C and photographed. H99 × KN99a, α pka1 (JHK7) × apka1 (JKH74), α gpa1 (YSB83) × KN99a, α pde1 (JKH63) × KN99a, α PGPD1-CRG2 (CDX154) × KN99a, α PGPD1-CRG2 pka1 (CDX186) × aPGPD1-CRG2 pka1 (CDX187), α PGPD1-CRG2 gpa1 (CDX188) × KN99a, α PGPD1-CRG2 pde1 (CDX189) × KN99a. B) capsule production was visualized by India ink staining after growth on DME medium for 3 days. The following strains were used in this assay; H99, α gpa1 (YSB83), α pka1 (JHK7), α pde1 (JKH63), α PGPD1-CRG2 (CDX154), α PGPD1-CRG2 gpa1 (CDX188), α PGPD1-CRG2 pka1 (CDX186), α PGPD1-CRG2 pde1 (CDX189). C) Melanin production was assayed on Niger seed media. The same set of strains as in B) were used for melanin assays. Plates were incubated at 37°C. D) Melanin production was assayed on Niger seed media containing 1% glucose and without (upper) or with (lower) addition of 1 mM cAMP. H99, crg2 (CDX50), gpa1 (YSB83), PGPD1-CRG2 (CDX154),PGPD1-GPA1Q284L(CDX156-2), crg1 crg2 (CDX115), and crg2 crg3 (CDX193) strains were used in this assay. Plates were incubated at 30°C. Melanin levels produced by the strains tested in panels C and D were photographed after incubation for 3 days.
Crg2 contains three transmembrane (TM) domains at its C-terminus. To examine the potential function of these TM domains, we generated a CRG2 allele in which the TM domains were deleted and a CRG2 allele in which the TM domains were replaced by the Cryptococcus Ras1 CAAX membrane anchor motif. These CRG2 alleles were expressed in a crg2 mutant background under the control of the CRG2 native promoter. Neither allele rescued the phenotype of crg2 mutants and these strains also produced enhanced mating hyphae, similar to crg2 mutants. But in strains expressing the full length CRG2 gene, mating efficiency was restored to the level of wild type strains (Figure 2B and Figure S2). These results indicate that the TM domains are important for the proper function of Crg2.
Besides increased dikaryotic mating filament production, a significant sporulation defect was also observed in a unilateral mating between a crg2 mutant and a wild type strain or a bilateral mating between two crg2 mutants (Figure 2B and C). Based on both light microscopy and scanning electron microscopy, basidia in α × a wild type crosses are decorated with four long spore chains whereas α crg2 × a crg2 mutant crosses yield predominantly four spores on each basidium. In some cases, basidia produced by α crg2 × a crg2 crosses were decorated with five spores. In some cases, basidia produced by α crg2 × a crg2 crosses were decorated with five spores. In some cases these appeared to emerge from five rather than the typical four points on the basidium surface, suggesting that Crg2 could play a role in regulating both the efficiency of sporulation and bud site selection for spore emergence. In addition, the spore length was elongated in the crg2 mutants and the shape of the distal spore tip was irregular and less rounded compared to wild type (Figure 2C). A normal pattern of nuclear localization (dikaryons with two paired nuclei per cell) was observed in hyphae produced in the α crg2 × a crg2 mating based on nuclear staining, similar to mating of wild type strains, excluding the possibility that the filaments are the result of monokaryotic fruiting in crg2 mutant crosses (Figure 2D). The observed sporulation defect is similar to unilateral mating of strains expressing the dominant active allele GPA1Q284L, based on a previous study (Xue et al., 2006). A strain that expresses the GPA1Q284L allele in a gpa1 mutant background was generated to investigate the Gpa1-cAMP signal pathway. In a unilateral mating between this Gpa1 dominant active strain and wild type, sporadic mating filament production with significantly reduced spore formation was observed (Xue et al., 2006). Here we generated strains expressing GPA1Q284L in a wild type strain background and found they exhibit similar mating defects (Figure 2B). This observation prompted us to investigate whether Crg2 also regulates Gpa1 in addition to its established role in regulating Gpa2 and Gpa3 (Hsueh et al., 2007).
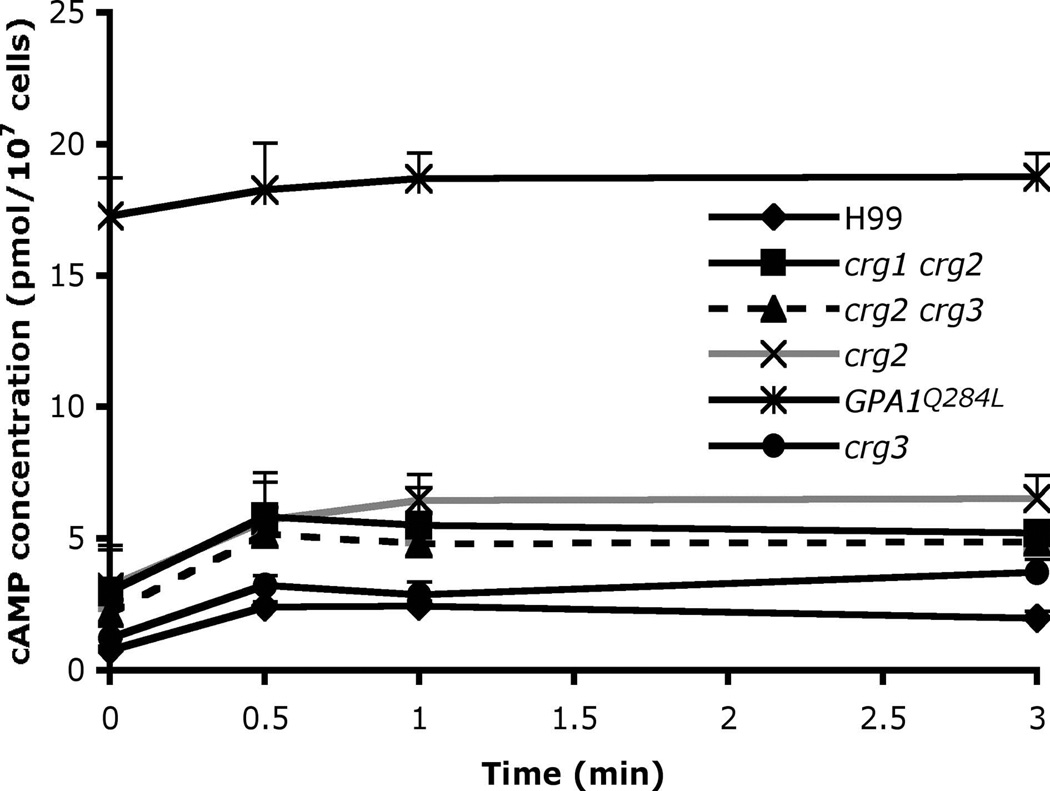
In the strains expressing GPA1Q284L, the basal level of cAMP production was over 10-fold higher than wild type strains (Figure 3). The defect in sporulation might be caused by the extremely high level of internal cAMP production in GPA1Q284L expressing strains. This connection suggested crg2 mutants might also have higher cAMP levels. The cAMP level was indeed found to be significantly higher in the crg2 mutant strain compared to wild type. Compared with the over 10-fold higher cAMP level produced in the Gpa1Q284L strain, the crg2 mutant strain showed ~2-fold increased basal cAMP levels compared to wild type, and it remained responsive to glucose induction (Figure 3). These results suggest that Crg2 plays a negative role in cAMP signaling, consistent with the hypothesis that Crg2 stimulates turnover of Gpa1-GTP. A crg3 mutant strain produced cAMP at wild type levels, indicating Crg3 may not be involved in Gpa1-cAMP signaling (Figure 3). We also tested the cAMP level in crg2 crg3 double mutants under glucose induction, and no significant difference compared to crg2 single mutants was observed, indicating that there is no functional redundancy between Crg2 and Crg3 on cAMP signaling regulation (Figure 3). cAMP production was also measured in a crg1 crg2 double mutant and no functional redundancy between Crg1 and Crg2 was observed (Figure 3). These results suggest that Crg1 and Crg3 are not important for Gpa1-cAMP signaling regulation.
Figure 3. Crg2 negatively regulates cAMP production in response to glucose.
The wild type strain H99, crg2 (CDX50), crg3 (CDX52), GPA1Q284L (CDX156), crg1 crg2 (CDX115), and crg2 crg3 (CDX193) strains were starved for glucose for 2 hr, 1 ml of cell culture for each strain was extracted, and cAMP levels were measured at the indicated time points after glucose re-addition. Each data point and error bar indicates the mean and standard error for three independent measurements.
Crg2 interacts with Gα proteins
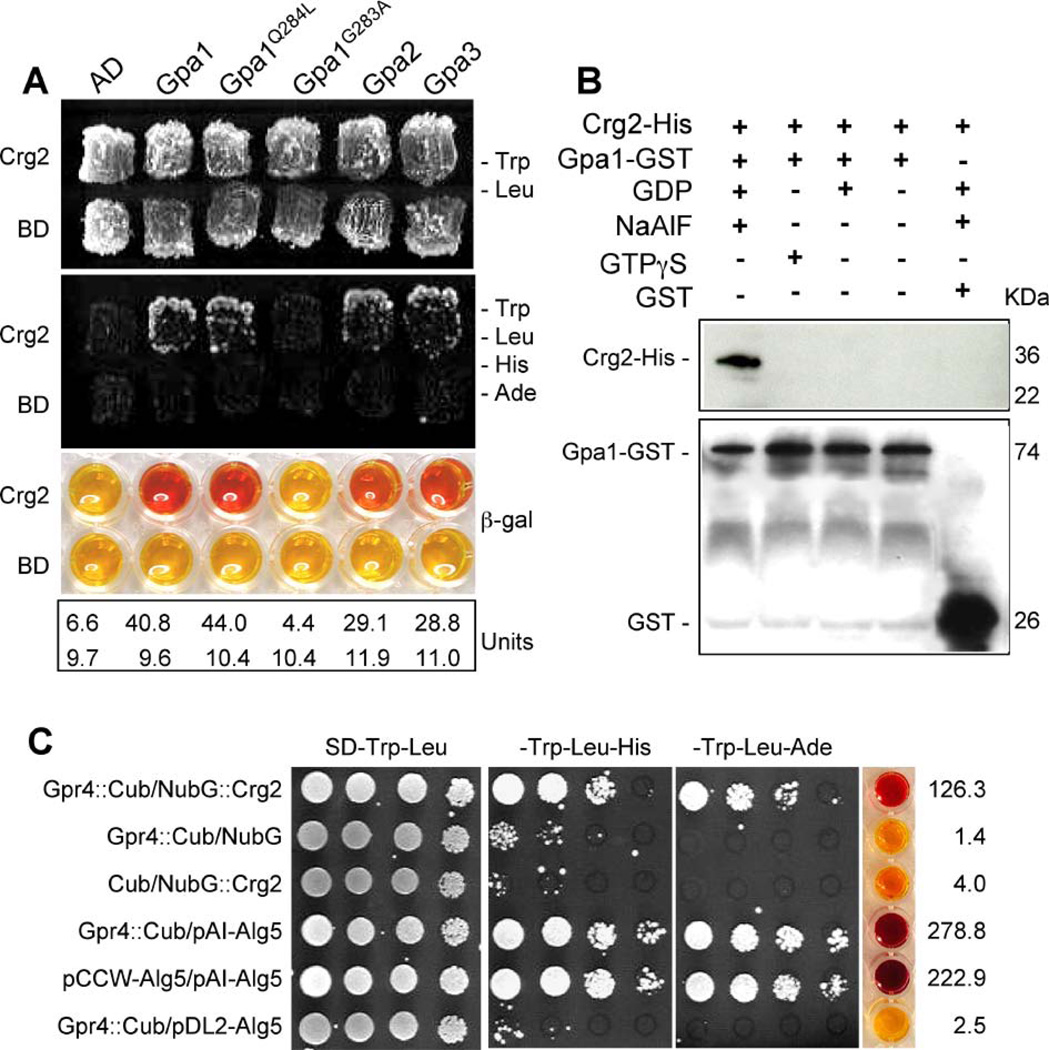
These phenotypic studies provide evidence that Crg2 is also a regulator of the Gpa1-cAMP signal pathway. We then tested whether Crg2 regulates this signaling via a direct association with Gpa1. Protein-protein interactions between Crg2 and Gpa1, Gpa2, and Gpa3 were evaluated in the yeast two-hybrid system. To test the potential interactions between Crg2 and G proteins, a truncated CRG2 cDNA including the first 909 nucleotides, which excludes the transmembrane domains, was expressed as the bait construct and a full-length cDNA of the GPA1 gene, the Gpa1 dominant active allele GPA1Q284L, and the Gpa1 dominant negative allele GPA1G283A were expressed as the prey constructs. The prey constructs containing full-length cDNA of GPA2 and GPA3 were also generated. Our results revealed that Crg2 interacts with both the wild type Gpa1 and the GTPase deficient Gpa1Q284L proteins in the yeast two-hybrid assay. No interaction was observed between Crg2 and Gpa1G283A, which remains in the GDP-bound conformation (Figure 4A). Interactions between Crg2 and Gpa2 or Gpa3 were also detected in these yeast two-hybrid interaction assays as expected based on previous phenotypic analysis (Hsueh et al., 2007). The phenotypic observations of crg2 mutants that link it to both Gpa1-cAMP pathway and pheromone response pathway suggest the interactions between Crg2 and Gα proteins are specific and biologically relevant, providing further evidence that Crg2 is a G protein regulator that coordinates both cAMP and pheromone responsive signaling.
Figure 4. Crg2 interacts with three Gα subunits.
A) Crg2 interacts with both Gpa1 and a Gpa1 dominant active mutant in the yeast two-hybrid interaction assay. The first 910 bp of the CRG2 cDNA was fused with the activation domain (AD), while the full-length cDNA of GPA1 and GPA1Q284L was fused with the binding domain (BD). Both fusion constructs were introduced into the yeast host strain PJ69-4A and colonies grown on SD medium without tryptophan and leucine were tested on medium also lacking histidine and adenine. β-galactosidase enzyme activity was measured for each strain. B) The interaction between Crg2 and Gpa1 was verified in an in vitro binding assay. Both Crg2-His fusion protein and Gpa1-GST fusion protein were expressed and purified from E. coli strain DL21 (DE3). Gpa1-GST protein extract was mixed with glutathione agarose to immobilize the Gpa1-GST protein. Crg2-His protein extract was added to the washed Gpa1-GST-agarose and the in vitro binding assay between the two tagged proteins was performed by incubating at 4°C for 1 h in the presence of NaAlF and GDP, GTPγS, GDP, or no nucleotide. After washing the agarose, protein mixtures were eluted and loaded on SDS-PAGE gel. Western blot was performed with His antibody (upper panel) and GST antibody (bottom panel). C) Interactions between Crg2 and Gpr4 were observed in the split-ubiquitin system. The C-terminal half of ubiquitin (Cub) was fused to the C-terminus of Gpr4 cDNA (Gpr4::Cub). The N-terminal half of ubiquitin (NubG) was fused to the N-terminus of Crg2 cDNA (NubG::Crg2). Gpr4::Cub interaction with the control vector pAI-Alg5 served as a control to ensure the correct topology of the Gpr4::Cub fusion protein. Gpr4::Cub interaction with the empty vector pDSL-NX and pDL2-Alg5, NubG::Crg2 interaction with empty vector pCCW served as negative controls, and pAI-Alg5 interaction with pCCW-Alg5 served as a positive control. β-galactosidase activity assays were performed to further verify the interactions. Each number in units was averaged from two independent experiments.
We further confirmed the interaction between Crg2 and Gpa1 via an in vitro binding assay. A Crg2–6xHis fusion protein and Gpa1-GST fusion protein were expressed in E. coli strain BL21(DE3), purified, and interaction assays were performed. As shown in Figure 4B, the Crg2-His protein directly bound to Gpa1-GST in the presence of GDP and AlF4-, indicating that the intermediate transition state for nucleotide hydrolysis on Gpa1 likely mediates its binding to Crg2. No binding of Crg2 to Gpa1-GDP or Gpa1-GTPγS was detected. These results provide further evidence that Crg2 physically and functionally interacts with the Gpa1-cAMP pathway.
Crg2 was also found to interact with Gpr4, the GPCR that functions upstream of the Gpa1 G protein and activates cAMP signaling (Xue et al., 2006). Because both Crg2 and Gpr4 contain transmembrane domains, the interaction assay between these two proteins was carried out in the split-ubiquitin system. Based on prediction of the protein topological structure, Gpr4 contains a long C-terminal tail localized in the cytosol, while the N-terminus of Crg2 protein has a cytoplasmic localization. An interaction was only observed when half of ubiquitin (Cub) was fused to the C-terminus of Gpr4, and the other half of ubiquitin (NubG) was fused to the N-terminus of Crg2 (Figure 4C). No interaction was detected when Cub was fused to the N-terminus of Gpr4, or NubG was fused to the C-terminus of Crg2 (data not shown). These results indicate that Crg2, Gpa1, and Gpr4 may function as a protein complex.
Crg2 plays a role in virulence factor development
There are several well-characterized virulence factors in C. neoformans, including melanin, the polysaccharide capsule, and the ability to grow at mammalian body temperature (37°C). To understand the potential role of Crg2 in virulence, melanin production and capsule formation assays were performed as described in materials and methods. Under standard conditions (Niger seed agar with 0.1% glucose), the levels of melanin produced by wild type and crg2 mutant strains were similar. But when melanin production was assayed under less favorable conditions (Niger seed agar with 1% glucose and incubation at 37°C), melanin production was repressed in wild type cells whereas colonies of crg2 mutants still produced melanin. These results reinforce the model that Crg2 stimulates turnover of Gpa1-GTP and negatively regulates Gpa1-cAMP signaling. Consistent with this model, crg2 gpa1 double mutants showed a significant reduction in melanin production, similar to gpa1 mutants (Figure 5A). However, strains expressing the Gpa1 dominant active allele GPA1Q284L did not show enhanced melanin production (Figure 5A). The reason for the difference in melanin production between crg2 mutants and Gpa1Q284L strains could be because of the differing levels of cAMP produced. Too much cAMP production inhibits cell melanization, as previously reported (D'Souza et al., 2001). We tested this hypothesis by conducting the melanin assay on Niger seed medium containing 1% glucose with or without 1 mM cAMP. As shown in Figure 6D, additional cAMP indeed inhibited melanin production in wild type and crg2 mutants, supporting this hypothesis. To elucidate the possibility of functional redundancy between Crg2 and other RGS proteins in cAMP signaling regulation, melanin production of crg1 crg2 and crg2 crg3 double mutants was tested and found to be similar to crg2 single mutants, suggesting that Crg1 and Crg3 are not important regulator of melanin production (Figure 6D).
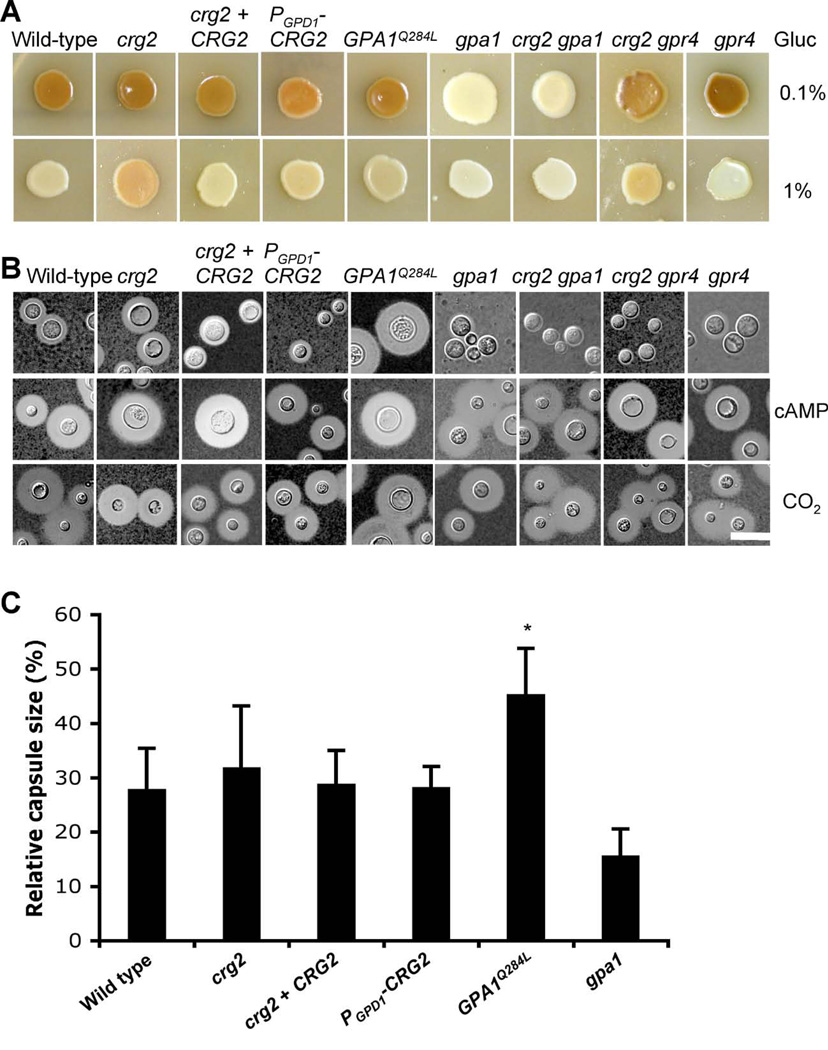
Figure 5. Melanin and capsule production increase in crg2 mutants.
A) Melanin production was assayed on Niger seed media containing 0.1% of glucose (upper panel), or 1% glucose (lower panel). Melanin levels produced by the strains tested on these two media were photographed after incubation for 3 days at 37°C. B) Capsule formation was assayed under three different conditions. Cells were incubated at 37°C on DME medium (upper panel), DME medium containing 10 mM cAMP (middle panel), and DME under 4 to 10% CO2 conditions (lower panel). Capsule production was visualized by India ink staining after growth on these media for 3 days. Bar, 10 µm. C) Quantitative measurement of the relative capsule diameter under standard conditions (See materials and Methods). Each data point and error bar indicates the standard error of the mean for more than 50 cells. Statistical analysis was performed based on a student’s t-test. * indicates statistical significance (P<0.05).
In capsule formation assays, strains expressing the GPA1Q284L allele produced larger capsules compared to wild type. crg2 mutants appeared to produce a slightly larger capsule size on average compared to the wild type, but this trend did not reach statistical significance based on the student’s t-test (Figure 5B and C). The difference in capsule size between the crg2 and GPA1Q284L strains may be also caused by differences in cAMP levels in the cells. Yeast cells may require a high concentration of cAMP to induce large capsules and typically the addition of 10 mM cAMP to DME medium is required to trigger wild type cells to produce capsule sizes similar to those produced by cells expressing GPA1Q284L. Exogenous cAMP also triggers larger capsule production in crg2, crg2 gpr4 and crg2 gpa1 strain backgrounds, indicating that all of these proteins function upstream of the point of cAMP action in the signaling pathway. It is also possible that Gpa1 may have additional targets that promote capsule production. When cells were incubated in high CO2 conditions (4 to 10%) during the capsule formation assay, all strains tested also produced large capsules. Because adenylyl cyclase (Cac1) has been identified as a direct sensor of CO2/HCO3- (Bahn et al., 2005a; Bahn and Muhlschlegel, 2006; Mogensen et al., 2006), the capsule results under high CO2 conditions further support that Crg2, Gpr4, and Gpa1 all function upstream of Cac1 (Figure 5B). Capsule sizes in a crg2 gpr4 double mutant were smaller than wild type, similar to gpr4 mutants. crg2 gpa1 double mutants showed a similar defect as the gpa1 single mutants, consistent with the model in which Crg2 regulates Gpa1. These results also indicate that Crg2 is a negative regulator of Gpa1, and plays a modest albeit detectable effect in virulence factor development. Both crg1 crg2 and crg2 crg3 double mutants were also assayed for capsule production and no significant difference was observed (data not shown).
Crg2 functions upstream of the Gpa1 Gα protein to regulate cAMP signaling
Further epistasis studies were conducted to understand how Crg2 regulates cAMP signaling. A CRG2 overexpression allele was introduced into several mutant strains lacking genes encoding proteins important for controlling cAMP signaling, including the Gα protein Gpa1, protein kinase A Pka1, and the phosphodiesterase Pde1. Overexpressing CRG2 in the gpa1 and pka1 mutant backgrounds did not suppress the mating, capsule, or melanin defect of these mutants (Figure 6A and B). These results are in accord with Crg2 functioning upstream of Gpa1 and Pka1 in cAMP signaling. Pde1 converts cAMP into AMP, and its deletion mutation only causes modest effects on capsule and melanin production. Overexpression of CRG2 also did not alter pde1 mutant phenotypes with respect to mating, capsule production, or melanin production (Figure 6).
The GPA1 dominant active allele (GPA1Q284L) was also introduced into a crg2 mutant background. Dominantly activated GPA1 yielded significantly enlarged capsule production in crg2 mutants, similar to when expressed in a wild type background, suggesting Gpa1 functions downstream of Crg2 (Figure 6B). crg2 gpa1 double mutants exhibit a severe defect in capsule and melanin production, similar to the gpa1 single mutant, also providing epistasis evidence that Crg2 functions upstream of Gpa1. Gpr4 is a G protein coupled receptor that activates cAMP signaling via Gpa1. crg2 gpr4 double mutants produced smaller capsules, similar to the gpr4 single mutant (Figure 5).
Crg2 positively regulates virulence
The involvement of Crg2 in cAMP signaling prompted us to investigate its possible role in virulence using a murine inhalation model of systemic C. neoformans infection. Female A/Jcr mice were intranasally inoculated with 105 yeast cells, and animals were monitored twice daily. All mice infected with wild type strain H99 survived between 18 to 25 days post-infection, in accord with previous results (Xue et al., 2006). In contrast, both crg2 mutants and the GPA1Q284L strain showed a significant virulence defect, and animals infected with either crg2 or GPA1Q284L strains survived between 30 to 50 days post-infection (Figure 7A). This virulence assay has been repeated three times with three different independent crg2 mutants (CDX50, CDX51, and CDX111) and three different independent GPA1Q284L expression strains (CDX156-2, CDX156-4, and CDX40) with consistent findings (Supplemental Figure 1).
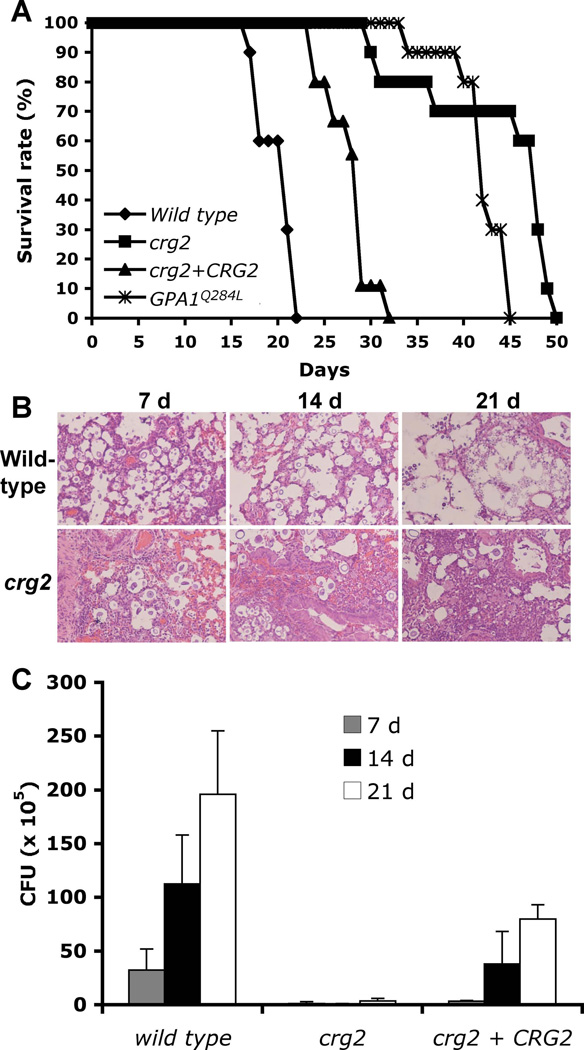
Figure 7. Crg2 regulates virulence in a murine inhalation model.
A) Female A/Jcr mice were intranasally inoculated with 105 cells of the following strains: wild type (H99), crg2 mutant (CDX50), crg2 + CRG2 complemented strain (CDX112), and GPA1Q284L overexpression strain (CDX156-2). Animals were monitored for clinical signs of cryptococcal infection and sacrificed at predetermined clinical end points that predict imminent mortality. The crg2 mutant and the GPA1Q284L expression strain are both significantly reduced virulence compared to wild type strain H99 (P<0.001). The crg2 + CRG2 complemented strain is also significantly more virulent than the crg2 mutant (P<0.001). The virulence difference between the crg2 mutant and the GPA1Q284L strain is not significant (P=0.0177). B–C) Progression of crg2 infection in vivo. Lungs from animals infected with crg2 mutant and wild type were isolated at 7, 14, and 21 days post-infection. H&E stained slides were prepared from lung cross sections and visualized by light microscopy. Colony forming units (CFU) were also measured in lung homogenates (C). Each data point and error bar indicates the standard error of the mean for values from three animals.
To understand why the crg2 mutant has a virulence defect, despite the relatively modest in vitro enhancement in virulence factor development, fungal burdens in organs of infected animals were examined at 7, 14, and 21 days post-infection. Colony forming units (CFU) counts and histopathology results indicate that while crg2 mutant strains could still cause infection in both the lung and brain, cell density and lesion development occurred later and were less extensive compared to animals infected with wild type or the crg2 + CRG2 complemented strains (Figure 7B and C, and data not shown). Because the regulatory subunit of protein kinase A, Pkr1, negatively controls virulence and virulence is enhanced by the activation of Pka1 in a pkr1 mutant (D'Souza et al., 2001), the virulence attenuation in both crg2 mutants and GPA1Q284L strains was not expected and suggests an alternative pathway may contribute to regulate virulence (Figure 7B). Defects in other cellular responses caused by the high level intracellular cAMP in these mutants that ultimately negatively regulate virulence in vivo are also possible.
Discussion
Crg2 plays important roles in both mating and cAMP signaling
Previous studies indicated that the RGS proteins Crg1 and Crg2 play redundant roles in regulating the pheromone response pathway (Hsueh et al., 2007). crg2 mutations significantly enhance dikaryotic mating hyphae production following cell-cell fusion, whereas crg1 mutations enhance conjugation tube formation prior to cell-cell fusion. Pheromone gene expression was significantly more induced in both crg1 and crg2 mutant strains compared to wild type under mating conditions. Also, Crg2 directly interacts with the pheromone receptor Ste3α in the split-ubiquitin yeast two-hybrid system (Hsueh et al., 2007).
Besides its role in regulating the pheromone response pathway, our results indicate that Crg2 also plays a role in Gpa1-cAMP signal transduction. The fact that the CRG2 gene is expressed constitutively under conditions tested, and is not pheromone induced, is another indication that this RGS protein might have a broader role than Crg1. The higher level of cAMP produced in a crg2 mutant strain under glucose induction conditions implicates that Crg2 serves as an important negative regulator of cAMP signaling. The direct interaction observed between Crg2 and Gpa1 provides further evidence that Crg2 is an RGS protein regulating Gpa1 Gα protein signaling. We also show by high resolution scanning electron microscopy (SEM) (Figure 2) that Crg2 plays a key role in promoting sporulation. Basidia in α × a wild type crosses are decorated with four long spore chains whereas α crg2 × a crg2 mutant crosses yield basidia decorated with predominantly only four spores, and in some cases five. This phenotype is reminiscent of crosses with strains expressing GPA1Q284L, or lacking the carbonic anhydrase Can2 (Bahn et al., 2005). Thus, hyperactive PKA signaling (via crg2 or dominant active Gpa1) or inhibition of PKA signaling (reduced cyclase activation in can2 mutants) impairs the production of long spore chains that normally occurs via mitotic division of post-meiotic nuclei.
It is not unprecedented that a single RGS protein can regulate multiple signal pathways. Rgs1 in M. grisea has recently been found to regulate all three G protein α subunits, controls important developmental events during asexual and pathogenic development, and physically interacts with all three Gα subunits (MagA, MagB, and MagC) (Liu et al., 2007). Rgs1 regulates MagA during pathogenesis and MagB during asexual development. In humans, RGS4 has been shown to interact with both Gαi and Gαq, but not with Gαs or Gα12 (Huang et al., 1997). In A. nidulans, three G proteins and five RGS proteins have been identified. In addition to FlbA, which regulates the FadA G protein (Lee and Adams, 1994; Yu, 2006), RgsA regulates the second Gα subunit GanB (Han et al., 2004) based on genetic analysis. The functions of three other RGS proteins remain to be understood (Yu, 2006), and it is still unclear whether there is crosstalk between these G protein signaling pathways through the regulation of RGS proteins. It is likely that similar G protein regulation paradigms also exist in other ascomycetous and basiomycetous fungi that remain to be investigated.
As a negative regulator of the Gpa1 protein, crg2 mutants would be hypothesized to exhibit similar phenotypes compared to cells expressing the Gpa1 dominant active allele GPA1Q284L. Interestingly, while crg2 mutants showed enhanced melanin production when incubated in a less favorable induction condition, such as Niger seed medium containing 1% glucose, they did not show a striking phenotype in capsule production, in contrast to GPA1Q284L expressing strains that have somewhat reduced melanin production and significantly enlarged capsule size. Capsule production in crg2 mutants was not statistically significantly larger compared to wild type cells (Figure 5C). The phenotypic differences between crg2 mutants and GPA1Q284L expressing strains in melanin and capsule production could be caused by the different basal level of cAMP production in these two different strain backgrounds. GPA1Q284L expressing strains have around 10-fold higher basal cAMP level compared to wild type strains, whereas crg2 mutant strains have a more modest 2-fold increased level of cAMP compared to wild type (Figure 3). The reason for this may be that in the Gpa1 dominant active strain, all Gpa1 molecules are locked in the GTP state, whereas in the crg2 mutant background a more limited pool of Gpa1-GTP accumulates because of continual spontaneous GTP hydrolysis by Gpa1. The difference in level of GTP-bound Gpa1 would result in different levels of adenylyl cyclase activity, and in turn affect cAMP production.
When 10 mM exogenous cAMP was added to capsule production medium, both wild type and crg2 mutants produced capsule sizes similar to those induced by GPA1Q284L, indicating that high cAMP levels may be required for larger capsule production, and crg2 mutants may lack sufficient cAMP levels to produce enlarged capsules even though the cAMP level is significantly higher than wild type strains. With respect to melanin production, the strain expressing the GPA1 dominant active allele showed no enhancement, consistent with the fact that high concentrations of cAMP inhibit melanin production. The negative regulation of melanin production by high concentrations of cAMP has been well documented. First, high levels of exogenous cAMP have been found to repress melanin production (D'Souza et al., 2001). Second, the deletion mutation of the catalytic subunit of PKA (pka1) eliminates a feedback loop that inhibits the activity of adenylyl cyclase (Cac1) and triggers dramatic elevation of cAMP level, but pka1 mutants have significant melanin defects (D'Souza et al., 2001; Hicks et al., 2005). The deletion mutation of the regulatory subunit of PKA, pkr1, also triggered enlarged capsule and reduced melanin production, consistent with the phenotype of the GPA1Q284L expressing strain. When we tested melanin production on Niger seed agar containing 1% glucose, addition of cAMP indeed further inhibited melanin production in wild type, crg2 mutants, and GPA1Q284L expressing strains, but increased melanin production of gpa1 mutants (Figure 6). These results provide direct evidence to support our hypothesis of the effects of cAMP levels on melanin production.
On the other hand, it is also possible there is another layer of regulation for Gpa1 signaling besides Crg2, such as additional negative regulators for Gpa1 Gα protein signaling that could exist and may contribute to inactivation of Gpa1 signaling. Crg1 and Crg3 are likely not important for regulation of cAMP signaling based on our studies of crg1 crg2 and crg2 crg3 double mutants. Recently, a fourth RGS protein has been found (Dr. Thomas Wilkie, personal communication), but its function remains to be investigated. It will be interesting to see whether this new RGS protein is also involved in Gpa1 Gα protein regulation.
Crg2, Gpr4, and Gpa1 form a protein complex to control cAMP signaling
Our epistasis analysis supports the hypothesis that Gpr4, Crg2, and Gpa1 function as a protein complex to control cAMP signaling (Figure 8). Gpr4 has been discovered as an amino acid receptor that regulates Gpa1-cAMP signaling (Xue et al., 2006). gpr4 mutants exhibit defects in capsule and mating, but no significant melanin difference was observed. crg2 gpr4 double mutants produced a level of melanin similar to crg2 single mutants. They also showed a reduction in capsule size, similar to that caused by the gpr4 mutation. A crg2 gpa1 mutant exhibited the phenotypes of gpa1 single mutants, consistent with the conclusion that Crg2 is a regulator of Gpa1.
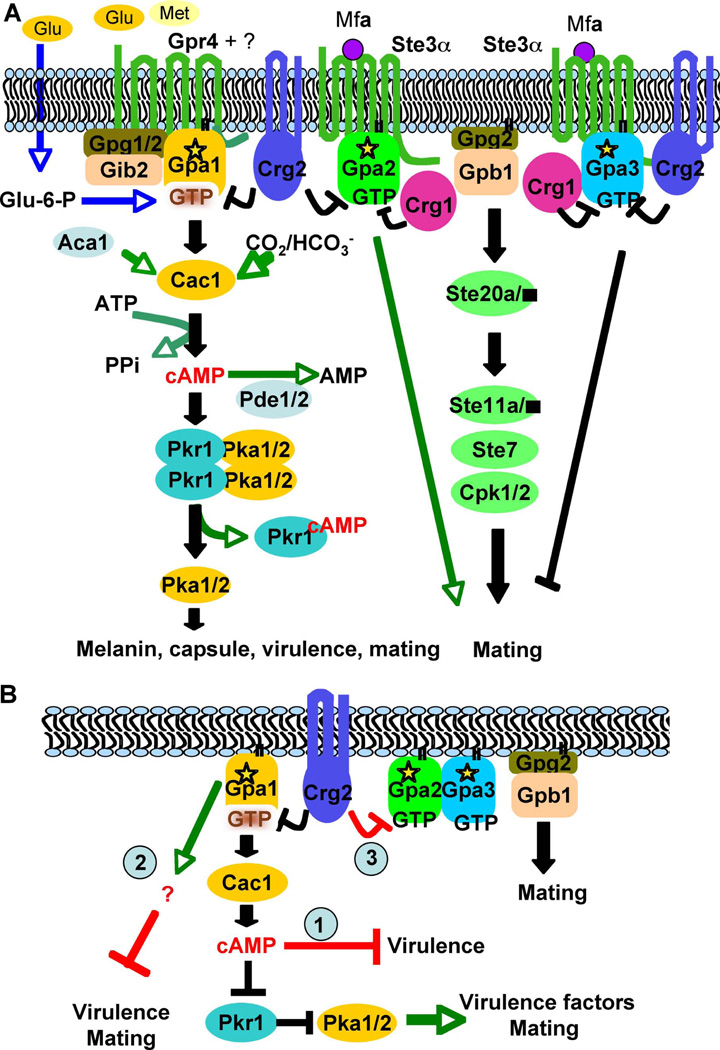
Figure 8. Model for the involvement of Crg2 in G protein complex regulation.
(A) In this model, the Gpr4 receptor activates Gpa1 and Crg2 functions as an RGS protein regulating Gpa1 inactivation. Crg2, Gpa1, and Gpr4 form a functional protein complex. Gib2 and Gpg1 function as βγ subunits that interact with the Gpa1 Gα subunit to govern Gpa1-cAMP signaling. Upon activation, the expression of Gpa2 is induced, and Gpa2 controls signaling by binding and releasing the Gβγ subunits. The active form of Gpa2 also plays a positive role in pheromone response that leads to mating, whereas the activated form of Gpa3 inhibits mating. Both Crg1 and Crg2 interact with the pheromone receptor Ste3 and the Gα subunits Gpa2 and Gpa3, and function to constrain Gpa2 and Gpa3 signaling by stimulating GTPase activity. (B) Three possible models on how Crg2 may be involved in the control of virulence. Model 1, crg2 mutants and GPA1Q284L expressing strains overproduce intracellular cAMP, which causes yeast cells to become more sensitive to stress responses than wild type and thus exhibit altered fitness in the host environment and overwhelm the impact of Pka1 activity. Model 2, the Ga protein Gpa1 also activates an alternative downstream signaling cascade in addition to the Cac1-cAMP-Pka1 signaling cascade, and this alternative target inhibits virulence and mating. Gpa1 activates both targets to maintain yeast cells in dynamic balance between virulence and mating. Model 3, interactions between Crg2 and Gpa2 and Gpa3 could also contribute to virulence.
A direct interaction between Crg2 and Gpa1 was observed in the yeast two-hybrid assay and in vitro protein binding assays. A direct interaction between Crg2 and the Gpr4 receptor was also observed in the split-ubiquitin system. A previous study also demonstrated that Gpr4 interacts with Gpa1 (Xue et al., 2006). Our results support the conclusion that these three proteins can form a protein complex. The interaction results suggest that Crg2 participates in two different G protein complexes: Crg1, Crg2, Ste3, Gpa2, and Gpa3 form protein complexes to regulate mating and monokaryotic fruiting, while Crg2, Gpa1, and Gpr4 form a protein complex to regulate cAMP signaling. These results support models in which Crg2 participates in regulation of both Gpa1-cAMP and pheromone signaling and may serve as a molecular interface between these two central signal transduction modules. Interactions between the RGS protein Sst2 and the pheromone receptor Ste2 have been reported in S. cerevisiae (Ballon et al., 2006), but there is no published data indicating that a dynamic interaction between RGS proteins and receptors also occurs in other G protein regulated pathways, such as the cAMP signaling pathway. Our findings suggest that the formation of protein complexes is more common than appreciated and may function to precisely control the activation and regulation of G protein signaling.
Crg2 plays a positive role in virulence
Strikingly, in the virulence assay using a murine inhalation model, both strains expressing the GPA1Q284L allele and crg2 mutants exhibited attenuated virulence, even though crg2 exhibited a moderate increase in melanin production, and GPA1Q284L strains produced larger capsules. We tested the growth rate of both strains at body temperature (37°C) and found they had similar growth rate as wild type strains H99 and KN99a. In vivo analysis indicated that crg2 mutants can still cause infection and lesion development, but infection progressed more slowly compared with wild type infection.
Previous studies in Cryptococcus revealed that intracellular cAMP binds to the protein kinase A regulatory subunit Pkr1 and promotes the release of the catalytic subunits Pka1 and Pka2 to activate downstream transcription factors (Bahn et al., 2004; D'Souza et al., 2001; Hicks et al., 2004; Hicks et al., 2005; Wang and Heitman, 1999). The virulence results from crg2 mutants and GPA1Q284L strains were unexpected because pkr1 mutants, which trigger high PKA activity, are hypervirulent (D'Souza et al., 2001). There are several possibilities on how Crg2 and Gpa1 are involved in virulence regulation based on our results. One possible explanation is that cells with a high internal cAMP level are more sensitive to stress responses and thus exhibit altered fitness in the host environment, outweighting the impact of increased PKA activity. rgs2 mutants in S. cerevisiae show a reduction in thermo-tolerance (Versele et al., 1999). We tested the heat resistance and the osmotic stress resistance of crg2 mutants, but no significant differences were observed for both stress responses comparing to the wild type (data not shown). Additional tests of other host-related stress factors are necessary to evaluate this model in further detail. The enlarged capsules in vivo may also impede the dissemination of yeast cells.
In addition to the Gα protein Gpa1 positively regulating virulence and mating via cAMP-PKA signaling pathway, it may also negatively regulate virulence via an unknown signaling pathway, independent of the Cac1-cAMP-PKA signaling cascade, that also partially inhibits mating. This model can explain our earlier observations why wild type and gpa1 crosses respond differently towards exogenous cAMP stimulation. When 1 mM cAMP is added to V8 mating medium, both wild type and gpa1 dikaryotic mating hyphae production were stimulated, while addition of 10 mM cAMP inhibits wild type mating but further induces gpa1 mating. To test this model, expression of the GPA1Q284L allele in the pkr1 mutant background should help dissect the existence of this additional signaling branch, which is beyond the scope of this study. It is also possible that consistent release of βγ subunits in a Gpa1Q284L mutant could trigger additional signaling events. Finally, recent studies reveal that the pheromone response pathway is also important for regulating virulence (Chang et al., 2003; Nielsen and Heitman, 2007), and the interactions between Crg2 and Gpa2 and Gpa3 may also alter virulence without a direct involvement of virulence factors (Figure 8B).
A more complicated signaling system than in S. cerevisiae
The functions of both RGS proteins in S. cerevisiae have been studied and provide a model for RGS regulation of both the pheromone response and cAMP pathways (Ballon et al., 2006; Versele et al., 1999). In this organism, Sst2 functions as a negative regulator of Gpa1 to control mating, while Rgs2 plays a role in the regulation of cAMP signaling via Gpa2. There is no indication that any RGS protein promotes cross talk between these two pathways. Compared to this simple model in S. cerevisiae, our results uncovered a more complicated scenario of G protein signaling in C. neoformans. Two RGS proteins, Crg1 and Crg2, play related but distinguishable roles in regulation of pheromone responses via two Gα proteins, Gpa2 and Gpa3, while Crg2 also enables cross talk with Gpa1-cAMP signaling (Figure 8A). S. cerevisiae has been developed as a very useful model system, but in many cases, the information obtained from studies in S. cerevisiae can not be directly translated to other systems. Our studies on GPCRs, G proteins, and RGS proteins (Hsueh et al., 2007; Xue et al., 2006) indicate that the signal transduction pathways in C. neoformans are more complicated than in S. cerevisiae, further indicating the importance of directed studies to understand this human pathogen and its interaction with the host and environment. Also most fungi studied have at least three Gα proteins and more than two RGS proteins, suggesting the outcome of these studies may provide an expanded model for understanding G protein signaling in other organisms.
Supplementary Material
Acknowledgements
We thank Sarah Covert (University of Georgia) and Thomas Wilkie (University of Texas Southwestern Medical Center) for providing unpublished RGS protein sequences. We thank Kirsten Nielsen and Andrew Alspaugh for assistance with animal experiments, Alisha Holtzhausen for technical assistance, Kasey Carroll for artistic assistance, and Valerie Knowlton for SEM assistance. We thank Hiroaki Matsunami, Andrew Alspaugh, and Michael Price for critical reading of the manuscript. We also acknowledge use of the C. neoformans serotype A project sequencing at Duke University and the Broad Institute. This work was supported by National Institute of Health R01 grant AI39115 and R21 grant AI070230 to C.X. and J.H.
References
- Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit Gpa1 and cAMP. Genes Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apanovitch DM, Slep KC, Sigler PB, Dohlman HG. Sst2 is a GTPase-activating protein for Gpa1: purification and characterization of a cognate RGS-Galpha protein pair in yeast. Biochemistry. 1998;37:4815–4822. doi: 10.1021/bi9729965. [DOI] [PubMed] [Google Scholar]
- Bahn YS, Hicks JK, Giles SS, Cox GM, Heitman J. Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryot Cell. 2004;3:1476–1491. doi: 10.1128/EC.3.6.1476-1491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, Cox GM, Perfect JR, Heitman J. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr Biol. 2005;15:2013–2020. doi: 10.1016/j.cub.2005.09.047. [DOI] [PubMed] [Google Scholar]
- Bahn YS, Kojima K, Cox GM, Heitman J. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans . Mol Biol Cell. 2005b;16:2285–2300. doi: 10.1091/mbc.E04-11-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, Muhlschlegel FA. CO2 sensing in fungi and beyond. Curr Opin Microbiol. 2006;9:572–578. doi: 10.1016/j.mib.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Ballon DR, Flanary PL, Gladue DP, Konopka JB, Dohlman HG, Thorner J. DEP-domain-mediated regulation of GPCR signaling responses. Cell. 2006;126:1079–1093. doi: 10.1016/j.cell.2006.07.030. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Perfect JR. Cryptococcus neoformans. Washington, DC: ASM Press; 1998. [Google Scholar]
- Chang YC, Miller GF, Kwon-Chung KJ. Importance of a developmentally regulated pheromone receptor of Cryptococcus neoformans for virulence. Infect Immun. 2003;71:4953–4960. doi: 10.1128/IAI.71.9.4953-4960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasse SA, Flanary P, Parnell SC, Hao N, Cha JY, Siderovski DP, Dohlman HG. Genome-scale analysis reveals Sst2 as the principal regulator of mating pheromone signaling in the yeast Saccharomyces cerevisiae . Eukaryot Cell. 2006;5:330–346. doi: 10.1128/EC.5.2.330-346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science. 2003;301:1728–1731. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]
- Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. Urease as a virulence factor in experimental cryptococcosis. Infect Immun. 2000;68:443–448. doi: 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza CA, Alspaugh JA, Yue C, Harashima T, Cox GM, Perfect JR, Heitman J. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans . Mol Cell Biol. 2001;21:3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Cruz MC, Sia RA, Allen B, Alspaugh JA, Heitman J. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans . Fungal Genet Biol. 2000;29:38–48. doi: 10.1006/fgbi.1999.1180. [DOI] [PubMed] [Google Scholar]
- Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, D'Souza C, Wang P, Heitman J. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148:2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- Dignard D, Whiteway M. SST2, a regulator of G-protein signaling for the Candida albicans mating response pathway. Eukaryot Cell. 2006;5:192–202. doi: 10.1128/EC.5.1.192-202.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG, Song J, Ma D, Courchesne WE, Thorner J. Sst2, a negative regulator of pheromone signaling in the yeast Saccharomyces cerevisiae: expression, localization, and genetic interaction and physical association with Gpa1 (the G-protein alpha subunit) Mol Cell Biol. 1996;16:5194–5209. doi: 10.1128/mcb.16.9.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohlman HG, Thorner JW. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- Fraser JA, Subaran RL, Nichols CB, Heitman J. Recapitulation of the sexual cycle of the primary fungal pathogen Cryptococcus neoformans var. gattii: implications for an outbreak on Vancouver Island, Canada. Eukaryot Cell. 2003;2:1036–1045. doi: 10.1128/EC.2.5.1036-1045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger DL, Perfect JR, Durack DT. Virulence of Cryptococcus neoformansRegulation of capsule synthesis by carbon dioxide. J Clin Invest. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KH, Seo JA, Yu JH. Regulators of G-protein signalling in Aspergillus nidulans: RgsA downregulates stress response and stimulates asexual sporulation through attenuation of GanB (Galpha) signalling. Mol Microbiol. 2004;53:529–540. doi: 10.1111/j.1365-2958.2004.04163.x. [DOI] [PubMed] [Google Scholar]
- Hicks JK, D'Souza CA, Cox GM, Heitman J. Cyclic AMP-dependent protein kinase catalytic subunits have divergent roles in virulence factor production in two varieties of the fungal pathogen Cryptococcus neoformans . Eukaryot Cell. 2004;3:14–26. doi: 10.1128/EC.3.1.14-26.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JK, Bahn YS, Heitman J. Pde1 phosphodiesterase modulates cyclic AMP levels through a protein kinase A-mediated negative feedback loop in Cryptococcus neoformans . Eukaryot Cell. 2005;4:1971–1981. doi: 10.1128/EC.4.12.1971-1981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Xue C, Heitman J. G protein signaling governing cell fate decisions involves opposing Galpha subunits in Cryptococcus neoformans . Mol Biol Cell. 2007;18:3237–3249. doi: 10.1091/mbc.E07-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Hepler JR, Gilman AG, Mumby SM. Attenuation of Gi- and Gq-mediated signaling by expression of RGS4 or GAIP in mammalian cells. Proc Natl Acad Sci U S A. 1997;94:6159–6163. doi: 10.1073/pnas.94.12.6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:e95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Baptiste G, Yang Z, Greenwood MT. Regulatory mechanisms involved in modulating RGS function. Cell Mol Life Sci. 2006;63:1969–1985. doi: 10.1007/s00018-006-6066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon A, Seo JA, Han KH, Yu JH, D'Enfert C. The heterotrimeric G-protein GanB(alpha)-SfaD(beta)-GpgA(gamma) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans . Genetics. 2005;171:71–80. doi: 10.1534/genetics.105.040584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon A, Han KH, Seo JA, Yu JH, D'Enfert C. G-protein and cAMP-mediated signaling in aspergilli: a genomic perspective. Fungal Genet Biol. 2006;43:490–502. doi: 10.1016/j.fgb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Lee BN, Adams TH. Overexpression of flbA, an early regulator of Aspergillus asexual sporulation, leads to activation of brlA and premature initiation of development. Mol Microbiol. 1994;14:323–334. doi: 10.1111/j.1365-2958.1994.tb01293.x. [DOI] [PubMed] [Google Scholar]
- Lengeler KB, Davidson RC, D'Souza C, Harashima T, Shen WC, Wang P, Pan X, Waugh M, Heitman J. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Shen G, Zhang ZG, Wang YL, Thompson JK, Wang P. Canonical heterotrimeric G proteins regulating mating and virulence of Cryptococcus neoformans . Mol Biol Cell. 2007a;18:4201–4209. doi: 10.1091/mbc.E07-02-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wright SJ, Krystofova S, Park G, Borkovich KA. Heterotrimeric G protein signaling in filamentous fungi. Annu Rev Microbiol. 2007b;61:423–452. doi: 10.1146/annurev.micro.61.080706.093432. [DOI] [PubMed] [Google Scholar]
- Liu H, Suresh A, Willard FS, Siderovski DP, Lu S, Naqvi NI. Rgs1 regulates multiple Galpha subunits in Magnaporthe pathogenesis, asexual growth and thigmotropism. EMBO J. 2007;26:690–700. doi: 10.1038/sj.emboj.7601536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, Miranda M, Anderson IJ, Fraser JA, Allen JE, Bosdet IE, Brent MR, Chiu R, Doering TL, Donlin MJ, D'Souza CA, Fox DS, Grinberg V, Fu J, Fukushima M, Haas BJ, Huang JC, Janbon G, Jones SJ, Koo HL, Krzywinski MI, Kwon-Chung JK, Lengeler KB, Maiti R, Marra MA, Marra RE, Mathewson CA, Mitchell TG, Pertea M, Riggs FR, Salzberg SL, Schein JE, Shvartsbeyn A, Shin H, Shumway M, Specht CA, Suh BB, Tenney A, Utterback TR, Wickes BL, Wortman JR, Wye NH, Kronstad JW, Lodge JK, Heitman J, Davis RW, Fraser CM, Hyman RW. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans . Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen EG, Janbon G, Chaloupka J, Steegborn C, Fu MS, Moyrand F, Klengel T, Pearson DS, Geeves MA, Buck J, Levin LR, Muhlschlegel FA. Cryptococcus neoformans senses CO2 through the carbonic anhydrase Can2 and the adenylyl cyclase Cac1. Eukaryot Cell. 2006;5:103–111. doi: 10.1128/EC.5.1.103-111.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect Immun. 2003;71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K, Heitman J. Sex and virulence of human pathogenic fungi. Adv Genet. 2007;57:143–173. doi: 10.1016/S0065-2660(06)57004-X. [DOI] [PubMed] [Google Scholar]
- Osman A. Yeast two-hybrid assay for studying protein-protein interactions. Methods Mol Biol. 2004;270:403–422. doi: 10.1385/1-59259-793-9:403. [DOI] [PubMed] [Google Scholar]
- Palmer DA, Thompson JK, Li L, Prat A, Wang P. Gib2, a novel Gbeta-like/RACK1 homolog, functions as a Gbeta subunit in cAMP signaling and is essential in Cryptococcus neoformans . J Biol Chem. 2006;281:32596–32605. doi: 10.1074/jbc.M602768200. [DOI] [PubMed] [Google Scholar]
- Perfect JR, Schell WA, Rinaldi MG. Uncommon invasive fungal pathogens in the acquired immunodeficiency syndrome. J Med Vet Mycol. 1993;31:175–179. doi: 10.1080/02681219380000211. [DOI] [PubMed] [Google Scholar]
- Pukkila-Worley R, Alspaugh JA. Cyclic AMP signaling in Cryptococcus neoformans . FEMS Yeast Res. 2004;4:361–367. doi: 10.1016/S1567-1356(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Segers GC, Regier JC, Nuss DL. Evidence for a role of the regulator of G-protein signaling protein CPRGS-1 in Galpha subunit CPG-1-mediated regulation of fungal virulence, conidiation, and hydrophobin synthesis in the chestnut blight fungus Cryphonectria parasitica . Eukaryot Cell. 2004;3:1454–1463. doi: 10.1128/EC.3.6.1454-1463.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siderovski DP, Willard FS. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 2005;1:51–66. doi: 10.7150/ijbs.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versele M, de Winde JH, Thevelein JM. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. EMBO J. 1999;18:5577–5591. doi: 10.1093/emboj/18.20.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Heitman J. Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans . Curr Opin Microbiol. 1999;2:358–362. doi: 10.1016/S1369-5274(99)80063-0. [DOI] [PubMed] [Google Scholar]
- Wang P, Cutler J, King J, Palmer D. Mutation of the regulator of G protein signaling Crg1 increases virulence in Cryptococcus neoformans . Eukaryot Cell. 2004;3:1028–1035. doi: 10.1128/EC.3.4.1028-1035.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland T, Lutz S, Chidiac P. Regulators of G protein signalling: a spotlight on emerging functions in the cardiovascular system. Curr Opin Pharmacol. 2007;7:201–207. doi: 10.1016/j.coph.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Xue C, Bahn YS, Cox GM, Heitman J. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans . Mol Biol Cell. 2006;17:667–679. doi: 10.1091/mbc.E05-07-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Tada Y, Dong X, Heitman J. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell Host & Microbe. 2007;1:263–273. doi: 10.1016/j.chom.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Yu JH. Heterotrimeric G protein signaling and RGSs in Aspergillus nidulans . J Microbiol. 2006;44:145–154. [PubMed] [Google Scholar]
- Zaragoza O, Fries BC, Casadevall A. Induction of capsule growth in Cryptococcus neoformans by mammalian serum and CO2. Infect Immun. 2003;71:6155–6164. doi: 10.1128/IAI.71.11.6155-6164.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Ma YC, Ostrom RS, Lavoie C, Gill GN, Insel PA, Huang XY, Farquhar MG. RGS-PX1, a GAP for GalphaS and sorting nexin in vesicular trafficking. Science. 2001;294:1939–1942. doi: 10.1126/science.1064757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.