Abstract
Single particle tracking is widely used to study protein movement with high spatiotemporal resolution both in vitro and in cells. Quantum dots, which are semiconductor nanoparticles, have recently been employed in single particle tracking because of their intense and stable fluorescence. Although single particles inside cells have been tracked in three spatial dimensions (X, Y, Z), measurement of the angular orientation of a molecule being tracked would significantly enhance our understanding of the molecule’s function. In this study, we synthesized highly polarized, rod-shaped quantum dots (Qrods) and developed a coating method that optimizes the Qrods for biological imaging. We describe a Qrod-based single particle tracking technique that blends optical nanometry with nanomaterial science to simultaneously measure the three-dimensional and angular movements of molecules. Using Qrods, we spatially tracked a membrane receptor in living cells in four dimensions with precision close to the single-digit range in nanometers and degrees.
Introduction
The combination of optical imaging with fluorescence has greatly advanced biological studies by enabling live investigation of protein expression, movement, and localization (1,2). Although the resolution of such optical imaging is constrained by the diffraction limit of light (3), the two-dimensional (2D) position of a single fluorophore can be determined by fitting its fluorescent image to a Gaussian function, thus overcoming the diffraction limit (4). The precision of the position depends on the number of photons emitted from the fluorophore, and can reach just a few nanometers (4,5). Individual fluorophores can be monitored to follow the movement of a protein in vitro, in cells, and even in whole animals (6–9). This method, single particle tracking, has now been expanded to three dimensions (3D; X, Y, Z), offering a powerful and popular tool for biophysical studies (10–13). In one of the techniques of 3D tracking of single fluorophores, bifocal plane microscopy, the differences between distinct optical pathways are used to estimate the Z-position of a fluorophore by contrasting the fluorescence intensities of two focal images (10–12). In 3D tracking using a photon-limited double-helix response system that has two twisting lobes along the optical axis in the image of a single fluorophore, the fluorophore appears as two fluorescent spots from which the Z-position can be determined (13). Here, we report the extension of the spatial dimensions of single particle tracking even further: to 3D tracking we added a fourth spatial component, the angular (θ) component, which gives information about a protein’s rotational orientation inside a living cell.
A critical aspect of successful single particle tracking is the labeling of a target with a fluorescent probe. Many types of fluorescent probes are available for this purpose, including fluorescent proteins from coelenterates that can be used easily for labeling molecules in living cells (14–16). Photobleaching is another property that often determines the choice of a fluorescent probe. Increasing illumination power can improve tracking precision (4), but this also increases the probability of photobleaching (17,18). Consequently, inorganic nanoparticles such as gold nanoparticles have become increasingly popular for precise and long-term tracking (19). However, even better than inorganic nanoparticles are quantum dots (Qdots), the semiconductor fluorescent nanocrystals that today are possibly the most preferred fluorescent probes because of their brightness, narrow spectral distribution, and low photobleaching (20–22). Qdots are used not only in basic research (23), but also in clinical studies (24,25). Using Qdots with bifocal plane microscopy, we achieved 3D single particle tracking with a spatial resolution of 2 nm in the XY plane and 4 nm in the Z axis and a temporal resolution of 2 ms, and were able to observe the behavior of motor proteins in detail during vesicle transport in a living cell (11).
The angular position of a fluorophore can be obtained using fluorescence anisotropy because fluorescence emissions are of unequal intensities along the P and S axes of polarization (P- and S-polarization) (26–28). Anisotropy is defined by r = (Ip-Is)/(Ip+Is), where Ip and Is are intensities in P- and S-polarization, respectively (26). Anisotropy measurements have successfully revealed the angular dynamics of protein movement in vitro and in cells (29–31). The anisotropy of an organic dye has been determined with a precision of <0.1, which translates to ∼10° when illuminated with circularly polarized light (26). However, photobleaching affects organic dyes, and therefore Qdots have been considered as an alternative for anisotropy studies. Because the fluorescence anisotropy of a Qdot depends on the aspect ratio of its shape (32), rod-shaped Qdots, called quantum rods (Qrods), have been synthesized (33,34), but to date the fluorescence anisotropy of Qrods has been applied in only a few biological studies and even fewer single particle tracking studies (35).
We previously used Qrods to observe motile myosin molecules in vitro by singe particle tracking in four dimensions (4D) (X, Y, θ, and φ coordinates), which provided information on the out-of-plane tilt angle (35). This method is applicable when the target protein displays planar movement, as in the case of a protein moving underneath a glass surface, but not when a protein moves dynamically in X-Y-Z coordinates in a living cell. Here, we report a modified single particle tracking approach that includes angular (θ) position in addition to the planar components of 3D single particle tracking, thus distinguishing this method from our previous method. We describe the synthesis of Qrods and the coating of their surface to make the Qrods suitable for long-term 4D spatial tracking of membrane proteins in living cells using our novel, to our knowledge, technique.
Materials and Methods
Chemicals and antibodies
The following reagents were purchased: cadmium oxide (CdO, 99.99%) and selenium (Se, powder, 99.999%) (Sigma-Aldrich); sulfur (S, crystalline, 99%) and hexylphosphonic acid (HPA, 99%) (Strem Chemicals); octadecylphosphonic acid (ODPA, 99%) (Alfa Aesar, UK); tri-n-octylphosphine (TOP) and tri-n-octylphosphine oxide (TOPO) (Tokyo Kasei, Japan); glutathione (GSH, reduced form), potassium t-butoxide (KOBut), and tetrahydrofuran (THF) (Wako, Japan); 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) and sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (sulfo-SMCC) (Thermo Fisher Scientific); 3-sulfo-N-hydroxysuccinimide sodium salt (sulfo-NHS) (Molecular Bioscience); thiol PEG (PEG-SH) and amino PEG (PEG-NH2) (NOF Corporation, Japan); poly(maleic anhydride-alt-1-octadecene) (PMAO) (Sigma-Aldrich); poly-D-lysine (Hycultec GmbH, Germany); and antimouse CD36 mAb (HM36), antimouse CD16/32 mAb (93), and American hamster IgG (HTK204) (BioLegend).
Synthesis of CdSe/CdS Qrods
CdSe/CdS (core/shell) Qrods were synthesized using a modified version of the procedure reported by Deka et al. (34). To synthesize the CdSe core, TOPO (3.00 g), ODPA (0.28 g), and CdO (0.06 g) were placed in a 3-necked flask and degassed at 150°C for 1 h under N2. After heating the reaction mixture to 300°C, TOP (1.8 mL) was injected into the flask, after which a solution of Se (0.058 g) in TOP (0.434 mL) at 370°C was rapidly injected in and the solution was annealed at 370°C for 30 s. After cooling the solution to room temperature, CdSe Qdots were precipitated by adding methanol and then redispersed in TOP (0.5 mL).
To synthesize CdSe/CdS Qrods, CdO (0.06 g) was mixed with TOPO (3.00 g), ODPA (0.290 g), and HPA (0.08 g) in the 3-necked flask and degassed at 150°C for 1 h under N2. After heating the mixture to 300°C, TOP (1.8 mL) was added in, after which a mixture of S (0.12 g) predissolved in TOP (1.8 mL) at 350°C and a CdSe/TOP solution (2 mM, 42 μL) were quickly injected into the flask. The resulting Qrods were allowed to grow at 350°C for 6 min and then annealed at 100°C overnight. After cooling the solution to 60°C, the reaction was halted by adding anhydrous toluene. Qrods were precipitated by adding methanol, and then separated and redispersed in cyclohexane (2.0 mL).
Preparation of glutathione-coated Qrods (GSH-Qrods) and polymer-coated Qrods (Poly-Qrods)
GSH-Qrods were prepared by a ligand-exchange method, in which the capping ligands TOPO, ODPA, and HPA are exchanged with GSH (36). An aqueous GSH solution (120 mg/mL, 3.0 mL) was mixed at room temperature with 6.0 mL of a THF solution containing 1.7 mM Qrods, and the mixture was heated to 70°C. After separating GSH-Qrod precipitates, deionized water (1.0 mL) and KOBut (10 mg) were added to the precipitates to deprotonate the GSH carboxyl groups. Excess GSH and KOBut were removed by dialysis (300 kDa cellulose acetate membrane, Harvard Apparatus, was used here and in all instances of dialysis mentioned below).
Poly-Qrods were prepared by the method of Yu et al. (37). To generate amphiphilic polymers (PMAO-PEG) for use as coating agents, PMAO (84 mg) was reacted with primary PEG-NH2 (300 mg) in 12 mL chloroform overnight at room temperature. The PMAO-PEG (0.8 mL) was mixed with CdSe/CdS Qrods in chloroform (4 mL) and stirred for 3 days at room temperature. Deionized distilled water (10 mL) was added to the solution, the chloroform was evaporated at room temperature, and the excess PMAO-PEG was removed by dialysis.
Preparation of antimouse CD36 mAb-conjugated Qrods (mAb-Qrods)
To block the nonspecific binding of GSH-Qrods to the plasma membrane, the GSH amino groups were reacted with PEG. First, GSH-Qrods (280 nM, 200 μL) were incubated with sulfo-SMCC (10 mM, 2.8 μL) for 20 min and then reacted with PEG-SH (4 mM, 14 μL) and gently stirred for 2 h at room temperature; unreacted reagents were removed by dialysis. Next, antibodies were conjugated to the carboxylic groups of GSH. The solution of PEG-conjugated GSH-Qrods was incubated with EDC (1 mM, 5.6 μL) and sulfo-NHS (1 mM, 11.2 μL) for 20 min at room temperature and then dialyzed to remove unreacted EDC and sulfo-NHS. Antimouse CD36 monoclonal antibodies (0.5 mg/mL, 84 μL) were incubated overnight with the dialyzed solution at 4°C. The reaction was quenched by adding PEG-NH2 (2 mM, 5.6 μL), and unreacted antibodies and PEG-NH2 were removed by dialysis.
Characterization of Qrods
All characterizations were conducted in phosphate buffered saline. Absorption and fluorescence spectra of Qdots and Qrods were measured using a U-1900 spectrometer (U-1900, Hitachi High-Technologies, Japan) and an FP-6200 spectrometer (FP-6200, Jasco, UK), respectively. The absolute quantum yield was determined using a QY measurement system (C10027, Hamamatsu Photonics, Japan). Qrod nanocrystals were examined using a transmission electron microscope (TEM) (H-800, Hitachi High-Technologies, Japan) at 200 kV. TEM samples were prepared by dropping a 2 mL solution of Qrods onto a carbon-coated copper grid and letting it stand overnight to evaporate the solvent.
The hydrodynamic diameters of GSH-Qrods and Poly-Qrods were determined by dynamic light scattering (DLS) at room temperature (Zetasizer Nano ZS, Malvern Instruments, UK). The conjugation of antimouse CD36 mAbs to GSH-Qrods was confirmed using fluorescence correlation spectroscopy (FCS) (38,39), as described in our previous reports (36,40). Fluorescence autocorrelation curves were collected at room temperature with a compact FCS apparatus (C9413-01MOD, Hamamatsu Photonics). The excitation power at the objective lens was 15 mW, and the measurement time and sample volume for FCS were 10 s and 5 mL, respectively. The confocal pinhole diameter was adjusted to 50 μm. Emission signals were detected at >568 nm. The FCS curves can be theoretically expressed as follows (36,39):
where N is the average number of Qrods in the confocal volume, ω0 and ωz are the width and length of the confocal volume, D is the diffusion coefficient, τd is the diffusion time of the Qrods (which depends on D and ω0), and i is the number of the components (for example, i = 1 assuming 1-component diffusion) (36).
Microscopy for 3D and angular single particle tracking
The microscopy system used consisted of an epi-fluorescence microscope (IX-71, Olympus, Japan), an objective lens (60× PlanApo, 1.45 NA, oil, Olympus, Japan), a light-resistant container for relay optics (GA01, G-angstrom, Japan), and an electron multiplier-type charge-coupled device (CCD) camera (EM-CCD, Ixon DV887, Andor Technology, UK). An ∼30 × 30 μm2 area was illuminated by a blue (473 nm) laser (J050BS-18-11-11, Showa Optronics, Japan). The excitation light was circularly polarized by introducing polarizing and λ/4 wave plates (26). The system included both polarized and cylindrical optics (Fig. 1). The optical pathway was split into two by a polarizing beam splitter (BS). Pairs of lenses, L1 and L3 or L2 and L3, relayed the fluorescence image to the camera and magnified the image 2.5 times. The focal distances of the lenses were 100 mm for L1 and L2, and 250 mm for L3. Moving either L1 or L2 along the optical axis adjusted the optical lengths between the two lenses. The pathways were projected side by side onto the camera by a prism mirror (Pr). Images obtained by the CCD camera were composed of 2 square images (256 × 256 pixels), one P-polarized and one S-polarized (see examples in Fig. 5 B, left and right, respectively). Thus, the CCD camera simultaneously acquired P-polarized and S-polarized images. Concave and convex cylindrical lenses (CnC and CvC) were set behind L3 for 3D measurements (−200 and 200 mm focal distances, respectively). For angular calibration, a 1/2 λ wave plate was set before the S. The objective lens was moved by a Piezo actuator (P-721.CDQ, Physik Instrumente, Germany) and a feedback loop (E-665.CR, Physik Instrumente). The microscopic stage was moved by another Piezo actuator (P-733.2CD, Physik Instrumente) and feedback loop (E-501.10, Physik Instrumente). The Piezo actuators were controlled using a computer to synchronize image acquisitions. The CCD camera obtained images (composed of 14-bit data for each pixel) that were imported into an analysis software program made using Visual C++ (Visual studio 2005, Microsoft) and OpenCV library (IBM).
Figure 1.
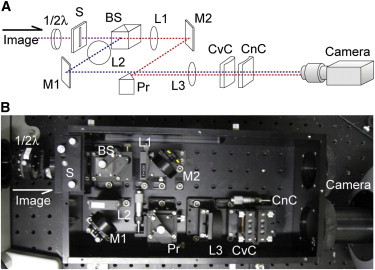
Optical system for simultaneous 3D and angular single particle tracking. Schematic drawing (A) and photograph (B) of the optical setup for simultaneous 3D and angular single particle tracking. 1/2 λ, 1/2-wave plate; S, slit; BS, beam splitter; L, lens; M, mirror; Pr, prism; CvC, convex cylindrical lens; and CnC, concave cylindrical lens. See Materials and Methods for details.
Figure 5.
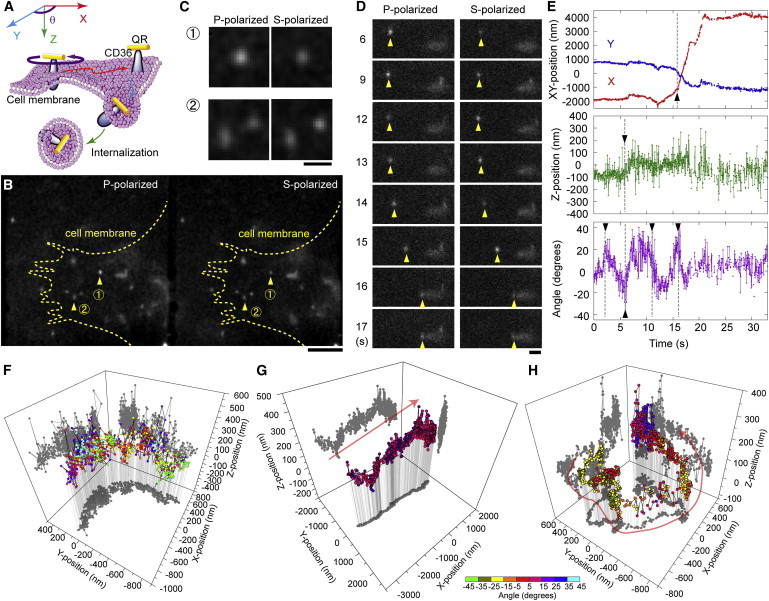
Single particle tracking in 4D of CD36 in macrophages using mAb-Qrods. (A) Schematic depiction of the internalization of mAb-Qrod-labeled CD36 from the cell membrane to the cytoplasm. (B) Fluorescence images of macrophages treated with 1 nM mAb-Qrods and simultaneously recorded in P- (left panel) and S-polarization (right panel). White spots indicate fluorescence signals from individual mAb-Qrods. Arrowheads indicate two typical views that are enlarged in C. Scale bar is 5 μm. (C) Enlarged images of the spots marked by arrowheads in B. Numbers on the left correspond to those in B. Scale bar, 1 μm. (D) Time-lapse fluorescence images in P- (left column) and S-polarization (right column). The white spot moved to the right over time (arrowheads). Scale bar, 1 μm. (E) Time course of the single mAb-Qrod from D along the X (red line, upper panel), Y (blue line, upper panel), and Z axis (middle panel) and the Qrod’s rotational motion (lower panel) at a temporal resolution of 30.28 ms. Arrowheads indicate notable changes in position and orientation. (F, G, H) Typical 4D traces of a single mAb-Qrod on the membrane (F), in the cytoplasm (G), and near the nucleus (H). The angle of the mAb-Qrod is indicated by the colored bar (bottom). Time courses of movement along individual axes for F, G, and H, are shown in Fig. S9, A, B, and C, respectively.
To measure the polarization of Qrods in vitro, individual GSH-Qrods were immobilized onto the surface of a glass chamber. The GSH-Qrod solution (0.2 nM) was loaded into a 5-μm-thick glass chamber whose surface was covered with nitrocellulose to fix the GSH-Qrods nonspecifically. After 5 min, the chamber was washed three times with 50 μL of 5 mM Hepes, pH 8.0, and sealed with enamel. With a camera exposure time of 30.28 ms, 100 images were acquired for each angle, giving a total of 1000 images.
For 4D observations of membrane proteins, isolated macrophages were incubated with American hamster IgG (10 mg/mL) and antimouse CD16/32 mAb (10 mg/mL) for 20 min at 37°C to block nonspecific binding by mAb-Qrods. Next, a 1 nM mAb-Qrod solution was added to the cells, and the chamber containing the cells was immediately transferred to the microscope stage incubator (37°C, 5% CO2) for measurements. Before experiments, the 1/2 λ wave plate was rotated 360° within 4 s for calibration.
Isolation of macrophages and confocal microscopy
The experimental protocol used for animal studies was approved by the Ethics Review Committee for Animal Experimentation of Osaka University Medical School. C57BL/6 mice (6–10 weeks old) were purchased from Japan SLC. After euthanizing the mice, macrophages were collected from the peritoneal lavage, plated in 8-well LabTek chambers (Nalge Nunc International) at 1.0 × 105 cells/well, and cultured overnight in RPMI 1640 containing 10% fetal calf serum (R10) at 37°C. Isolated macrophages were incubated with American hamster IgG and antimouse CD16/32 mAb before adding mAb-Qrods to block nonspecific binding (see above). For anti-CD36 antibody blocking-control experiments, macrophages were pretreated with antimouse CD36 monoclonal antibodies (25 mg/mL) or American hamster IgG (25 mg/mL) for 1 h on ice before incubation with mAb-Qrods (1 nM). After washing with R10, the cells were immediately transferred to the microscope stage incubator (37°C, 5% CO2) and then examined 25 min later using a confocal microscope (FV1000, Olympus, Japan). The fluorescence signals of the mAb-Qrod were detected through a band-pass filter at 575–675 nm.
Results
Synthesis of GSH-coated Qrods
Qrods were synthesized by elongating the CdS shell along the c axis of the CdSe core (34). The CdS shell produced a large Stokes shift that substantially changed both the absorbance and fluorescence spectra (Fig. 2, A and B, green and yellow) and increased the quantum yield from 6.2% to 49% at an emission peak of 596 nm. Both the absorbance and the fluorescence spectra depended on the size of the CdS shell when the same CdSe core was used. (see the Supporting Material, Fig. S1). We used the tripeptide GSH as a surface coating to promote water-solubilization, because GSH contains two reactive groups (amino and carboxyl) that enable easy conjugation with proteins and because GSH can coat the surface of Qdots thinly (36,40). An added benefit for biological studies is that the GSH coat is not cytotoxic (40). We coated the Qrod with GSH by ligand exchange (36): GSH-Qrods were easily prepared by mixing hydrophobic Qrods with the GSH solution at 70°C, a temperature at which the Qrods readily transferred into the aqueous phase. A transparent GSH-Qrod solution was obtained by deprotonating carboxylic groups with KOBut, which altered neither the absorbance nor the fluorescence spectra (Fig. 2, A and B, red), suggesting that the Qrod surface was unaffected by the procedure. Moreover, the GSH coating did not lower the quantum yield of the Qrods, which was 48% in the aqueous solution compared with 49% for Qrods in chloroform. Upon investigating the effects of GSH coating at other wavelengths, we found a high quantum yield of 39% at 612 nm (Fig. S1 B).
Figure 2.
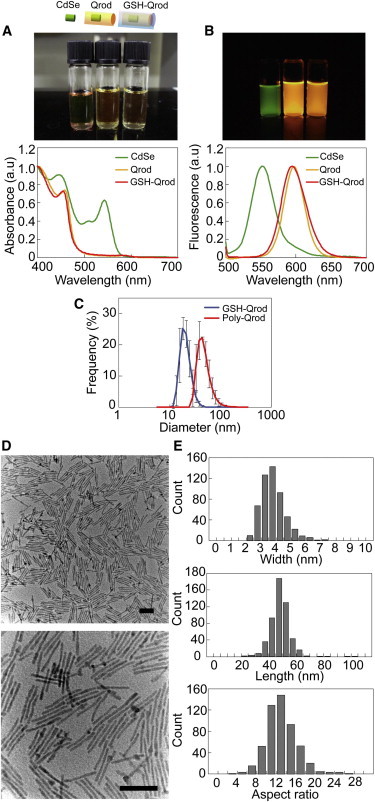
GSH-coated Qrods. (A) Photographs (upper panel) and normalized absorbance spectra (lower panel) of CdSe (green), CdSe/CdS (Qrod, yellow), and GSH-coated CdSe/CdS (GSH-Qrods, red). (B) Fluorescence images (upper panel) and normalized fluorescence spectra (lower panel) of CdSe (green), Qrods (yellow), and GSH-Qrods (red). (C) DLS histograms of GSH-Qrods (blue line) and polymer-coated Qrods (Poly-Qrod, red line). Error bars represent standard deviations. (D) TEM images of Qrods without GSH coating. Scale bars, 50 nm. (E) Distributions of the width (upper panel), length (middle panel), and aspect ratio (lower panel) of Qrods.
When designing a fluorescent probe, steric hindrance that may impair the function of a target protein must be considered. Polymer coating, which enlarges Qrods, has been widely used to prepare water-soluble Qdots and Qrods (34). We compared the difference in the hydrodynamic sizes of Qrods coated with GSH and amphiphilic polymers (PMO-PEG), as previously reported (34). Assuming a spherical shape and applying the DLS method, the apparent hydrodynamic diameter of GSH-Qrods was determined to be ∼18 nm, which is equal to that of noncoated Qrods (Fig. 2 C, blue). In contrast, the diameter of polymer-coated Qrods was 44 nm (Fig. 2 C, red). Thus, GSH coating can generate a water-soluble Qrod without increasing the Qrod size.
The Qrod polarization factor, which describes the maximum value of the anisotropy, depends on the aspect ratio and saturates at ∼0.65 at an aspect ratio of 5 (32). To reduce the variation in the polarization factor, we elongated the Qrods so that their aspect ratio was >5: TEM revealed that the Qrods were ∼4 ± 0.5 nm wide and 50 ± 10 nm long (Fig. 2, D and E), and the mean aspect ratio was 13 for 99.6% of Qrods with an aspect ratio ≥5 (Fig. 2 E, lower panel). The average polarization factor of Qrods measured using a fluorescence polarimeter (FP-715, JASCO, Japan) was 0.63, which is consistent with a previous report (32).
Polarization measurements of individual Qrods
We examined how the fluorescence of individual Qrods depends on polarization. The polarization angle of a GSH-Qrod was defined as the rotation angle against the 1/2 λ wave plate placed in front of the polarizing BS in the microscope (Fig. 1). Fig. 3 A shows the fluorescence images of a single GSH-Qrod fixed on a coverslip that were recorded using two perpendicular polarizations at detection angles ranging from 0° to 180°. The intensities of P- and S-polarization (Ip and Is, respectively) changed symmetrically with the angle (Fig. 3 B, upper graph), although the total fluorescence intensity did not change (Fig. 3 B, middle graph). The blinking properties of single GSH-Qrods were similar to those of Qdots (24). Moreover, the anisotropy changed with the detection angle, indicating that the GSH-Qrod displayed polarized fluorescence (Fig. 3 B, lower graph) that can be described as a sine function with an amplitude of 0.74 (Fig. 3 C). The calculation precision of the anisotropy, ∼0.06–0.12, was defined as the standard deviation during a 10 s observation (100 data points; Fig. 3 C). Plotting anisotropy versus the detection angle generated a calibration curve (Fig. S2). We replotted the anisotropy-angle relationship data by adjusting the phase so that 0 anisotropy corresponded to 0° angle (Fig. S2 A), and extracted the plots from −45° to 45° as a calibration curve (Fig. S2 B). Although we synthesized the Qrods to have large aspect ratios (Fig. 2 E), the polarization factor described by the amplitude of the sine function showed a wide distribution, with a mean value of 0.68 ± 0.09 (Fig. 3 D); this value is consistent with the measurement using the fluorescence polarimeter (0.63). Because the variation in the polarization factor hampered efforts to prepare a universal calibration curve, especially for large angles (Fig. S2 C), we generated calibration curves for individual Qrods that enabled anisotropy-angle conversion with high reproducibility (Fig. S3 A). The tracking precision of the angle, which was unaffected by the polarization factor but depended on the angle of the Qrods, was <6° (Fig. S3 B).
Figure 3.
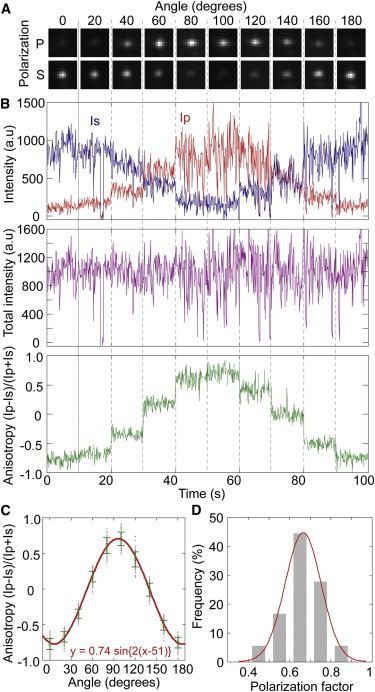
Polarization characteristics of individual Qrods. (A) Fluorescence images of a single GSH-Qrod on a coverslip acquired by simultaneously recording the P-polarization (upper images) and S-polarization (lower images). (B) The rotation angle of the 1/2 λ wave plate was incrementally changed from 0° to 180° every 10 s in 20° intervals. Fluorescence intensities of P-polarization (Ip, red line in upper panel) and S-polarization (Is, blue line in upper panel), total fluorescence intensity (middle panel), and anisotropy (defined as r = (Ip-Is)/(Ip+Is); lower panel) are shown as functions of time. These data correspond to the fluorescence images in A. (C) The anisotropy of a single GSH-Qrod as a function of the rotation angle of the 1/2 λ wave plate. Gray, raw data; green circles, mean values; error bars, standard deviation. The mean values and standard deviations were calculated from the fluorescence intensity ratios (B, lower panel) every 10 s (100 data points). The red line shows the fitting with a sine function (y = a1·sin{2·(x- a2)}). (D) Histogram of the polarization factor, which was defined as the amplitude (a1) of the sine functions used to fit the anisotropy of individual Qrods. The red line is the fitted single Gaussian distribution with a peak of 0.68 ± 0.09 nm.
3D position measurements of individual Qrods with polarized optics
Out of the many methods available for 3D single particle tracking, we chose one that exploits the astigmatism produced by cylindrical lenses with long focal lengths (41,42). The cylindrical lenses generated astigmatic aberration, which was created by using different optical path lengths along the X and Y axes and resulted in a measurable relationship between the Z-position of a single fluorophore and the ellipticity of its image (Fig. 4 A and Fig. S4 A). The X- and Y-positions were determined by fitting the image with a 2D ellipsoidal Gaussian function, and the Z-position of the fluorophore was determined from the ellipticity, which was defined as the ratio of the full width at half-maximum (FWHM) values of the Gaussian function in the X and Y axes generated by the different focal lengths (Fig. S5). The relationship between the Z-position and the ellipticity of the image of a single fluorophore was measured while the objective lens was moved in 100 nm discrete steps by a Piezo actuator to prepare a calibration curve for Z-positions (Fig. S4 B). We optimized the astigmatism using concave and convex cylindrical lenses (CnC and CvC in Fig. 1), rather than a cylindrical lens of long focal length (41), and we used a fluorescent bead with 100 nm in diameter instead of Qrods to avoid the effects of the blinking of the dye (Fig. S4, C and D). The optimal distance between CnC and CvC was found to be 5 mm. The calculated precision of the 3D position, defined as the standard deviation of the tracking time course, depends on the number of photons that the camera receives from the fluorophore (4) and the Z-position of the fluorophore (42). The precision here was 2 nm along the X and Y axis and 5 nm along the Z axis when the camera received 15,000 photons from a fluorescent bead at a Z-position near zero (Fig. S4 D), consistent with theoretical values (42). When 1500 photons were received from a Qrod, the precision decreased to 6, 7, and 17 nm in the X, Y, and Z axis, respectively. A 20 nm artificial step was detectable in all axes (Fig. S4 E).
Figure 4.
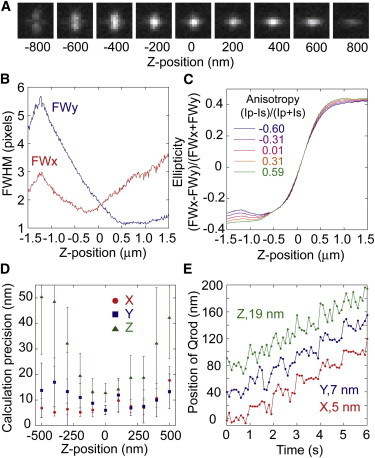
Single particle tracking in 3D of a GSH-Qrod using polarized optics. (A) Fluorescence images of a single GSH-Qrod at various Z-positions ranging from −800 to 800 nm. Each image is the sum of the P- and S-polarization images at one Z-position. (B) FWHM values of the images in A as a function of the Z-position along the X axis (red line) and Y axis (blue line). (C) Ellipticity as a function of the Z-position at various anisotropies (−0.60 to 0.59). (D) Calculated precision in the position of Qrods along the X, Y, and Z axis (red, blue, and green symbols) as a function of the Z-position. Error bars represent standard deviations. (E) Test tracking with 20 nm steps when the Z-position of the Qrod was near zero. A GSH-Qrod fixed on a coverslip was moved (using two Piezo actuators) at discrete 20 nm steps once every 1 s simultaneously along the X (red line), Y (blue line), and Z axis (green line). The frame rate of the CCD camera was 10 frames/s. Standard deviations of the tracking for 10 s (excluding the stepping moments) were 5, 7, and 19 nm along the X, Y, and Z axis, respectively.
For polarized optics, we summed the P- and S-polarized images before calculating the 3D position (Fig. 4 A). This process created a small gap while merging the images, leading to an asymmetrical relationship between the respective FWHM values of the X and Y axis (Fig. 4 B). Because the relationship between the ellipticity and Z-position varied with anisotropy, we performed 3D tracking over a spatial range where the variation was negligible (−500 to 500 nm) (Fig. 4 C). We investigated the calculated precision of the 3D position of Qrods (the standard deviation of the tracking time course) with both polarized and cylindrical optics. When the camera received ∼1500 photons per 100 ms from a single Qrod in one polarization pathway within a frame (total 3000 photons in the merged image), and when the Z-position was near zero, the calculated precisions for the X-, Y-, and Z-positions were at maximum 5, 5, and 12 nm, respectively, but decreased to as much as 20 nm for the X- and Y-positions and 50 nm for the Z-position depending on the Z-position of the Qrod (Fig. 4 D). A 20 nm artificial step was detectable in all axes (Fig. 4 E), much as when only cylindrical optics was used (Fig. S4 E). Thus, we could simultaneously measure the angular position (Fig. 3) and 3D position (Fig. 4) of a single Qrod.
4D tracking of a membrane protein in living cells
To demonstrate the applicability of 4D single particle tracking in biophysical studies, we examined the movement of CD36, a membrane protein involved in phagocytosis, in freshly isolated macrophages (Fig. 5). CD36, a member of the class B scavenger receptor family (43), is preferentially expressed in phagocytes and used for the uptake of multiple ligands including lipoproteins. We tracked the internalization of surface CD36 in macrophages at the single-molecule level using the anti-CD36 monoclonal antibody-conjugated GSH-Qrods (mAb-Qrods), which were prepared using the amino and carboxylic groups of GSH. The conjugation of mAbs to GSH-Qrods was confirmed by FCS, which determines the relative sizes of fluorophores by measuring diffusion times through a tiny confocal volume (38). The FCS curve of the Qrods was closely fitted by the function assuming 1-component diffusion (see Methods) (Fig. S6, blue); if the Qrods had aggregated, the FCS curve would have been the summation of 2-component or more (36,38–40). We therefore concluded that Qrods were monodispersed in solution. The diffusion time of Qrods, estimated to be 0.82 ± 0.09 ms, yielded an apparent hydrodynamic radius r = 16.4 nm, because the diffusion time of a fluorescent bead with a radius of 20 nm is 1.0 ms (36). The 16.4 nm value is consistent with the 18 nm estimated by the DLS histogram showing a single peak (Fig. 2 C) and further indicates the monodispersion of Qrods. Following antibody conjugation, the FCS curve was again closely fitted by the theoretical function of 1-component diffusion, and the diffusion time of the Qrods increased to 1.69 ± 0.20 ms (Fig. S6, red). This result indicated that our conjugation procedure successfully linked mAbs to Qrods (see Methods), and that antibody conjugation did not affect the dispersion of Qrods in an aqueous solution or trigger the aggregation of Qrods. The fluorescent characteristics of the Qrods, including the spectra and the quantum yield, also did not change after mAb conjugation.
Isolated macrophages were incubated with American hamster IgG and antimouse CD16/32 mAb to block nonspecific binding before adding mAb-Qrods (1 nM), and after adding mAB-Qrods the cell-containing chamber was transferred immediately to the microscope incubator stage for measurements (see Methods). Distinct Qrods drifting on the membrane were identified under the fluorescence microscope (Fig. 5, A and B, and Movie S1). The white spots in Fig. 5 B are the fluorescence signals of individual mAb-Qrods. The enlarged images (marked by arrowheads in Fig. 5 B) present two typical views: one shows different intensities in the P- and S-polarized images, indicating that the Qrod was inclined against the orthogonal axes (Fig. 5 C, upper panels); the other shows two spots, one circular and one elliptical, indicating two Qrods at distinct Z-positions (Fig. 5 C, lower panels). The binding of mAb-Qrods to CD36 was inhibited by pretreatment with excess anti-CD36 mAb, but not with an isotype control (American hamster IgG) (Fig. S7), suggesting that mAb-Qrods bound specifically to CD36 on the cell surface.
The large variability of the polarization factor of mAb-Qrods (Fig. 3 D) diminished the accuracy of anisotropy-angle conversion and hindered the preparation of a universal calibration curve (Fig. S2) (see above). However, because the anisotropy-angle relationship of the Qrods follows an arcsine function (Fig. 3 C and Fig S2) as for other organic dyes (26–31), only the information on the amplitude of the sine function (polarization factor) is required to generate a calibration curve. Therefore, before tracking, we obtained angular data on mAb-Qrods bound to a protein by rotating the 1/2 λ wave plate 360° within 4 s to determine the polarization factor (Fig. S8 A). We calculated the polarization factor by fitting a sine function to the data that clearly included the half pitch of a sine curve (Fig. S8 B). Thus, we prepared anisotropy-angle calibration curves (θ = arcsin(r/a1)/2, where a1 = polarization factor and r = anisotropy) with a1 as the only fitting parameter for individual Qrods. We were, however, unable to pursue the movements of all Qrods: because of the procedure used, the polarization factor could be obtained for the Qrods that moved slowly or when the average displacement of a Qrod was 0 during a 1-s period including the time when the anisotropy was maximum or minimum (with the Qrod rotating during the calibration procedure). Thus, we could not prepare the calibration table only in the situation wherein the tracking trace for 4 s excluded the maximum or minimum anisotropy, for example when the Qrods rotated in the same direction as the 1/2 λ wave plate at a comparable speed. However, such a scenario is rare, because Qrods transported by motor protein did not rotate during linear movements (Fig. 5 G), whereas those on the membrane often changed their direction (Fig. 5 F). Accuracy of calibration can be enhanced by increasing the speed of calibration, which will be implemented in the future.
One of the spots observed moved dynamically over time (Fig. 5, B and C, arrowhead 1) and showed different P- and S-polarization fluorescence intensities with a polarization factor of 0.63 (Fig. 5 D), providing evidence of lateral and angular motion of the particle in the cell. Using the intensity ratio between the P- and S-polarization and the Gaussian shape of the spot, we obtained the time course for the four spatial dimensions of this single mAb-Qrod (Fig. 5 E). The mAb-Qrod rotated at 2.5, 6, 11, and 16 s, and moved substantially along the Z axis at 6 s and the X axis at 16 s (Fig. 5 E, arrowheads). These observations suggest that the CD36 receptor labeled by the Qrod entered the cytoplasm at 6 s with an angular motion and was then transported linearly along the cytoskeleton at 16 s. One mAb-Qrod showed half-moon-like motion along the X and Y axes together with highly fluctuating movement along the Z axis and fast angular motion before endocytosis (Fig. 5 F, and Fig. S9 A), suggesting that this receptor rotated freely on the plasma membrane but diffused laterally only within the domain immediately beneath the membrane. Another mAb-Qrod moved unidirectionally along tracks in the cytoplasm (most likely microtubules) in 3D and rotated slightly (Fig. 5 G, and Fig. S9 B), whereas one mAb-Qrod near the nucleus appeared to drift and slowly rotate helically (Fig. 5 H, and Fig. S9 C). Thus, our imaging system enabled 4D single particle tracking of Qrods bound to biomolecules in real time in living cells.
Discussion
We have designed a new, to our knowledge, fluorescent probe, GSH-Qrods, to track single membrane proteins over time in 4D (x, y, z, θ). We previously used Qrods for 4D tracking with a different set of spatial coordinates (x, y, θ, φ) (35). Although combining these methods might be considered to achieve five-dimensional tracking (x, y, z, θ, φ), this would be feasible only when the density of the Qrods is low (as during in vitro measurements) because single fluorophores are imaged as four crowded fluorescent spots by dividing the beam path using a BS and two Wollaston prisms (35). The present observation systems will require considerable modifications to allow comprehensive observation of all polar and Euclidean movements of a biomolecule in a living cell.
Gold nanorods, which are another example of inorganic nanorods, were considered as an alternative to Qrods because, with the gold nanorods, scattered light and not fluorescence is detected, meaning that photobleaching can be ignored (44–47). Qrods, however, are more suitable for multicolor imaging required to detect both molecular orientations and molecular interactions; like Qdots, Qrods have broad absorption spectra and narrow, symmetric emission spectra. To use our Qrods as biological probes, we coated their surface to make them soluble in water. Organic polymers are generally used for surface coating, but they increase the size of the Qrods and enhance the risk of steric effects compromising the functions of the protein targeted by the Qrod.
The polarization factor of individual Qrods (Fig. 3 D) showed a Gaussian distribution because of the large variability of the aspect ratio (with a mean of ∼13; Fig. 2 E). This Gaussian distribution complicated the design of an orientation-polarization calibration curve (Fig. S2), necessitating the preparation of calibration curves before experimental measurements for each Qrod individually to determine the angle precisely (Fig. S8). Nevertheless, with improved purification methods that result in a narrower aspect ratio distribution, better tracking can be achieved. Because the reactive groups of GSH were uniformly distributed on the surface of Qrods, the exact part of a Qrod that binds to the target molecule cannot be established. For example, a membrane protein may bind to the center of a Qrod, whereas another may bind to the poles. Moreover, oblique rotations of a protein-bound Qrod would decrease the polarity in the 2D plane, a caveat that applies to all inorganic nanorods (44–47).
Using the novel, to our knowledge, method described here, we observed the diffusion and transport of CD36 in macrophages in 4D (Fig. 5). We obtained additional information about movement dynamics by calculating the mean square displacements (MSD) of the 4D trajectories (Fig. 5, F, G, and H; Fig. S10) (48–50); for comparison, the MSD of free diffusion (Fig. S11) was also plotted (Fig. S10, A–C, black). The MSD of the XY plane in Fig. 5 F shows a linear function, indicating that the mAb-Qrod diffused freely on a lateral plane (Fig. S10 A, red), whereas the MSD in Fig. 5 G is quadratic, indicating unidirectional movement (Fig. S10 A, blue). In the Z axis, all MSD plots were linear in the short term, but saturated over longer times (Fig. S10 B), suggesting that the mAb-Qrod fluctuated along the Z axis and diffused in a limited area, being confined in a thin volume. The MSD along the angular axis (Fig. 5 F) also showed saturation (Fig. S10 C, red), possibly because of the dynamic range of the anisotropy-angle conversion, but before saturation the MSD plots followed a linear function, indicating free angular diffusion of the mAd-Qrod. The angular diffusion coefficients of the mAb-Qrods on the membrane and in the cytoplasm were respectively calculated to be (344°)2/s and (1278°)2/s by fitting the data until 0.66 s (Fig. S10 C, red and black lines). These values agree with the coefficients of polytopic membrane proteins on artificial cell membranes ((1000°∼10,000°)2/s) measured by optical and electron paramagnetic spectroscopy (51). The movement of mAb-Qrods shown in Fig. 5, G and H, is potentially driven by motor proteins; the MSD plots (Fig. S10 C, blue and green) suggest that the angular movements were restricted in these cases, because the slope of the logarithmic plot was <1 (49).
In conclusion, we synthesized highly fluorescent, highly polarized, small water-soluble Qrods, and constructed an optical microscope based on polarized and cylindrical optics. By combining these technologies, we performed simultaneous 3D and angular (4D) single particle tracking of the membrane receptor CD36 in freshly isolated macrophages. Our technique has the potential to enable studies of biological phenomena such as receptor internalization, ligand-receptor interactions, receptor complex associations, and signal transduction to help reveal the relationship between the intracellular orientation of biomolecules and their functions.
Acknowledgments
We gratefully acknowledge Peter Karagiannis (Riken, QBiC) for critically reading this manuscript and Eiji Taguchi (Ultrahigh Vacuum Electron Microscopy Center (UHVEM), Osaka University) for assisting with TEM.
F.F. was supported by The Ministry of Education, Science, Sport and Culture of Japan (Grant-in-Aid for Scientific Research, No, 22700491), the Cosmetology Research Foundation, and Japan Science and Technology. T.M.W. was supported by PRESTO of Japan Science and Technology.
Supporting Material
References
- 1.Ellinger P. Fluorescence microscopy in biology. Biol. Rev. Camb. Philos. Soc. 1940;15:323–347. [Google Scholar]
- 2.Lichtman J.W., Conchello J.A. Fluorescence microscopy. Nat. Methods. 2005;2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 3.Abbe E. Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung. Arch. Mikr. Anat. 1873;9:413–468. [Google Scholar]
- 4.Thompson R.E., Larson D.R., Webb W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yildiz A., Forkey J.N., Selvin P.R. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 6.Park H., Toprak E., Selvin P.R. Single-molecule fluorescence to study molecular motors. Q. Rev. Biophys. 2007;40:87–111. doi: 10.1017/S0033583507004611. [DOI] [PubMed] [Google Scholar]
- 7.Kural C., Kim H., Selvin P.R. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science. 2005;308:1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 8.Watanabe T.M., Higuchi H. Stepwise movements in vesicle transport of HER2 by motor proteins in living cells. Biophys. J. 2007;92:4109–4120. doi: 10.1529/biophysj.106.094649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonda K., Watanabe T.M., Higuchi H. In vivo nano-imaging of membrane dynamics in metastatic tumor cells using quantum dots. J. Biol. Chem. 2010;285:2750–2757. doi: 10.1074/jbc.M109.075374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toprak E., Balci H., Selvin P.R. Three-dimensional particle tracking via bifocal imaging. Nano Lett. 2007;7:2043–2045. doi: 10.1021/nl0709120. [DOI] [PubMed] [Google Scholar]
- 11.Watanabe T.M., Sato T., Higuchi H. Three-dimensional nanometry of vesicle transport in living cells using dual-focus imaging optics. Biochem. Biophys. Res. Commun. 2007;359:1–7. doi: 10.1016/j.bbrc.2007.04.168. [DOI] [PubMed] [Google Scholar]
- 12.Dalgarno P.A., Dalgarno H.I., Greenaway A.H. Multiplane imaging and three dimensional nanoscale particle tracking in biological microscopy. Opt. Express. 2010;18:877–884. doi: 10.1364/OE.18.000877. [DOI] [PubMed] [Google Scholar]
- 13.Pavani S.R., Thompson M.A., Moerner W.E. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc. Natl. Acad. Sci. USA. 2009;106:2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimomura O., Johnson F.H., Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962;59:223–239. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 15.Tsien R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 16.Nifosí R., Amat P., Tozzini V. Variation of spectral, structural, and vibrational properties within the intrinsically fluorescent proteins family: a density functional study. J. Comput. Chem. 2007;28:2366–2377. doi: 10.1002/jcc.20764. [DOI] [PubMed] [Google Scholar]
- 17.Song L., Hennink E.J., Tanke H.J. Photobleaching kinetics of fluorescein in quantitative fluorescence microscopy. Biophys. J. 1995;68:2588–2600. doi: 10.1016/S0006-3495(95)80442-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song L., Varma C.A.G.O., Tanke H.J. Influence of the triplet excited state on the photobleaching kinetics of fluorescein in microscopy. Biophys. J. 1996;70:2959–2968. doi: 10.1016/S0006-3495(96)79866-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murcia M.J., Naumann C.A. Biofunctionalization of fluorescent nanoparticles. Nanotechnologies for the Life Sciences. 2007;1:1–39. [Google Scholar]
- 20.Pierobon P., Cappello G. Quantum dots to tail single bio-molecules inside living cells. Adv. Drug Deliv. Rev. 2012;64:167–178. doi: 10.1016/j.addr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Bruchez M., Jr., Moronne M., Alivisatos A.P. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 22.Chan W.C.W., Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 23.Warshaw D.M., Kennedy G.G., Trybus K.M. Differential labeling of myosin V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys. J. 2005;88:L30–L32. doi: 10.1529/biophysj.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barroso M.M. Quantum dots in cell biology. J. Histochem. Cytochem. 2011;59:237–251. doi: 10.1369/0022155411398487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi H.S., Frangioni J.V. Nanoparticles for biomedical imaging: fundamentals of clinical translation. Mol. Imaging. 2010;9:291–310. [PMC free article] [PubMed] [Google Scholar]
- 26.Harms G.S., Sonnleitner M., Schmidt T. Single-molecule anisotropy imaging. Biophys. J. 1999;77:2864–2870. doi: 10.1016/S0006-3495(99)77118-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weber G. Rotational Brownian motion and polarization of the fluorescence of solutions. Adv. Protein Chem. 1953;8:415–459. doi: 10.1016/s0065-3233(08)60096-0. [DOI] [PubMed] [Google Scholar]
- 28.Albrecht A. Polarizations and assignments of transitions: the method of photoselection. J. Mol. Spectrosc. 1961;6:84–108. [Google Scholar]
- 29.Sase I., Miyata H., Kinosita K., Jr. Axial rotation of sliding actin filaments revealed by single-fluorophore imaging. Proc. Natl. Acad. Sci. USA. 1997;94:5646–5650. doi: 10.1073/pnas.94.11.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forkey J.N., Quinlan M.E., Goldman Y.E. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature. 2003;422:399–404. doi: 10.1038/nature01529. [DOI] [PubMed] [Google Scholar]
- 31.Mizuno H., Higashida C., Watanabe N. Rotational movement of the formin mDia1 along the double helical strand of an actin filament. Science. 2011;331:80–83. doi: 10.1126/science.1197692. [DOI] [PubMed] [Google Scholar]
- 32.Hu J.T., Li Ls, Alivisatos A.P. Linearly polarized emission from colloidal semiconductor quantum rods. Science. 2001;292:2060–2063. doi: 10.1126/science.1060810. [DOI] [PubMed] [Google Scholar]
- 33.Peng X.G., Manna L., Alivisatos A.P. Shape control of CdSe nanocrystals. Nature. 2000;404:59–61. doi: 10.1038/35003535. [DOI] [PubMed] [Google Scholar]
- 34.Deka S., Quarta A., Manna L. CdSe/CdS/ZnS double shell nanorods with high photoluminescence efficiency and their exploitation as biolabeling probes. J. Am. Chem. Soc. 2009;131:2948–2958. doi: 10.1021/ja808369e. [DOI] [PubMed] [Google Scholar]
- 35.Ohmachi M., Komori Y., Yanagida T. Fluorescence microscopy for simultaneous observation of 3D orientation and movement and its application to quantum rod-tagged myosin V. Proc. Natl. Acad. Sci. USA. 2012;109:5294–5298. doi: 10.1073/pnas.1118472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin T., Fujii F., Yoshioka Y. Preparation and characterization of highly fluorescent, glutathione-coated near infrared quantum dots for in vivo fluorescence imaging. Int. J. Mol. Sci. 2008;9:2044–2061. doi: 10.3390/ijms9102044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu W.W., Chang E., Colvin V.L. Forming biocompatible and nonaggregated nanocrystals in water using amphiphilic polymers. J. Am. Chem. Soc. 2007;129:2871–2879. doi: 10.1021/ja067184n. [DOI] [PubMed] [Google Scholar]
- 38.Magde D., Elson E.L., Webb W.W. Fluorescence correlation spectroscopy. I. Conceptual basis and theory. Biopolymers. 1974;13:1–27. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 39.Heuff R.F., Swift J.L., Cramb D.T. Fluorescence correlation spectroscopy using quantum dots: advances, challenges and opportunities. Phys. Chem. Chem. Phys. 2007;9:1870–1880. doi: 10.1039/b617115j. [DOI] [PubMed] [Google Scholar]
- 40.Tiwari D.K., Tanaka S., Jin T. Synthesis and characterization of anti-HER2 antibody conjugated CdSe/CdZnS quantum dots for fluorescence imaging of breast cancer cells. Sensors (Basel) 2009;9:9332–9364. doi: 10.3390/s91109332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kao H.P., Verkman A.S. Tracking of single fluorescent particles in three dimensions: use of cylindrical optics to encode particle position. Biophys. J. 1994;67:1291–1300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holtzer L., Meckel T., Schmidt T. Nanometric three-dimensional tracking of individual quantum dots in cells. Appl. Phys. Lett. 2007;90:053902. [Google Scholar]
- 43.Silverstein R.L., Febbraio M. CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci. Signal. 2009;2:1–8. doi: 10.1126/scisignal.272re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sönnichsen C., Alivisatos A.P. Gold nanorods as novel nonbleaching plasmon-based orientation sensors for polarized single-particle microscopy. Nano Lett. 2005;5:301–304. doi: 10.1021/nl048089k. [DOI] [PubMed] [Google Scholar]
- 45.Failla A.V., Qian H., Meixner A.J. Orientational imaging of subwavelength Au particles with higher order laser modes. Nano Lett. 2006;6:1374–1378. doi: 10.1021/nl0603404. [DOI] [PubMed] [Google Scholar]
- 46.Chang W.S., Ha J.W., Link S. Plasmonic nanorod absorbers as orientation sensors. Proc. Natl. Acad. Sci. USA. 2010;107:2781–2786. doi: 10.1073/pnas.0910127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G., Sun W., Fang N. Resolving rotational motions of nano-objects in engineered environments and live cells with gold nanorods and differential interference contrast microscopy. J. Am. Chem. Soc. 2010;132:16417–16422. doi: 10.1021/ja106506k. [DOI] [PubMed] [Google Scholar]
- 48.Kusumi A., Sako Y., Yamamoto M. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 1993;65:2021–2040. doi: 10.1016/S0006-3495(93)81253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin D.S., Forstner M.B., Käs J.A. Apparent subdiffusion inherent to single particle tracking. Biophys. J. 2002;83:2109–2117. doi: 10.1016/S0006-3495(02)73971-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saxton M.J. Single-particle tracking: the distribution of diffusion coefficients. Biophys. J. 1997;72:1744–1753. doi: 10.1016/S0006-3495(97)78820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cherry R.J. Membrane protein dynamics: rotational dynamics. In: Yeagle P.L., editor. The Structure of Biological Membranes. CRC Press; Boca Raton, FL: 2005. pp. 389–410. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


