Abstract
In enteric bacteria, the kinase LsrK catalyzes the phosphorylation of the C5-hydroxyl group in the linear form of 4,5-dihydroxy-2,3-pentanedione (DPD), the precursor of the type II bacterial quorum sensing molecule (AI-2). This phosphorylation is required for AI-2 sequestration in the cytoplasm and subsequent derepression of AI-2 related genes necessary for quorum development. While LsrK is a critical enzyme within the DPD quorum sensing relay system, kinetic details of this kinase have yet to be reported. A continuous UV-vis spectrophotometric assay was developed, which allowed steady-state kinetic analysis of LsrK to be undertaken with the substrates ATP and DPD. The data was most consistent with a rapid equilibrium ordered mechanism with ATP binding first: kcat (7.4 ± 0.6 s−1), Km,ATP (150 ± 30 µM) and Km(app),DPD (1.0 ± 0.2 mM). The assay also allowed a DPD substrate profile to be conducted, which provided an unexpected biochemical disconnect between the previous agonist/antagonist cell based reporter assay and the LsrK assay presented herein. Together these findings raise the importance of LsrK and lay the foundation not only for further understanding of this enzyme and its critical biological role but also the rational design of regulatory molecules targeting AI-2 quorum sensing in pathogenic bacteria.
Quorum sensing (QS) is known as one type of bacterial cell-to-cell communication, which co-ordinates bacterial behaviors within a population.1 The communication is mediated by a plethora of diffusible signaling molecules, called autoinducers. Through QS, each bacterial cell can produce, secrete and detect particular autoinducers and translate the signal to a programmed gene expression pattern in a population-wide synchronous manner. Through concerted behavior, bacterial communities adapt their responses to environmental challenges in a manner similar to multi-cellular organisms. Quorum enabling responses can be symbiotic as well as pathogenic. The latter includes the formation of biofilms and the production of virulence factors once the colony has reached a critical cell density.2 Autoinducer-2 (AI-2) is a class of signaling molecules, derived from the common precursor, (4S)-4,5-dihydroxy-2,3-pentanedione (DPD). Based largely on the prevalence of the DPD synthase, LuxS, AI-2 is hypothesized as a “universal bacterial language” in which AI-2 mediates cross-species communication among more than 80 Gram-positive and Gram-negative strains, and elicits a variety of phenotypes in different bacterial species. As such, the modulation of AI-2 signaling could serve to alter individual bacterial phenotypes across a broad spectrum of species.3
In the enteric bacteria Escherichia coli and Salmonella enterica serovar Typhimurium, AI-2 signaling is carried out by the Lsr proteins.4,5 When encountered extra-cellularly, AI-2 is recognized by the receptor protein LsrB and internalized through the trans-membrane channel LsrA/C.6 Once inside the cell, AI-2 is phosphorylated by the kinase family protein LsrK. Phosphorylation serves to trap and activate AI-2 within the cell;7 while phosphorylated AI-2 (phospho-DPD) binds and inactivates the transcriptional repressor protein LsrR thereby inducing the expression of the lsr operon and potentially other LsrR-regulated, QS-related genes.8,9 Recently, several reports have detailed the successful development of AI-2 analogues, which are structurally similar to DPD, to modulate AI-2 quorum sensing. The identified molecules agonize or antagonize the AI-2 related gene expression in reporter strains, although the targeting mechanisms and regulatory mode are poorly understood.10–14
The function of LsrK is essential to the AI-2 signaling pathway, since it serves as the prerequisite activating step for AI-2 downstream signaling. Bacterial strains mutated to carry a dysfunctional lsrK are unable to sequester AI-2 in the cytoplasm and/or activate lsr transcription.4,5,15 In addition, the derepression of LsrR is directly correlated to cytoplasmic phospho-DPD levels,16 while AI-2 phosphorylation serves as the starting point for AI-2 catabolism and subsequent quorum quenching.16 Given its critical role in the AI-2 system, the LsrK kinase may represent an attractive anti-infective target whose selective modulation should ablate AI-2 related pathogenesis.
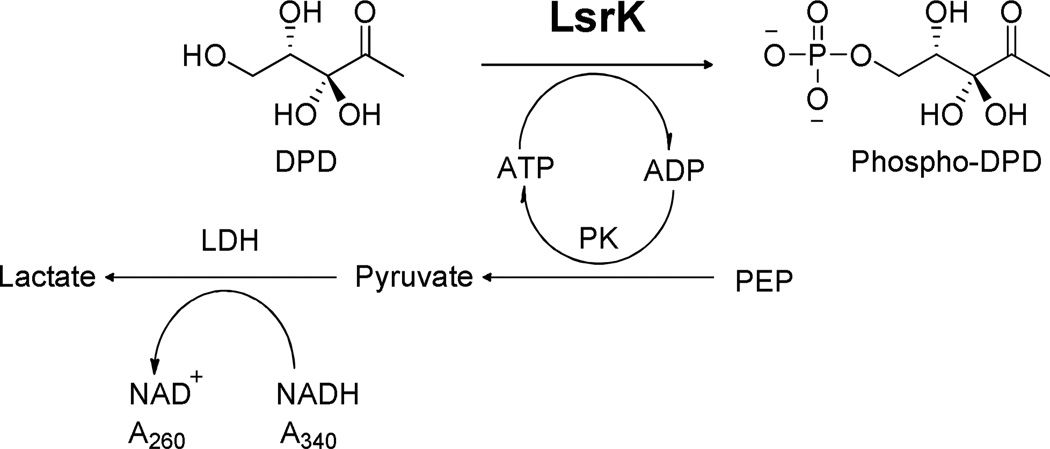
In vitro purified LsrK catalyzes the transfer of the phosphate group from ATP to the linear form of AI-2 at the C5 position (Figure 1), supporting the involvement of LsrK in QS.4,5,7 Previously, the reaction was qualitatively characterized by isotope labeling via 1D thin layer chromatography (TLC) or ATP related luminescence.5,17 However, more detailed kinetic evaluation of LsrK has been hampered by the lack of a reliable source of LsrK, DPD/DPD-related alternative substrates, and a highly reproducible/sensitive detection method.
Figure 1.
LsrK catalyzed phosphorylation of DPD and the coupled PK/LDH reaction.
Our laboratory has reported a detailed structural analysis of synthetically prepared DPD in aqueous solution via NMR spectroscopy.18 This study demonstrated that DPD is a rather stable molecule compared to other autoinducers (such as AHLs and oligopeptides) although it exists in a complex equilibrium of great structural diversity.19–21 From this latter standpoint, phosphorylation by LsrK drives the equilibrium to one tautomeric form in the linear conformation.16 Recently, we reported a purification procedure for the Salmonella LsrK from an E. coli expression system.22 The purified protein is monomeric and highly active, which rendered it suitable for mechanistic and inhibition studies. Thus, to overcome the limited knowledge of LsrK-DPD and provide a foundation for the design of potent phosphorylation modulators, we herein present a continuous assay, which has allowed a detailed mechanistic pattern to emerge on the interaction between DPD and related congeners with LsrK.
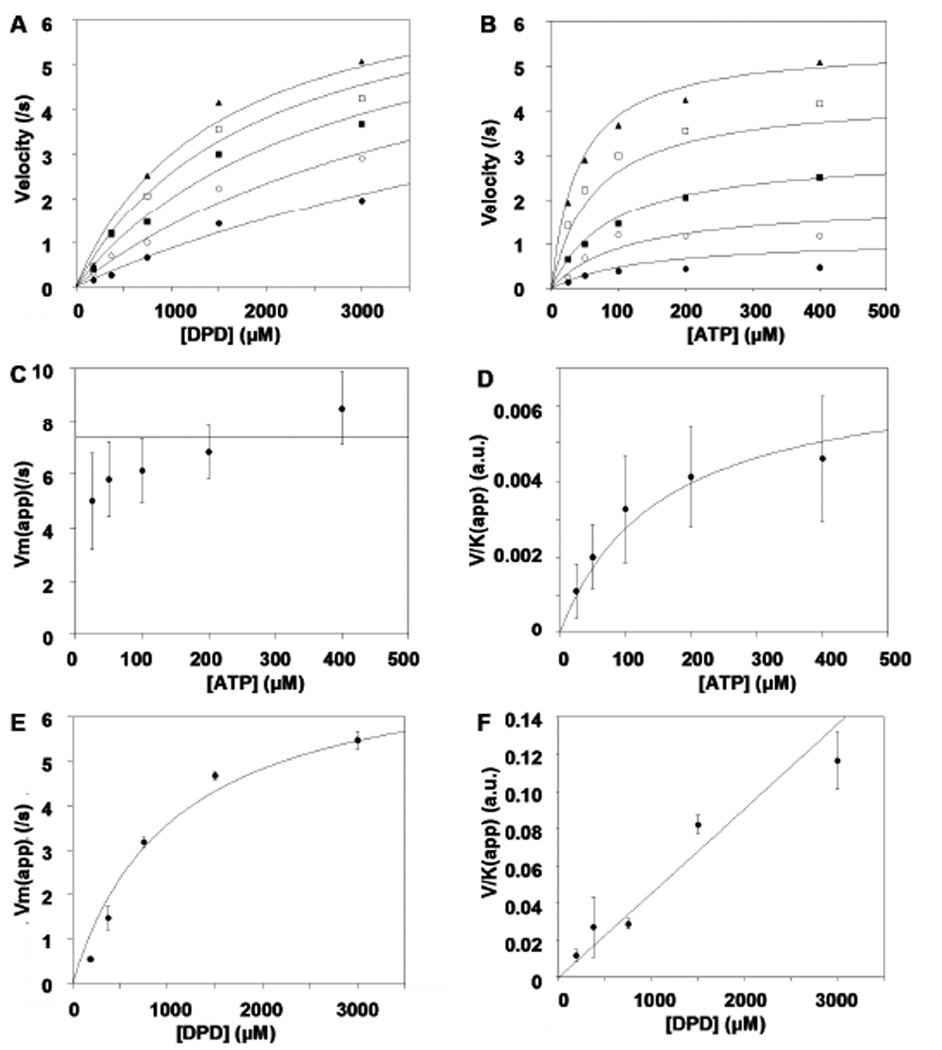
Our assay was designed to couple the production of ADP with the oxidation of NADH via the reactions catalyzed by pyruvate kinase (PK) and lactate dehydrogenase (LDH), as shown in Figure 1. The consumption of NADH, measured at 340 nm (εNADH=6.22×104 M−1 cm−1) was used to calculate the initial velocity of LsrK catalysis. The assay was conducted at 24.0 ± 1.0 °C in 10 mM Tris, pH 7.5, 25 mM KCl, 3 mM MgCl2, 4 U/mL PK, 6 U/mL LDH, 1 mM NADH, 1 mM PEP and 1 mg/mL bovine serum albumin (BSA). From the generated kinetic studies, the initial velocities were determined as a function of both substrates, DPD (0–3 mM) and ATP (0–400 µM) using a 5×5 matrix of differing concentrations.
Commonly encountered bi-substrate kinetic mechanisms such as rapid equilibrium ordered & random, steady state ordered & random, and ping-pong were evaluated. The velocity equations describing each of the various kinetic mechanisms were fit globally to the initial rates of the substrate concentration matrix. Relevant plots and re-plots of the results are presented in Figure 2. A rapid equilibrium ordered mechanism with ATP binding as the first substrate was most consistent with the kinetic data based on global fitting and upon re-plot analysis of individual saturation plots to the Michaelis-Menten equation. There are two visual diagnostics that distinguish a rapid equilibrium ordered mechanism from rival kinetic mechanisms. These are illustrated by replots of (Vmax)apparent and (V/K)apparent versus concentration of the opposite substrate (Panels C–F of Figure 2). The first diagnostic is that (Vmax)apparent for the second substrate, DPD, is independent of the concentration of the first substrate, ATP (Figure 2C). This behavior results in a rapid equilibrium ordered model, in which the saturating second substrate “traps” the first substrate in the Michaelis complex making the chemical step (or products release) always rate limiting. The second diagnostic is that the (V/K)apparent for the first substrate, ATP, is linear with respect to the second substrate, DPD concentration (Figure 2F). Under limiting concentrations of ATP, DPD can only drive the reaction forward but not provide an alternate path (DPD cannot bind to the enzyme without ATP binding first), thus the observation of a linear response.
Figure 2.
Global fit of a rapid equilibrium ordered mechanism with ATP binding first to LsrK. Panels A & B present the velocity saturation curves versus [DPD] at varying [ATP] (circle, 25 µM; solid circle 50 µM; square 100 µM; solid square 200 µM; solid triangle 400 µM) (A) and [ATP] at varying [DPD] (circle, 187.5 µM; solid circle 375 µM; square 750 µM; solid square 1500 µM; solid triangle 3000 µM) (B) to the matrix of substrate concentrations. Panels C–F present re-plots of parameters (Vmax)apparent and (V/K)apparent obtained from individually fit saturation curves to one substrate at fixed concentrations of the second substrate. Curves depicted are the best fit results using the parameters from the global fit. Error bars represent standard deviation.
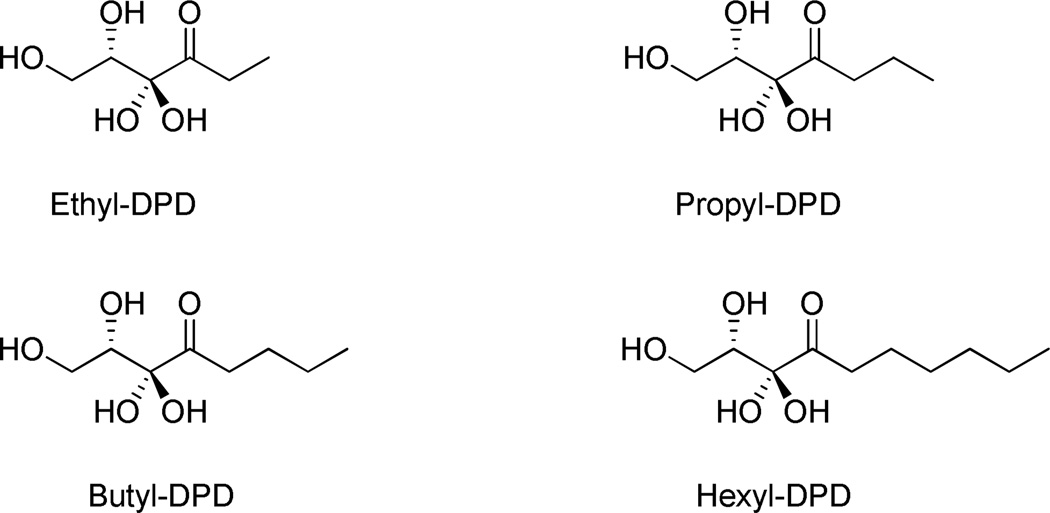
A focused substrate specificity profile was conducted using a series of C1-alkyl DPD analogues which are known potent antagonists or weak agonists (Ethyl-DPD) for Salmonella AI-2 signaling cascade (Figure 3).13 Initial velocities of LsrK were measured at varied concentrations of DPD analogues with saturating ATP (500 µM) and the Michaelis-Menten equation was fit to the initial rates to obtain substrate kinetic constants. Interestingly, we observed that the impact on specificity parameter kcat/Km(app) versus alkyl chain was minimal (less than 2-fold) with the exception of hexyl-DPD, whose activity was undetectable under the experimental conditions. It is tempting to speculate that with hexyl-DPD, a resulting steric clash is the cause for abolished enzymatic activity. Also noteworthy was that butyl-DPD possessed a depressed kcat, yet, its Michaelis constant Κm indicated significantly tighter binding compared to DPD, suggesting a better fit of the C1 butyl group in the binding cavity but a compromised orientation of the C5 hydroxyl group with active site residues for the phosphoryl-transfer event. This was further enunciated when butyl-DPD was varied with three fixed concentrations of DPD, again measuring the production of total NAD+ as a readout. A competing substrate model fits well to the resulting data (see supporting information, Figure S1).
Figure 3.
Structures of C1-alkyl DPD analogues used in the kinetic assays.
Comparison of the kinetic parameter of substrate specificity (kcat/Κm(app)) with the formerly reported DPD analogue profiling reveals a previously unappreciated structure-activity relationship as it relates to each homologue’s mechanism of action. First, it is evident that as an activating enzyme within the AI-2 signaling pathway, LsrK is a promiscuous kinase with a broad substrate spectrum since it phosphorylates DPD and DPD analogues with similar catalytic efficiency, i.e. kcat/Km(app). Second, while propyl- and butyl-DPD present similar biochemical profiles as seen in both the Salmonella β-gal reporter assay and the current LsrK activity assay, another equally phosphorylated analogue, ethyl-DPD, acts as a weak agonist in the cell-based reporter assay. As such, these findings support a model in which the closely structured DPD-analogues are phosphorylated by LsrK and the subsequent interaction of the phospho-analogue with LsrR is responsible for the observed agonist/antagonist profiles.11,12
These substrate specificity studies present an apparent biological/biochemical dichotomy seen with hexyl-DPD. Curiously, this DPD analogue displayed slightly diminished QS antagonism in Salmonella, yet, it is not phosphorylated by purified LsrK under our assay conditions. The possibility that hexyl-DPD might disrupt the coupling enzyme system was excluded as no inhibition with the PK and LDH enzymes was observed (See supporting information for details). The fact that hexyl-DPD was not readily phosphorylated by in vitro purified LsrK has also been seen using a semi-quantitative 1D TLC assay.12 We interpret these findings such that either phosphorylation is not necessary for this analogue to function as a QS modulator or there is an alternative promiscuous kinase phosphorylating DPD analogues. In either case, the analogue or phospho-analogue likely competes with phospho-DPD for the binding site on LsrR, and it is the structural variations embedded within the DPD-scaffolding that are responsible for the differential activity in stabilizing or destabilizing the LsrR-DNA complex.
Thus, we posit that LsrK does not serve as the critical juncture in determining QS agonist or antagonist activity in cell-based reporter assays as shown through examined DPD analogues. However, in making this statement we would also submit that since LsrK activity is required for QS activation,5 this enzyme is a suitable target for the development of kinase inhibitors as antagonists of QS provided these kinase inhibitors do not interact with the LsrR repressor.
A number of recent reports demonstrate LsrK to be a critical enzyme in type II bacterial quorum sensing in enteric bacteria including pathogenic strains such as S. typhimurium; to further investigate the function of this enzyme a quantitative assay for the LsrK kinase was developed and with it we determined that the enzyme operates under a rapid equilibrium ordered mechanism with ATP binding first. Furthermore, substrate specificity studies support the potential of developing C1-alkyl-DPD analogues as LsrK inhibitors based on preliminary structure-activity relationships. These findings also expose the danger of relying on a simple cell-based reporter assay as mentioned for analyzing QS activity, which does not necessarily reflect the complete mechanism of QS agonism/antagonism as seen from the reporter readout.
In sum, understanding the kinetic mechanism for the LsrK kinase is a first step in the path towards the establishment of LsrK as a distinct molecular target in AI-2 signaling and for the rational design of potent mechanism based inhibitors. Additional comprehensive studies of each component in the AI-2 signaling pathway, including signal recognition, transduction, and degradation will lead to a better understanding of the AI-2 signaling and regulation cascade.
Supplementary Material
Table 1.
Catalytic properties of LsrK toward C1-Alkyl DPD analogues.
| Compound | kcat (s−1) | Κm(app) (mM) |
kcat/Κm(app) (s−1 µM−1) |
IC50 (µM)a |
|---|---|---|---|---|
| DPD | 7.6 ± 0.6 | 1.0 ± 0.2 | 7.5×103 | |
| Ethyl-DPD | 5.8 ± 0.3 | 1.1 ± 0.1 | 5.3×103 | >50 |
| Propyl-DPD | 5.8 ± 0.5 | 1.4 ± 0.3 | 4.2×103 | 5.3 |
| Butyl-DPD | 1.5 ±0.1 | 0.27 ±0.03 | 5.5×103 | 5.0 |
| Hexyl-DPD | N/A | N/A | N/A | 25 |
IC50 value for QS antagonism.13
ACKNOWLEDGMENT
We gratefully acknowledge financial support from the NIH (Grant AI077644 to K.D.J.), the Skaggs Institute for Chemical Biology, and a postdoctoral fellowship from the German Academic Exchange Service (DAAD) to D.G.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Synthetic protocols, characterization of compounds, protein purification, assay methods, and kinetics fitting methods. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Fuqua WC, Winans SC, Greenberg EP. J Bacteriol. 1994;176:269. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waters CM, Bassler BL. Annual review of cell and developmental biology. 2005;21:319. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 3.Lowery CA, Dickerson TJ, Janda KD. Chem Soc Rev. 2008;37:1337. doi: 10.1039/b702781h. [DOI] [PubMed] [Google Scholar]
- 4.Xavier KB, Bassler BL. J Bacteriol. 2005;187:238. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taga ME, Miller ST, Bassler BL. Mol Microbiol. 2003;50:1411. doi: 10.1046/j.1365-2958.2003.03781.x. [DOI] [PubMed] [Google Scholar]
- 6.Pereira CS, de Regt AK, Brito PH, Miller ST, Xavier KB. J Bacteriol. 2009;191:6975. doi: 10.1128/JB.00976-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xavier KB, Miller ST, Lu W, Kim JH, Rabinowitz J, Pelczer I, Semmelhack MF, Bassler BL. ACS Chem Biol. 2007;2:128. doi: 10.1021/cb600444h. [DOI] [PubMed] [Google Scholar]
- 8.Xue T, Zhao L, Sun H, Zhou X, Sun B. Cell Res. 2009;19:1258. doi: 10.1038/cr.2009.91. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Attila C, Wang L, Wood TK, Valdes JJ, Bentley WE. J Bacteriol. 2007;189:6011. doi: 10.1128/JB.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy V, Meyer MT, Smith JA, Gamby S, Sintim HO, Ghodssi R, Bentley WE. Appl Microbiol Biotechnol. 2013;97:2627. doi: 10.1007/s00253-012-4404-6. [DOI] [PubMed] [Google Scholar]
- 11.Gamby S, Roy V, Guo M, Smith JA, Wang J, Stewart JE, Wang X, Bentley WE, Sintim HO. ACS Chem Biol. 2012;7:1023. doi: 10.1021/cb200524y. [DOI] [PubMed] [Google Scholar]
- 12.Roy V, Smith JA, Wang J, Stewart JE, Bentley WE, Sintim HO. J Am Chem Soc. 2010;132:11141. doi: 10.1021/ja102587w. [DOI] [PubMed] [Google Scholar]
- 13.Lowery CA, Park J, Kaufmann GF, Janda KD. J Am Chem Soc. 2008;130:9200. doi: 10.1021/ja802353j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganin H, Tang X, Meijler MM. Bioorg Med Chem Lett. 2009;19:3941. doi: 10.1016/j.bmcl.2009.03.163. [DOI] [PubMed] [Google Scholar]
- 15.Pereira CS, Santos AJ, Bejerano-Sagie M, Correia PB, Marques JC, Xavier KB. Mol Microbiol. 2012;84:93. doi: 10.1111/j.1365-2958.2012.08010.x. [DOI] [PubMed] [Google Scholar]
- 16.Marques JC, Lamosa P, Russell C, Ventura R, Maycock C, Semmelhack MF, Miller ST, Xavier KB. J Biol Chem. 2011;286:18331. doi: 10.1074/jbc.M111.230227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy V, Fernandes R, Tsao CY, Bentley WE. ACS Chem Biol. 2010;5:223. doi: 10.1021/cb9002738. [DOI] [PubMed] [Google Scholar]
- 18.Globisch D, Lowery CA, McCague KC, Janda KD. Angew Chem Int Ed Engl. 2012;51:4239. doi: 10.1002/anie.201109149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaufmann GF, Sartorio R, Lee SH, Rogers CJ, Meijler MM, Moss JA, Clapham B, Brogan AP, Dickerson TJ, Janda KD. Proc Natl Acad Sci U S A. 2005;102:309. doi: 10.1073/pnas.0408639102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horswill AR, Stoodley P, Stewart PS, Parsek MR. Anal Bioanal Chem. 2007;387:371. doi: 10.1007/s00216-006-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park J, Jagasia R, Kaufmann GF, Mathison JC, Ruiz DI, Moss JA, Meijler MM, Ulevitch RJ, Janda KD. Chemistry & biology. 2007;14:1119. doi: 10.1016/j.chembiol.2007.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsuchikama K, Zhu J, Lowery CA, Kaufmann GF, Janda KD. J Am Chem Soc. 2012;134:13562. doi: 10.1021/ja305532y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.