Abstract
In the analysis of biological systems, it is of interest to identify the components of the system and to monitor their changes in abundance under different conditions. The AQUA (for ‘absolute quantification’) method allows sensitive and specific targeted quantification of protein and post-translational modifications in complex protein mixtures using stable isotope–labeled peptides as internal standards. Each AQUA experiment is composed of two stages: method development and application to a biological scenario. In the method development stage, peptides from the protein of interest are chosen and then synthesized with stable isotopes such as 13C, 2H or 15N. The abundance of these internal standards and their endogenous counterparts can be measured by mass spectrometry with selected reaction monitoring or selected ion monitoring methods. Once an AQUA method is established, it can be rapidly applied to a wide range of biological samples, from tissue culture cells to human plasma and tissue. After AQUA peptide synthesis, the development, optimization and application of AQUA analyses to a specific biological problem can be achieved in ~1 week. Here we demonstrate the usefulness of this method by monitoring both Polo-like kinase 1 (Plk1) protein abundance in multiple lung cancer cell lines and the extent of Plk1 activation loop phosphorylation (pThr-210) during release from S phase.
INTRODUCTION
Protein and protein post-translational modification abundance undergo dynamic changes as cells traverse the cell cycle, during cellular differentiation and in response to intra- and extracellular stimuli. Advances in mass spectrometric instrumentation and methodologies allow accurate measurements of these changes in a high-throughput manner. Relative quantification of protein and post-translational modification abundance changes is commonly carried out by in vivo labeling in tissue culture using stable isotope-labeled amino acids (SILAC)1 or by chemical labeling using isotope-coded affinity tag (ICAT)2 and isobaric tag for relative and absolute quantitation (iTRAQ)3 reagents. These labeling strategies are often coupled with shotgun sequencing platforms and are ideally suited for discovery experiments designed to investigate many potentially unknown or undiscovered analytes and their differences between samples. Thus, these ‘hypothesis-generating’ experiments are invaluable for defining a narrower subset of dynamic protein targets to explore further in subsequent experiments that target them explicitly.
To this end, targeted quantification strategies such as AQUA4 and QconCAT5, which are based on stable isotope dilution of synthetic peptides enriched in 18O, 13C, 2H or 15N in the sample, are extremely powerful. Synthetic AQUA peptides are chemically and physically indistinguishable from their endogenous counterparts with respect to retention time, ionization efficiency and fragmentation mechanism. Therefore, they are both ideal internal standards that can be readily introduced to a sample during or after protease digestion as well as template analytes for developing methods that target them and their native counterparts. The heavy AQUA peptide and its endogenous light cognate are detected by a selected reaction monitoring (SRM) experiment in a mass spectrometer. On the basis of the known amount of the AQUA peptide added and the intensity ratio of both peptides, the amount of endogenous peptide can be calculated.
AQUA analyses are limited in the number of proteins that can be measured in a single liquid chromatography-mass spectrometry (LC-MS) run by the duty cycle of the mass spectrometer and the distribution of AQUA peptides across the LC elution profile. For some proteins of interest, this may result in only one or a few proteins being analyzed in a single run; for others, dozens of proteins can be targeted. An advantage of the approach is that it allows highly accurate and selective measurements of analyte abundance across many samples and conditions with moderate to high throughput. The high sensitivity of the AQUA approach reduces the sample amount necessary for analyte detection, and thus allows the identification and quantification of peptides that may be undetectable by shotgun proteomics. The simple addition of internal standards eliminates time-consuming in vivo or in vitro labeling of the samples and makes results easily transferable between instruments and laboratories.
In contrast to AQUA peptides, which are generated by chemical synthesis, QconCAT5,6 peptides are derived from artificial proteins by chemical or proteolytic digestion. Although the synthesis of AQUA peptides may be more expensive, it allows the introduction of amino acid modifications such as phosphorylation, ubiquitination, methylation and acetylation into the peptide; these are more difficult to selectively introduce into QconCAT-generated standards.
We and others have successfully used the AQUA strategy to determine absolute amounts of proteins and post-translational modifications such as phosphorylation and ubiquitination in complex mixtures. Recently, Langenfeld et al.7 described the quantification of cytochrome P450 2D6 expression in 30 human liver samples. The cell cycle–dependent expression of the human protein separase and its in vivo phosphorylation on serine 1126 in HeLa cells was measured using AQUA4. The same publication also identified the kinase that phosphorylates separase on a second residue, serine 1501, using AQUA in an in vitro kinase assay. Mayya et al.8 used AQUA to determine multisite phosphorylation of cyclin-dependent kinases on threonine T14 and tyrosine Y15 in a cell cycle–dependent manner in Jurkat cells. With an in vitro reconstruction system, Kirkpatrick et al.9 deciphered the topology of conjugated ubiquitin chains on cyclin B1. In an exhaustive in vivo analysis in yeast, Xu et al.10 monitored polyubiquitin chains of different linkage by AQUA and determine their function in proteasomal destruction. In this protocol, we describe peptide selection, sample preparation and methods development for an AQUA experiment, in which we determine the site occupancy of Plk1 T-loop phosphorylation on threonine 210 across the cell cycle.
Experimental design
Once you have selected the protein of interest, the following steps and considerations should be incorporated in the design of an AQUA experiment:
Peptide design
First, the peptide sequence for the synthesis of the internal standard is selected. If the absolute amount of a protein is to be quantified, theoretically, any peptide of the protein produced by a proteolytic digest (e.g., trypsin, chymotrypsin, Glu-C, Lys-C) can be selected. Ideally, two or three peptides are chosen per target protein to ensure accurate quantification and exclude artifacts that can originate from unknown modifications of the endogenous protein. In cases in which a specific splice variant, isoform or post-translational modification is targeted, the choice of peptides is limited by the site of modification and can only be marginally modified by the use of alternative proteases for digestion. In any case, it is essential that the chosen peptide is unique to the protein of interest. Ideally, peptides that have been previously sequenced in a shotgun experiment are chosen to ensure optimal chromatographic behavior, ionization, fragmentation and SRM transitions. Information on sequenced peptides can be found in published proteomics data sets and repositories such as PeptideAtlas11 (http://www.peptideatlas.org/), Human Proteinpedia12 (http://www.humanproteinpedia.org/) or PRIDE13 (http://www.ebi.ac.uk/pride/). These databases can be queried by protein ID, accession number or peptide sequence. If a peptide of interest (or peptides of the protein of interest) is in these databases, the search results return detailed information characterizing the peptide, previously recorded mass spectra of modified and unmodified versions of it, and information on the samples in which it was identified. If the peptide has not yet been sequenced, bioinformatic prediction programs14 can be used to choose potential peptides, or unlabeled peptides can be generated by peptide synthesis and inspected for their ability to function as a reliable AQUA peptide. In these cases, it is important to take into consideration that certain amino acid sequences or modifications can change the cleavage pattern of the selected protease. For instance, the protease trypsin normally cleaves at the carboxylic side of arginine and lysine. However, if proline is at the carboxylic side, the bond is resistant to trypsin cleavage. Similarly, if the amino acid on the amino-terminal side is phosphorylated, trypsin cleavage may be inhibited. If the protein contains a series of arginines and lysines, trypsin might cleave after the first arginine or lysine, or after any one following those, creating so-called ‘ragged ends’. For absolute quantification it is important that the abundance of all forms of the peptide be measured. If all the resulting peptide sequences can be established by discovery experiments, several AQUA peptides can be synthesized to account for them, or one AQUA peptide that spans the cleavage site can be generated and introduced into the sample before digestion, assuming that it will be digested in a manner similar to its native counterparts. Methionines in the peptide may be differentially oxidized during sample preparation; therefore, it is advisable to avoid methionine-containing peptides, but if that is not possible, to chemically oxidize all methionines to reduce the number of peptide species. In the case of post-translational modifications, not only the absolute amount of modified peptide but also the site occupancy can be determined by measuring the amount of unmodified peptide using a second AQUA peptide.
Peptide synthesis
AQUA peptides with a peptide sequence corresponding to the one generated during digestion of the endogenous protein are synthesized. During peptide synthesis, amino acids containing stable isotopes (18O, 13C, 2H or 15N) are incorporated into the peptide, leading to a peptide with the same chemical and physical characteristics as the endogenous one, but with a defined mass difference. Most commonly, 13C or 15N are used as stable isotopes, because they do not lead to chromatographic retention shifts (although deuterated peptides do). Usually, one heavy isotope–labeled leucine, proline, valine, phenylalanine or tyrosine is incorporated into an AQUA peptide, leading to a mass shift of 6–8 Da. For tryptic peptides, the C-terminal arginine or lysine is often heavy isotope–labeled such that the resulting y-ion series can be used for monitoring. The peptide is purified, and the exact amount of peptide is determined by amino acid analysis or total nitrogen detection. Today, there are commercial vendors that conduct peptide synthesis (including incorporation of a stable isotope-labeled amino acid; e.g., Sigma-Aldrich, Thermo Fisher Scientific or Cell Signaling Technologies). The cost of an AQUA peptide is currently about $400, although with increasing interest in the method, AQUA peptide libraries15 can be assembled, allowing faster and cheaper orders of previously made and tested peptides.
Peptide validation
After synthesis, the AQUA peptide is analyzed by LC-MS/MS to verify its chromatographic behavior and fragmentation spectra. If they correspond to the previously detected or predicted characteristics, the peptide is ready for use.
Choice of MS instrument and analysis method
There are two major instrumentation platforms that are used to develop and deploy AQUA methods: trapping instruments (e.g., ion trap, Orbitrap, etc.) and quadrupole-based instruments (e.g., triple quad, quadrupole-TOF (time of flight), etc.). Layered on top of instrument selection are two methods for quantification: selected ion monitoring (SIM) or SRM methods. SIM methods rely only on the identification and quantification of intact peptide ions in full scan (MS1) spectra, whereas SRM methods fragment both the native and internal standard peptides to produce greater selectivity in quantification in MS2 spectra. Thus, instruments that are capable of high mass accuracy in MS1 spectra are better suited to SIM analyses than lower resolution instruments, because of the application of accurate mass in discriminating between peptide ions of interest and co-eluting chemical noise. On the contrary, quadrupole-based instruments are much better suited to SRM methods by virtue of their ability to generate a continuous ion beam in the SRM transition. In general, trapping instruments are easier to set up and operate, whereas quadrupole-based instruments often require more expertise to optimize fully. However, well-designed SRM methods on a triple quadrupole MS instrument are capable of significantly lower limits of quantification in mixtures of very high complexity, such as whole-cell lysate or unfractionated serum. Ultimately, the choice of instrument platform will be guided by a combination of the target application, user experience and instrument availability.
Method optimization
An MS spectrum of the peptide (or peptides) is first collected, either by infusion or by LC-MS. Typically, the initial charge-state distribution is interrogated to establish the most sensitive charge state for further monitoring. Note that the actual charge state distribution in a complex mixture may be different than that observed from purified peptide samples; when you first deploy an AQUA method in a real biological matrix, it is advisable to sample from multiple available charge states to ensure that the most sensitive form of the analyte is ultimately used. A SIM method with a narrow m/z scan range for the charge state with the highest intensity that covers both the AQUA peptide and the endogenous peptide is established from this MS1 spectrum. For SRM-based methods, the MS/MS spectra of the most intense precursor ions of the AQUA peptide are collected and inspected. Fragment ions at m/z ratios higher than the precursor ion are often more suitable for monitoring because of reduction of noise compared with the lower m/z space. Ion intensity and retention time can be optimized by varying the amount of organic solvent in the peptide loading buffer and in the column equilibration phase of an LC-MS method. In addition, software tools are now available to assist scientists in developing scheduled SRM methods and interpreting their data16.
Sample preparation
The biological samples are collected and lysed. AQUA has been successfully applied to many different types of biological samples such as yeast4,10, mice17 and human7 tissues, in vitro systems9 and human cell lines4,8,18. The sample can be directly protease digested, or, to reduce complexity, the sample can be fractionated before digestion by gradient centrifugation, fast protein liquid chromatography (FPLC) or SDS–polyacrylamide gel electrophoresis (PAGE), or other protein fractionation methods. Measurement of protein abundance by AQUA is indirect and based on the abundance of the resulting peptides; therefore, complete proteolysis is essential, and care should be taken to digest the target mixture with increasing amounts of protease and/or for longer time periods until no additional endogenous peptide signal can be observed. Furthermore, tryptic digestion generates peptides with chemical properties (e.g., charge states) ideally suited for MS/MS analysis. If other proteases are used for digestion, efficiency of digestion should be determined by careful titration of the amount of enzyme against a known amount of protein standard. The AQUA peptide is introduced during digestion to account for all potential peptide losses in the following steps.
MS analysis
The peptide mixture containing the endogenous and the AQUA peptide is analyzed on the mass spectrometer by a SIM or a SRM method, and the amount of endogenous peptide is determined. In contrast to a full MS scan, in a SIM experiment, only a very narrow mass range is scanned, often by selectively injecting or trapping ions from the narrow scan range to increase the target ion signal-to-noise ratio. In an SRM experiment, a fragment ion or set of fragment ions is monitored. Typically, triplicate measurements for well-designed AQUA experiments produce coefficients of variation between 8% and 15% relative standard deviation.
MATERIALS
REAGENTS
SDS (Pierce, cat. no. 62202)
Glycerol (Fisher)
Tris-HCl (Sigma-Aldrich, cat. no. T5941)
NaCl (Sigma-Aldrich, cat. no. S6191)
Sodium fluoride (Sigma-Aldrich, cat. no. 71519)
Sodium orthovanadate (Sigma-Aldrich, cat. no. S6508)
Sodium molybdate (Sigma-Aldrich, cat. no. M1651)
Sodium tartrate (Sigma-Aldrich, cat. no. 228729)
β-Glycerophosphate (Sigma-Aldrich, cat. no. G6251)
Complete mini EDTA-free protease inhibitor (Roche, cat. no. 11846170001)
dl-DTT (Sigma-Aldrich, cat. no. D0632)
Iodoacetamide (Sigma-Aldrich, cat. no. I1149)
NuPAGE LDS SDS-PAGE sample buffer (4×; Invitrogen, cat. no. NP0008)
NuPAGE Novex Bis-Tris gel (4–12%; Invitrogen, cat. no. NP0335BOX)
Mark12 unstained protein molecular weight marker (Invitrogen, cat. no. LC5677)
NuPAGE MOPS SDS running buffer (Invitrogen, cat. no. NP0001)
Coomassie Blue R-250 (Pierce, cat. no. 20278)
Acetic acid (Fisher) ! CAUTION It is an acid and is hazardous.
Methanol (Fisher)
Acetonitrile (HPLC-MS grade; Honeywell Burdick and Jackson, cat. no. AH015-4)
Ammonium biocarbonate (Fluka, cat. no. 09830)
Water (HPLC-MS grade; Honeywell Burdick and Jackson, cat. no. AH365-4)
Trifluoroacetic acid (TFA; EMD, cat. no. TX1276) ! CAUTION It is an acid and is hazardous.
AQUA peptides (Cell Signaling Technology, Sigma-Aldrich)
Sequencing-grade trypsin (Promega, cat. no. V5111)
High-purity formic acid (EMD, cat. no. 11670) ! CAUTION It is an acid and is hazardous.
Sodium periodate
Samples (tissues, cell lines, yeast or other appropriate cell types) ! CAUTION Any experiments with human or animal tissues must adhere to all relevant institutional and governmental guidelines.
EQUIPMENT
Water bath
Bench-top centrifuge
SDS-PAGE gel running system
Glass plate
Vortexer
SpeedVac (or similar vacuum concentrator)
Microcentrifuge
Microcentrifuge tubes
Column puller (P-2000, Sutter Instrument)
LTQ-Orbitrap mass spectrometer (Thermo Fisher)
Agilent 1200 binary HPLC pump (Agilent) with degasser
FAMOS autosampler (LC Packings)
Reprosil-Pur (C18, 3 μm, 200 Å, Dr. Maisch)
Kimwipes
Scalpel (for cutting gel bands)
Syringe with 27-gauge needle
Qual Browser software (or equivalent)
REAGENT SETUP
HeLa cell lysis buffer
Combine SDS (1% (wt/vol)), 15% (wt/vol) glycerol, 50 mM Tris-HCl (pH 8.7), 150 mM NaCl, phosphatase inhibitor (1 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM sodium molybdate, 1 mM sodium tartrate, 1 mM β-glycerophosphate), and complete mini EDTA-free protease inhibitor. ▲ CRITICAL Prepare fresh before use.
Sodium orthovanadate
To prepare 100 mM stock, dissolve 1.83 g in 100 ml of water. Adjust the solution to pH 10 and boil until the solution turns from orange to translucent, indicating that monovanadate is present. Aliquot and store at –20 °C for up to 1 year.
Coomassie Blue dye
Add Coomassie Blue R-250 (2.5 g) to 300 ml methanol. Stir for 15 min, let sit for 30 min and allow the particles to settle, and then decant into a 1-liter bottle. Add 600 ml water and 100 ml glacial acetic acid. Store at room temperature (RT, 20–25°C) for up to 3 months.
Coomassie destain solution
Combine glacial acetic acid (10% (vol/vol)) and 30% (vol/vol) methanol. Store at RT for up to 6 months.
Coomassie clarifying solution
Glacial acetic acid (7% (vol/vol)). Store at RT for up to 6 months.
Gel destaining solution
Ammonium bicarbonate (50 mM; no pH adjustment) in 50% (vol/vol) HPLC-MS-grade acetonitrile/50% (vol/vol) HPLC-MS-grade water. Store at RT in clean glass vials for up to 6 months.
Peptide extraction solution
HPLC-MS-grade acetonitrile (47.5% (vol/vol))/HPLC-MS-grade water (47.5% (vol/vol)) containing 5% (vol/vol) high-purity formic acid. Store at RT for up to 6 months.
Peptide loading buffer
HPLC-MS-grade acetonitrile (5% (vol/vol))/high-purity formic acid (1% (vol/vol)) in HPLC-MS-grade water. Store at RT for up to 3 months.
AQUA peptide storage solution
HPLC-MS-grade acetonitrile (5% (vol/vol))/high-purity formic acid (1% (vol/vol)) in HPLC-MS-grade water. Store at RT for up to 3 months.
AQUA peptide diluent
HPLC-MS-grade acetonitrile (5% (vol/vol))/high-purity formic acid (1% (vol/vol)) in HPLC-MS-grade water. Store at RT for up to 3 months.
LC-MS/MS solvents
Buffer A: 3% (vol/vol) HPLC-MS-grade acetonitrile and 0.125% (vol/vol) high-purity formic acid in HPLC-MS-grade water. Buffer B: 95% (vol/vol) HPLC-MS-grade acetonitrile and 0.125% (vol/vol) high-purity formic acid in HPLC-MS-grade water. Store both buffers at RT for up to 3 months.
Autosampler chase solution
HPLC-MS-grade acetonitrile (5% (vol/vol))/high-purity formic acid (1% (vol/vol)) in HPLC-MS-grade water. Store at RT for up to 3 months.
EQUIPMENT SETUP
LC-MS/MS equipment
FAMOS autosampler with 8-μl loop; Agilent 1200 capillary HPLC pumps operating between 2 and 100 μl min–1 set up with a double-split system to provide an in-column flow rate of ~0.5 μl min–1 on a microcapillary column (125 μm inner diameter and 16 cm length) packed with C18 reverse-phase material (Reprosil-Pur C18, 3 μm, 200 Å, Dr. Maisch GmbH); and an LTQ-Orbitrap mass spectrometer equipped with a nanospray source.
PROCEDURE
AQUA peptide validation and MS method development
Dilute AQUA peptide in peptide loading buffer to a final concentration of 10–100 fmol μl–1, depending on the sensitivity of your LC-MS platform.
- Analyze the diluted AQUA peptide using a standard LC-MS/MS method.
- ▲ CRITICAL STEP Optimize the LC-MS method by varying peptide loading conditions, LC gradient and peptide amount to achieve optimal and reproducible retention time, chromatographic peak shape and ion intensity. For example, overloading of the column with too much sample will cause broadening of the elution profile and shifts in retention time. Injecting a dilution series of a sample is useful to establish the maximal loading capacity of a column setup (the point at which the retention time and peak shape begins to change). Note that more hydrophobic peptides are often recovered to a greater degree when the reverse-phase column is equilibrated in higher concentrations of organic solvent. Tailor the LC gradient to the peptide or peptides of interest.
- Develop an LC-SIM method using option A and a LC-SRM method using option B.
- LC-SIM method
- Extract the exact mass of the AQUA peptide from the full-scan LC-MS spectrum and note the retention time.
- Calculate the mass (m/z) of the endogenous peptide. This can also be accomplished by calculating the mass of the heavy isotope label and subtracting this from the observed mass of the AQUA peptide. For high mass accuracy instruments, calculate the exact monoisotopic mass (m/z) of the peptide using standard mass tables (http://physics.nist.gov/cgi-bin/Compositions/stand_alone.pl?ele=&all=all&ascii=html&isotype=some).
- Write an MS method for a SIM scan, with the observation window centered between the m/z of the AQUA and endogenous peptides and an m/z width covering both peptide forms and isotope distributions. Schedule the SIM as needed by bracketing the elution profile of the AQUA peptide with SIM start and SIM end time windows based on the reproducibility of your particular LC platform. A typical LC system requires 0.25 minutes of padding before and after the peptide retention time. Note that it is common for peptides to elute at different time periods when they are injected as pure standards (versus when they are diluted in a complex peptide matrix).
- Reanalyze the diluted AQUA peptide sample by the LC-SIM method to confirm the performance. Adjust the schedule windows if necessary.
- LC-SRM method
- Generate tandem mass spectra for the AQUA peptides of interest by infusion or by LC-MS/MS.
- Inspect MS/MS spectra for high-intensity and informative fragment ions. For example, neutral losses (ammonia, water, phosphoric acid, etc.) are not generally informative. Experience suggests that while mass spectrometers can be tuned to improve the sensitivity of peptides (in general) over other analytes, there are few distinct differences between most proteotypic peptides that require specific tuning parameters. Collision energy is likely the most important parameter with respect to optimizing sensitivity of a given peptide SRM over that of a different peptide.
- Raster the collision energy while recording data to establish the optimum fragmentation conditions for your peptide SRM, or use instrument vendor-specific linear equations based on m/z values to establish the most sensitive collision energy for your peptide(s).
- Write an MS method for an SRM scan of the selected fragment ion(s). Experience suggests that multiple fragments may not be required to achieve maximum sensitivity. Although additional fragment ion transitions will contribute positive signal to the SRM, each additional transition is also an opportunity for nonspecific chemical noise to contaminate the method. Expect to return to this section of the protocol after deploying the SRM in a real biological matrix to discern which transitions ultimately produce meaningful and informative results for your particular analysis setup.
- Schedule the SRM as needed by bracketing the elution profile of the AQUA peptide with SRM start and SRM end time windows based on the reproducibility of your particular LC platform. A typical LC system requires 0.25 minutes of padding before and after the peptide retention time. Note that it is common for peptides to elute at different time points when injected as pure standards versus when diluted in a complex peptide matrix.
- Reanalyze the diluted AQUA peptide sample using the LC-SRM method. Adjust the schedule windows if necessary.
- Develop a corresponding LC-SRM method for the endogenous peptide, keeping the method parameters developed for the AQUA peptide consistent between the two. Keep in mind that not only the precursor mass but also only certain fragment ions will change in mass between the AQUA and endogenous peptide MS/MS spectra. Knowledge of which amino acid is labeled in the AQUA peptide and the specific type of fragment ion to be monitored is essential in developing the endogenous peptide SRM.
Cell lysis, reduction and alkylation
-
4
Prepare lysis buffer, add protease and phosphatase inhibitors, and chill it on ice.
-
5
If the sample was frozen, thaw it quickly under a warm water stream or in a 37 °C bath. If the sample is freshly harvested, place it on ice to chill.
-
6
Add lysis buffer to sample (any cell type can be used; we use HeLa cells here) Add 200 μl buffer to 106 cells; this yields a final protein concentration of ~2 mg ml–1.
-
7
Pass the lysate three times though a syringe with a 27-gauge needle to shear the DNA. For larger volumes, syringing can be replaced by sonication using a microtip-equipped sonicator. Remove cell debris by centrifuging the lysate at 13,000g in a microcentrifuge at 4 °C for 15 min.
-
8Transfer the supernatant to a clean tube.
- ■ PAUSE POINT Snap-freeze the lysate in liquid nitrogen; most sample types can be stored at –80 °C for up to 1 year.
-
9
To ensure the highest efficiency of protease digestion, cysteines should be reduced and alkylated. This procedure guarantees access of the protease to all sites, increases the number of proteolytic peptides and ensures complete digestion. Furthermore, some target sequences of interest may contain cysteine residues: AQUA peptides are readily available with acetamide-capped cysteines. Adjust the pH of the lysate to 8–8.25, add DTT to a final concentration of 5 mM, vortex and then heat the sample to 65 °C for 20 min.
-
10
Carefully and completely cool the sample to RT. Add iodoacetamide to a final concentration of 15 mM, vortex and incubate for 1 h at RT in the dark. A bench drawer works fine for this.
-
11
Quench the alkylation reaction by adding more DTT to a final concentration of 10 mM (add the same amount as in Step 9), and then incubate for 15 min.
SDS-PAGE
-
12
Add one volume of NuPAGE LDS 4× SDS-PAGE sample buffer to three volumes of lysate.
-
13
Heat to 90 °C for 5 min, then allow the sample to cool to RT.
-
14
Load 10 μl Mark12 protein molecular weight marker into the first well of a 4–12% NuPAGE Novex Bis-Tris gel and sample (between 20 and 30 μg of total protein) into the remaining wells.
-
15
Fill the gel running chamber with NuPAGE MOPS SDS running buffer. Start the gel at 80 V for 30 min, and then increase the voltage to 200 V until the end.
-
16
Remove the gel from the cassette and stain it for 30 min in Coomassie Blue dye.
-
17
Decant the Coomassie stain, rinse the gel with ddH2O, and add destaining solution. The addition of a Kimwipe to the Coomassie destain solution speeds up the destain process. Destain the gel until the molecular weight marker is clearly visible.
-
18
Remove the destaining solution and completely destain the background of the gel by adding Coomassie clarifying solution.
In-gel digestion
-
19Remove the gel to a clean glass plate and excise the gel region containing the protein of interest with a clean scalpel. Dice the gel slice in 1-mm cubes.
- ▲ CRITICAL STEP It is important to cleanly and completely capture the target protein in the gel slice during this step. Use the molecular weight marker as a guide, bearing in mind that western blot markers do not always correspond to similar molecular weight ranges for unstained markers. If you are unsure of where your target protein resides in the gel, remove multiple slices in the general vicinity of the target protein molecular weight, taking care to note the relative position of each section (scanning or photographing the gel after excision is useful in this regard).
- ■ PAUSE POINT Gel cubes can be stored at –20 °C for up to 1 month.
-
20
Transfer the gel cubes to a clean tube. Add 1 ml in-gel destain solution to completely destain them. Vortex and incubate at 37 °C for 3 h.
-
21
Remove the destain solution and repeat Step 20 until the cubes are completely clear. This process can take several hours, depending on the amount of protein and the intensity of the stain.
-
22
When the cubes are completely destained, dehydrate them by adding 1 ml of acetonitrile to the tubes, vortex and incubate at RT for 15 min.
-
23Remove acetonitrile and repeat Step 22 to ensure complete dehydration. Break up aggregated cubes with a pipette tip and completely dry cubes in a SpeedVac for 15 min.
- ■ PAUSE POINT Dehyrated gel cubes can be stored at –20 °C for up to 1 month.
-
24Dilute stock AQUA peptides to a final concentration of 5–50 fmol μl–1 in 50 mM ammonium bicarbonate. Dilute only the amount of AQUA peptides required to conduct the experiment.
- ▲ CRITICAL STEP Note that peptide solutions at < 1 pmol μl–1 are not quantitatively stable for longer than 24 h at RT and should not be frozen for storage. Prepare these dilute solutions fresh for use.
-
25
Place 50 mM ammonium bicarbonate solution on ice until chilled.
-
26
Thaw trypsin stock solution.
-
27
Place tubes containing dehydrated gel cubes on ice.
-
28
Dilute trypsin stock solution in 50 mM ammonium bicarbonate to 5–15 ng μl–1 and vortex to mix.
-
29
Rehydrate the gel cubes by adding the trypsin solution. A typical gel piece will require 25 μl trypsin solution.
-
30
Add AQUA peptides to the gel cubes. The appropriate amount of AQUA peptides added depends on their absolute ion intensity as well as their relative intensity compared with the endogenous peptides, which should not diverge more than tenfold. Titration of this ratio might be necessary with a starting point in the 5–50 fmol range.
-
31
Incubate on ice for 20–30 min or until the gel cubes are completely rehydrated and clear. If necessary, add additional 50 mM ammonium bicarbonate for complete rehydration.
-
32
Incubate gel cubes overnight at 37 °C.
Peptide extraction and in-gel digestion
-
33
Extract peptides by adding 100 μl extraction buffer, vortex and incubate for 10 min at RT. Centrifuge gel cubes at 16,000g for 30 s.
-
34
Transfer the supernatant to a clean tube and repeat Step 33 once.
-
35Dry the extracted peptides in a SpeedVac. If the peptides of interest contain methionines, they can be oxidized to sulfoxide by resuspension in 2 mM freshly prepared sodium periodate solution at RT for 20–30 min, followed by quenching with an equal volume of 5 mM freshly prepared lead acetate solution, desalting on STAGE tips or similar reverse-phase desalting device, and drying in a SpeedVac.
- ■ PAUSE POINT Store the samples until use at –20 °C for up to 1 month.
SIM or SRM experiment with tandem MS
-
36
Resuspend the peptides in peptide loading buffer, vortexing or sonicating well to mix, to achieve ~1 μg μl–1 peptide concentration. Peptide mass amounts can be estimated from the determination of total protein in the initial steps of lysis or from visualization steps such as Coomassie staining in cases in which SDS-PAGE is employed.
-
37
Analyze the peptides using the previously established LC-SIM or LC-SRM method. Note that the maximum loading capacity of most microcapillary LC-MS columns is about one μg. Note that overloading the column can cause significant retention time shifts of peptides, poor chromatographic peak shapes and memory effects on subsequent injections.
-
38Carry out data analysis with either option A (LC-SIM method) or option B (LC-SRM method).
- LC-SIM method
- Extract masses of the AQUA and the endogenous peptide from the SIM scan. Note that for high-mass-accuracy instruments, you should use the exact masses observed for the AQUA peptide and calculated for the endogenous peptide in Step 3.
- Determine the peak area with data processing software supplied by the instrument manufacturer or other postprocessing software.
- Calculate the amount of endogenous peptide.
- ? TROUBLESHOOTING
- LC-SRM method
- Extract masses of the fragment ions from the SRM channel(s).
- Determine the peak area with data processing software supplied by the instrument manufacturer or other postprocessing software.
- Calculate the amount of endogenous peptide.
- ? TROUBLESHOOTING
? TROUBLESHOOTING
Troubleshooting advice can be found in table 1.
TABLE 1.
Troubleshooting table.
| Problem | Solution |
|---|---|
| Peptides generated by the sample protein have ragged ends | Synthesize all occurring peptide forms or synthesize extended AQUA peptides |
| Peptide contains cysteine residues | Reduce with DTT and alkylate with iodoacetamide |
| Peptide contains methionine | Oxidize with sodium periodate |
| Incomplete digestion | Use a different protease |
| Peptide elutes late or in a wide peak | Optimize HPLC gradient and peptide loading buffer by changing the percentage of acetonitrile |
| Cannot detect endogenous peptide | Check gel slices above and below the original location |
● TIMING
AQUA peptide selection: 30 min
Steps 1 and 2, AQUA peptide validation: 1 h
Step 3, Development and validation of LC-SIM or LC-SRM method: 3 h
Steps 4–11, Cell lysis, reduction and alkylation: Steps 4–8, 30 min; Steps 9–11, 105 min
Steps 12–18, SDS-PAGE: Steps 12–15, 90 min; Step 16, 30 min; Step 17, 1 h; Step 18, overnight
Steps 19–32, In-gel digestion: Steps 19–21, 3 h; Steps 22 and 23, 45 min; Steps 24–31, 30 min; Step 32, 6 h to overnight
Steps 33–35, Peptide extraction and in-gel digestion: Steps 33 and 34, 20 min; Step 35, 1.5 h
Steps 36 and 37, SRM experiment via tandem MS: 45 min per run
Steps 36 and 37, SIM experiment via tandem MS: 45 min per run
Step 38, Data analysis: 1–2 h
ANTICIPATED RESULTS
Peptide selection for Plk1 abundance and T-loop phosphorylation measurements
To demonstrate the utility of the AQUA method for measuring abundance levels of proteins as well as post-translational modifications, we determined the expression levels of Plk1 in seven lung cancer cell lines and the site occupancy of Plk1 T-loop phosphorylation on threonine 210 during release from S phase and entry into mitosis. Plk1 mRNA expression has been previously shown to negatively correlate with prognosis in patients with lung cancer19, and the activation of this essential kinase by Aurora A–dependent phosphorylation on threonine 210 is crucial for cells to enter and navigate a successful mitosis20,21.
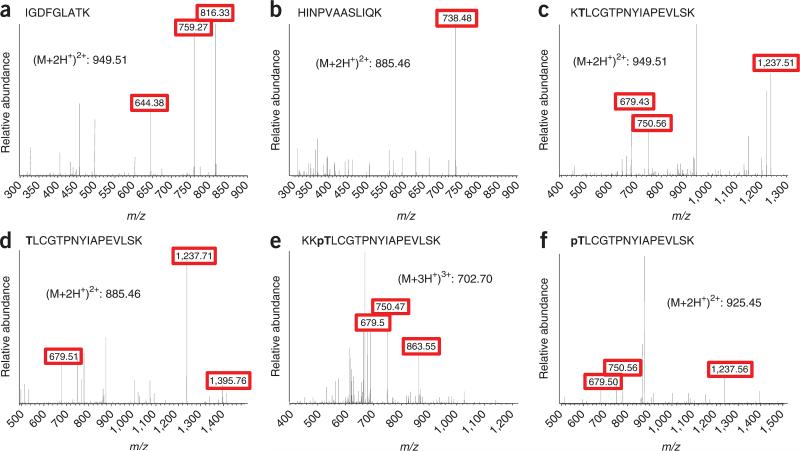
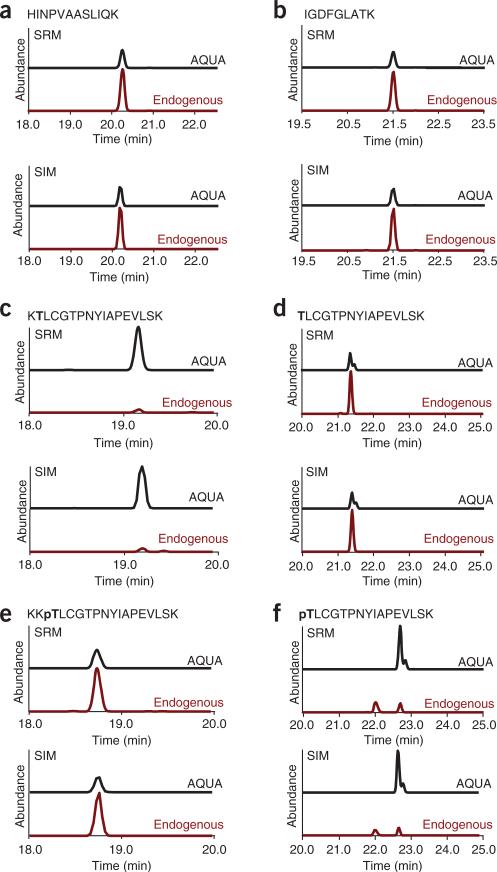
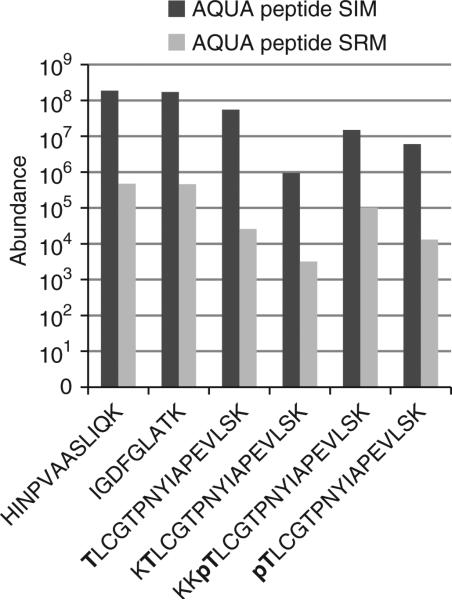
Figure 1 is a depiction of the general workflow. For peptide selection, Plk1 was digested in silico with trypsin. Two peptides outside the T-loop, which did not contain the potentially problematic residues cysteine or methionine (IGDFGLATK and HINPVAASLIQK), were chosen for AQUA peptide synthesis for total protein abundance measurements. We confirmed, for both peptides, that no phosphorylation of the serine or threonine was previously observed by consulting published large-scale mitotic phosphoproteomics data sets22,23 and online resources (http://www.uniprot.org/). In silico digestion generated peptides with ragged ends that spanned the activation site T210. To conclusively determine which end-forms of phosphorylated and unphosphorylated peptides are generated by trypsin, we immunoprecipitated Plk1 from nocodazole-arrested HeLa cells. Treatment with nocodazole arrests cells in mitosis by depolymerizing microtubules and activating the mitotic checkpoint, which leads to hyperphosphorylation of Plk1. On the basis of this analysis, we found two forms of the phosphorylated (KKpTLCGTPNYIAPEVLSK and pTLCGTPNYIAPEVLSK) and two forms of the unphosphorylated (KTLCGTPNYIAPEVLSK and TLCGTPNYIAPEVLSK) peptides generated during trypsin digestion that spanned T210. To determine the abundance of all peptide forms, one can synthesize AQUA peptides corresponding to all these forms or to one extended peptide which is then digested into the different forms during incubation with trypsin. An extended peptide may be useful when you are unable to analyze the intact protein obtained from native sources to generate information on the in vitro–produced peptide forms. In this analysis, we synthesized all four peptides covering the T-loop. After receiving the peptides, we generated a 50 fmol μl–1 stock solution containing the four peptides and analyzed it by LC-MS/MS. To optimize retention time and chromatographic behavior for these relatively hydrophobic peptides, we increased the percentage of acetonitrile to 8% (vol/vol) in the peptide loading buffer during peptide loading and at the beginning of the elution gradient. The analysis was performed on an LTQ-Orbitrap. We have previously detected Plk1 protein in shotgun sequencing experiments from nocodazole-arrested HeLa whole-cell lysates fractionated by SDS-PAGE and analyzed by LC-Orbitrap-MS/MS; thus, we were assured of observing Plk1 peptides by full-scan MS1 spectra on our Orbitrap mass spectrometer platform. For inexperienced users, this is a good rule of thumb—if the protein of interest can be sequenced in a shotgun sequencing experiment on any MS platform, there is a high likelihood of success in developing an AQUA method for that protein. Here, we describe the development of SRM- and SIM-based methods for the characterization of Plk1 expression and threonine 210 phosphorylation. For the SIM methods, we chose scan ranges centered between the parent masses of the AQUA and endogenous peptides, with a width of 10 m/z at 30,000 resolutions to detect both ions in the same SIM scan. On the basis of the MS/MS spectra obtained in the initial analysis of the AQUA peptide, we chose fragment ions (red boxes) for each peptide for SRM transitions (Fig. 2). To guarantee comparable ion fill times in the trap and, thereby, comparable ion intensity of the AQUA and endogenous peptides, we monitored both fragment ions from the AQUA and the endogenous peptides in a single, wide isolation SRM scan. Comparison of peptide abundance measured by SRM and SIM methods yielded highly similar results for both the AQUA and endogenous peptides from immunoprecipitated Plk1 (Fig. 3). However, the measured peak area/ion intensity for a respective peptide was, on average, 400-fold higher with the SIM method than the SRM method (Fig. 4). This is generally true: full-scan (SIM) quantification is more sensitive than MS2 scan quantification, whereas MS2 scan quantification can be much more selective than quantification by SIM scan.
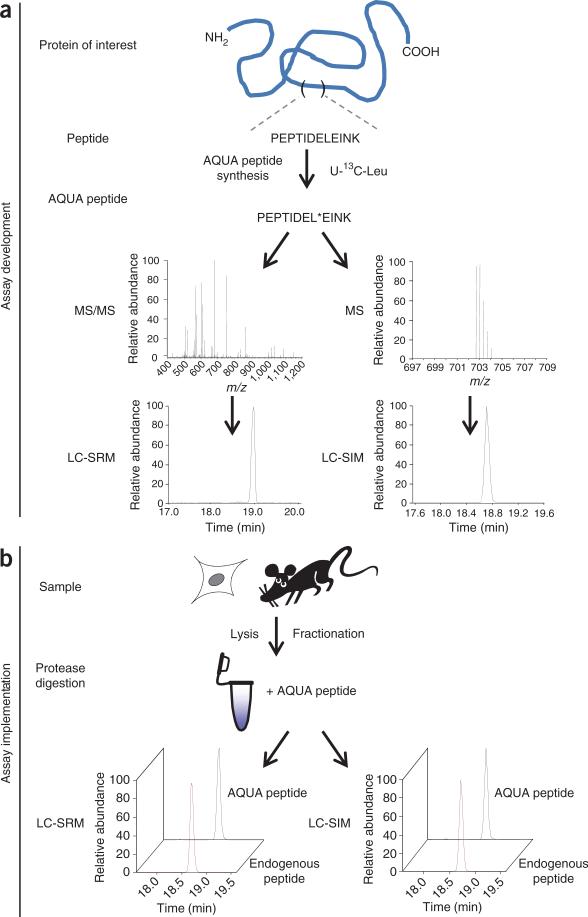
Figure 1.
Scheme of AQUA workflow. (a) Assay development: the protein of interest is inspected for suitable peptides for AQUA analysis. The selected peptide is synthesized as an AQUA peptide by incorporating a heavy isotope–labeled amino acid (e.g., U-13C-leucine). The AQUA peptide is analyzed by LC-MS/MS to determine the retention time, full scan (MS1) and fragmentation spectra. LC-SRM and LC-SIM methods are developed and implemented for the AQUA peptide. (b) Assay implementation: a sample (primary tissue, yeast or tissue culture cells) is lysed. At this step, proteins can be fractionated by SDS-PAGE or other methods. During protease digestion, AQUA peptides are added to the sample. Peptides are extracted and analyzed by LC-SRM or LC-SIM.
Figure 2.
MS/MS spectra of the AQUA peptides for measuring Plk1 abundance and Plk1 T210 T-loop phosphorylation. (a–f) Unmodified peptides for the measurement of total Plk1 protein abundance (a,b); unphosphorylated peptides at the T-loop (c,d); and phosphorylated versions of the T-loop peptides (e,f). Fragment ions that were selected for monitoring are boxed in red.
Figure 3.
Comparison of LC-SRM and LC-SIM traces for 10 fmol of each AQUA peptide as well as their endogenous counterparts from a Plk1 immunoprecipitation. (a–f) Unphosphorylated peptides outside the T-loop (a,b); unphosphorylated versions at the T-loop (c,d); and phosphorylated versions of the T-loop peptides (e,f).
Figure 4.
Comparison of the abundance measurements by LC-SRM and LC-SIM for the AQUA peptides. The signal intensity of the LC-SIM method is ~400-fold higher than the LC-SRM method.
Determining Plk1 abundance in lung cancer cell lines
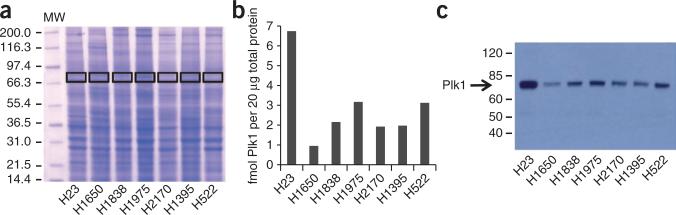
To determine expression levels of Plk1 in unsynchronized, growing H23, H1650, H1838, H1975, H2170, H1395 and H522 cells, they were collected, lysed at 2 mg ml–1 concentration, reduced and alkylated. Twenty micrograms of protein was loaded onto a SDS-PAGE gel and separated. The gel was stained with Coomassie blue dye, and a region of ~70 kDa was excised from the gel (Fig. 5a). The gel region was diced in cubes, completely destained, dehydrated and rehydrated using trypsin solution. The AQUA peptide mixture was added to the rehydrated gel cubes. For complete digestion, gel pieces were incubated overnight at 37 °C. The next day, peptides were extracted and analyzed by LC-SRM. Data analysis was performed using LTQ-Orbitrap instrument data viewing software (Qual Browser). In this analysis, we found that H23 contained the highest amount of Plk1 (6.7 fmol per 20 μg total protein), whereas H1650 contained the lowest amount of Plk1 (0.95 fmol per 20 μg total protein) (Fig. 5b). Western blot analysis of these cell lines using an antibody to Plk1 showed similar relative expression levels (Fig. 5c).
Figure 5.
Plk1 expression in lung cancer cells lines. (a) Coomassie gel of lysates of unsynchronized lung cancer H23, H1650, H1838, H1975, H2170, H1395 and H522 cell lines. MW, molecular weight (in kDa). (b) Plot depicting Plk1 abundance in lung cancer cell lines, as measured by LC-SRM of HINPVAASLIQK and IGDFGLATK. (c) Western blot of Plk1 expression from the same lysates.
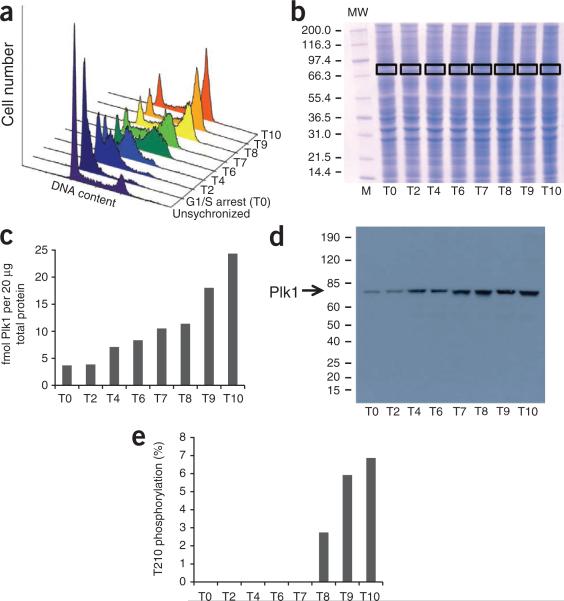
Monitoring of Plk1 T-loop phosphorylation during release from S phase by the AQUA method
To monitor Plk1 T210 phosphorylation, HeLa cells were arrested at the G1/S transition by a double thymidine block and then released; samples were collected 2, 4, 6, 7, 8, 9 and 10 h after release (Fig. 6a). Lysates were prepared at 2 mg ml–1 concentration, reduced and alkylated. Twenty micrograms of protein was loaded onto a SDS-PAGE gel and separated. The gel was stained with Coomassie blue dye, and a region of ~70 kDa was excised from the gel (Fig. 6b). The gel region was diced in cubes, completely destained, dehydrated and rehydrated with trypsin solution. The AQUA peptide mixture was added to the rehydrated gel cubes. For complete digestion, gel pieces were incubated overnight at 37 °C. The next day, peptides were extracted and analyzed by both LC-SRM and LC-SIM. Data analysis was performed with LTQ-Orbitrap instrument data viewing software (Qual Browser).
Figure 6.
Plk1 threonine T210 phosphorylation during release from S phase. (a) Flow cytometry analysis of unsynchronized HeLa cells, HeLa cells in thymidine G1/S arrest (T0), and 2 (T2), 4 (T4), 6 (T6), 7 (T7), 8 (T8), 9 (T9) and 10 h (T10) after release. (b) Coomassie gel of HeLa cell lysates from the indicated time points. MW, molecular weight (in kDa). (c) Plot depicting Plk1 abundances at indicated time points, as measured by LC-SIM. (d) Western blot of Plk1 expression in HeLa cell lysates from thymidine arrest and release. (e) Plot depicting occupancy of Plk1 T210 phosphorylation at indicated time points.
Although we were able to detect all four peptides by SRM in digests of immunoprecipitated Plk1, only the SRM of the main form of the unphosphorylated peptide TLCGTPNYIAPEVLSK had sufficient intensity to be reproducibly and accurately detected during analyses of Plk1 directly from whole-cell lysates. The improved sensitivity obtained by the SIM method, however, allowed us to detect both unphosphorylated versions of the peptide and the main form of the phosphorylated peptide KKpTLCGTPNYIAPEVLSK. However, we were not able to detect a specific signal for the endogenous peptide pTLCGTPNYIAPEVLSK from SDS-PAGE–separated lysates. On the basis of our analysis of immunoprecipitated Plk1, only 3% of the total amount of phosphorylated peptide is produced in this form during trypsin digest. Inspection of the SIM spectra revealed a contaminating ion with high intensity at a very similar retention time, which precluded the SIM-based analysis of this peptide. This is not uncommon—there may be cases when complications arise (in this case, interfering chemical noise in the SIM and poor sensitivity in the SRM) that preclude useful data for that particular peptide. In this example, our up-front analysis with immunoprecipitated Plk1 established that an analysis without this digestion product would reduce our calculations by 3% of the amount we observe as phosphorylated.
In our analysis, we found a steady increase in Plk1 protein expression levels in HeLa cells after thymidine release. At 10 h after release, we detected 6.6-fold more Plk1 than during thymidine arrest (Fig. 6c). A similar increase in Plk1 expression was detected by western blotting (Fig. 6d). Phosphorylation of threonine 210 on Plk1 was undetectable under thymidine arrest. We calculated the occupancy of phosphorylation at T210 by dividing the amount of phosphorylation by the amount of total Plk1 protein determined by the average of the two unmodified peptides (HINPVAASLIQK and IGDFGLATK). At 8 h after thymidine release, 2.7% of all detected Plk1 was phosphorylated at T210. This increased to 5.9% and 6.9% at 9 and 10 h, respectively (Fig. 6e). The increase in T210 phosphorylation correlated with an increased number of cells entering mitosis, as observed by flow cytometry analysis (Fig. 6a).
ACKNOWLEDGMENTS
We thank S. Cullati and D. Schweppe for helpful discussion and comments on the manuscript. This work was supported by the National Institutes of Health Grant P20-RR018787 from the IDeA Program of the National Center for Research Resources and the American Cancer Society Grant IRG-82-003-24 (to S.A.G.).
Footnotes
AUTHOR CONTRIBUTIONS A.N.K. designed and conducted the experiments for Plk1 AQUA analysis, interpreted the data and wrote the paper; J.R. synthesized the Plk1 AQUA peptides used in this study; and S.A.G. conceived of the experiments for Plk1 AQUA analysis, interpreted the data and wrote the paper.
COMPETING FINANCIAL INTERESTS The authors declare competing financial interests (see the HTML version of this article for details).
References
- 1.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics. 2002;1:376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Gygi SP, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 1999;17:994–9. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 3.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beynon RJ, Doherty MK, Pratt JM, Gaskell SJ. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat. Methods. 2005;2:587–589. doi: 10.1038/nmeth774. [DOI] [PubMed] [Google Scholar]
- 6.Pratt JM, et al. Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat. Protoc. 2006;1:1029–1043. doi: 10.1038/nprot.2006.129. [DOI] [PubMed] [Google Scholar]
- 7.Langenfeld E, Zanger UM, Jung K, Meyer HE, Marcus K. Mass spectrometry-based absolute quantification of microsomal cytochrome P450 2D6 in human liver. Proteomics. 2009;9:2313–2323. doi: 10.1002/pmic.200800680. [DOI] [PubMed] [Google Scholar]
- 8.Mayya V, Rezual K, Wu L, Fong MB, Han DK. Absolute quantification of multisite phosphorylation by selective reaction monitoring mass spectrometry: determination of inhibitory phosphorylation status of cyclin-dependent kinases. Mol. Cell. Proteomics. 2006;5:1146–1157. doi: 10.1074/mcp.T500029-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 10.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathivanan S, et al. Human Proteinpedia enables sharing of human protein data. Nat. Biotechnol. 2008;26:164–167. doi: 10.1038/nbt0208-164. [DOI] [PubMed] [Google Scholar]
- 13.Jones P, et al. PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res. 2006;34:D659–D663. doi: 10.1093/nar/gkj138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallick P, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 15.Picotti P, et al. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods. 2010;7:43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 16.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papaioannou MD, et al. Loss of Dicer in Sertoli cells has a major impact on the testicular proteome of mice. Mol. Cell Proteomics. doi: 10.1074/mcp.M900587-MCP200. Published online; doi:10.1074/mcp.M900587-MCP200 (13 May 2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stemmann O, Zou H, Gerber SA, Gygi SP, Kirschner MW. Dual inhibition of sister chromatid separation at metaphase. Cell. 2001;107:715–726. doi: 10.1016/s0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- 19.Wolf G, et al. Prognostic significance of polo-like kinase (PLK) expression in non-small cell lung cancer. Oncogene. 1997;14:543–549. doi: 10.1038/sj.onc.1200862. [DOI] [PubMed] [Google Scholar]
- 20.Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora a cooperatively activate the kinase Plk1 and control mitotic entry. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macurek L, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- 22.Olsen JV, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 23.Beausoleil SA, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]