Abstract
Obesity is a calorie excessive state that is associated with high risk of diabetes, atherosclerosis and certain types of tumors. Obesity may induce inflammation and insulin resistance. We found that the expression of interferon regulatory factor 9 (IRF9), a major transcription factor mediating interferon (IFN) responses, was lower in the livers of obese mice than in those of their lean counterparts. Furthermore, whole-body IRF9 knockout (KO) mice were more obese and had aggravated insulin resistance, hepatic steatosis and inflammation after chronic high-fat diet (HFD) feeding. In contrast, adenoviral-mediated hepatic IRF9 overexpression in both diet-induced and genetically (ob/ob) obese mice showed markedly improved hepatic insulin sensitivity and attenuated hepatic steatosis and inflammation. We further employed a yeast two-hybrid screening system to investigate the interactions between IRF9 and its cofactors. Importantly, we identified that IRF9 interacts with peroxisome proliferator-activated receptor α (PPARα), an important metabolism-associated nuclear receptor, to activate PPARα target genes. In addition, liver-specific PPARα overexpression rescued insulin sensitivity and ameliorated hepatic steatosis and inflammation in IRF9 KO mice. Taken together, our results indicate that IRF9 attenuates hepatic insulin resistance, steatosis and inflammation through interaction with PPARα
Keywords: high-fat diet, diabetes, metabolism, inflammation, PPARα
Introduction
Metabolic disorders, including obesity, non-alcoholic fatty liver disease (NAFLD), metabolic syndrome and diabetes, are global public health issues and are increasingly severe owing to an aging population, urbanization and associated lifestyle changes (1, 2). Obesity is recognized as a chronic low-grade systemic inflammatory state (3). In obesity, IKKβ/NFκB and JNK1/AP1 pathways are activated in multiple tissues (4). Consequently, inflammatory cells infiltrate into adipose tissue. M1-like macrophages secrete proinflammatory cytokines (e.g., TNF-α and IL-1β), which impair insulin action (5, 6). Additionally, ectopic lipid accumulation (7) and endoplasmic reticulum (ER) stress (8) may contribute to insulin resistance. Nuclear receptors and their cofactors play essential roles in glucose and lipid metabolism and insulin sensitivity, among which peroxisome proliferator-activated receptors (PPARs) (9) and peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) (10) have been intensively studied. However, the underlying mechanisms of obesity related metabolic disorders still remain elusive.
Interferon regulatory factors (IRFs) are a family of nine transcription factors (IRF1 to IRF9) in mammals (11). IRFs are involved in cytosolic pattern-recognition receptor (PRR)-mediated and Toll-like receptor (TLR)-mediated signal transduction and regulate type I IFN expression(12). IRFs play central roles in gene expression regulation in innate immunity and immune cell differentiation (13). IRFs were also involved in malignant transformation through regulating cell growth and apoptosis (14). Moreover, we newly observed that cardiovascular diseases, such as cardiac hypertrophic, can be regulated by IRF family members (15). Besides the above mentioned, metabolic roles of IRFs have also emerged. IRF3 was reported to regulate metabolism related nuclear receptors, such as liver X receptor (LXR) and retinoid X receptor α (RXR-α) (16, 17). Another group found that IRFs regulate adipogenesis and adipocyte lipid metabolism (18, 19). However, the roles of IRFs in hepatic and whole-body metabolism are unclear.
IRF9, an IRF family member, has previously been characterized as mediating innate immunity by activating IFN-mediated transcription (20–22). In the present study, we discovered a protective role for IRF9 against metabolic disorders. IRF9 KO mice fed a high-fat diet (HFD) had higher levels of obesity-induced inflammation, lower insulin sensitivity and more severe hepatic steatosis than did wild-type (WT) controls, whereas liver-specific IRF9 overexpression ameliorated these phenotypes in both diet-induced and genetically (ob/ob) obese mice. Furthermore, we determined that IRF9 upregulated the expression of PPARα target genes. These results suggest that IRF9 improves hepatic lipid metabolism and insulin sensitivity.
Materials and methods
Mice and diets
All protocols were approved by the Animal Care and Use Committee of Renmin Hospital of Wuhan University. IRF9 knockout (KO) mice were kindly provided by Dr. Tadatsugu Taniguchi (Department of Immunology, Graduate School of Medicine and Faculty of Medicine, University of Tokyo). Ob/ob mice were purchased from Jackson Laboratory (stock number: 000632). 9-week old female and 8-week old male ob/ob mice were used. 8-week old male C57BL/6 mice were fed with either NC (protein 18.3%, fat 10.2%, carbohydrates 71.5%, D12450B, Research Diets) or an HFD (protein 18.1%, fat 61.6%, carbohydrates 20.3%, D12492, Research Diets) ad libitum for up to 26 weeks. Detailed protocols for animal experiments were described in the Supporting materials and methods.
Recombinant adenoviral vectors and in vivo adenovirus-mediated gene transfer
To overexpress IRF9 and PPARα we used replication-defective adenoviral vectors encompassing the entire coding region of Flag-tagged IRF9 (obtained from OriGene) and Flag-tagged PPARα (ordered from Seajet Scientific Inc.) under the control of the cytomegalovirus promoter. A similar adenoviral vector encoding GFP was used as a control. Adenovirus was injected via jugular vein. Please find the animal perform procedures in Supporting materials and methods.
Yeast two-hybrid analysis
For yeast two-hybrid screening, we used a Matchmaker Gold Y2H system according to the manufacturer’s instruction (Clontech). The bait vector, pGAKT7-IRF9, was constructed by cloning encoding region of IRF9 gene of human into pGAKT7 to create an in-frame fusion with Gal4 DNA-binding domain. pGAKT7-IRF9 was transformed into yeast strain Y2H Gold on SD/–Trp according to a standard PEG/ssDNA/LiAc procedure. Y2H Gold [pGADT7-IRF9] strain were mated with Y187 [Mate & Plate Library] strain by mixing 4–5 ml Bait Strain and 1 ml Library Strain in 45 ml of 2xYPDA liquid medium and incubating at 30°C for 20–24 hr, slowly shaking (30–50 rpm). Then we centrifuged to pellet the cells and discarded the supernatant. Pelleted cells were then resuspended in 10 ml of 0.5X YPDA/Kan liquid medium. 100Tl of 1/10 dilutions was plated onto SD/–Leu/–Trp (DDO) to select for mated colonies. Plates were incubated at 30°C for 5 days.
Statistical analysis
The data are presented as the mean ± SEM. Statistical analysis was performed with the Student’s 2-tailed t test or one-way ANOVA. P < 0.05 was considered statistically significant.
Methods for histological analysis, serum examination, western blot and real-time PCR analysis, plasmid construction, immunoprecipitation, GST pull-down assay, confocal microscopy were described in Supporting materials and methods.
Results
Diet-induced and genetically obese mice have lower hepatic IRF9 levels than the normal controls
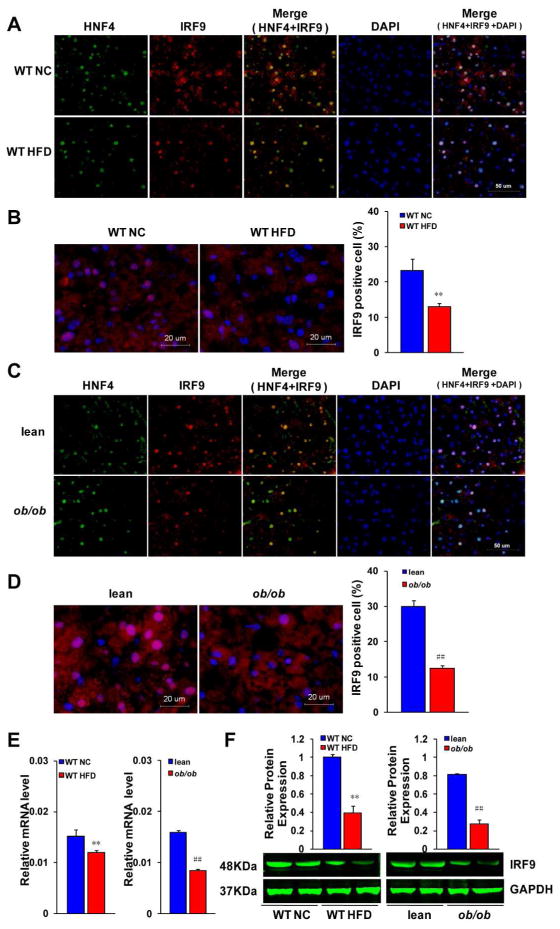
To investigate whether IRF9 is involved in metabolic diseases, we used HFD-induced and genetic (ob/ob) obesity models. We stained liver section slides with antibodies against hepatic nuclear factor 4 (HNF4), a molecular marker of hepatocytes, and IRF9. Almost all IRF9 was localized in HNF4 positive cells, which indicates that IRF9 was mainly expressed in hepatocytes rather than other types of cells in the liver (Fig 1A and 1C). We calculated the proportion of IRF9 positive hepatocytes. We observed that hepatocytes expressed IRF9 decreased after 26 weeks of high-fat diet (Fig. 1B). Consistently, the proportion of IRF9-expressing cells in the livers of ob/ob mice was lower than in the WT mice (Fig. 1D). The mRNA and protein expression levels of IRF9 were significantly lower in the livers of the HFD-fed obese mice than in the normal chow (NC) controls (Fig. 1E and 1F). In agreement with these results, ob/ob mice also had lower IRF9 expression levels than WT mice (Fig. 1E and 1F). All these data indicate that IRF9 expression in the liver is downregulated in obesity, which suggests an important role for IRF9 in metabolic disorders.
Figure 1. IRF9 expression decreases in the liver of obese mice.
(A and C) Representative immunofluorescent images of liver section slides, which were stained with antibodies against hepatic nuclear factor 4 (HNF4, green) and IRF9 (red). DAPI (blue) was used to show the nuclei. Scale bar indicates 50 μm. (B and D) Representative immunofluorescence expression of IRF9 in livers of C57BL/6 mice (WT) and lean, ob/ob mice. For (A) to (D), WT mice were on the normal chow diet (NC) or high fat diet (HFD)-feeding for 26 weeks. ob/ob mice were fed with the chow diet for 9 weeks. Each group contained four sections for staining analysis, scale bar indicates 20 μm. Quantitative analysis of percents of IRF9 positive cells in liver is also shown. (E) Real-time PCR analysis of mRNA levels of IRF9 in liver. n=6–12 each group. (F) Protein expression of IRF9 in liver was detected by western blot. Quantification of protein expression levels was normalized to the GAPDH loading control, All value shown as mean ± SEM, n=3 per group. For all the statistical significance is indicated and compared with the WT NC group, **p < 0.01; compared with the lean group, ##p < 0.01.
IRF9 deficiency aggravates obesity and impaired glucose metabolism
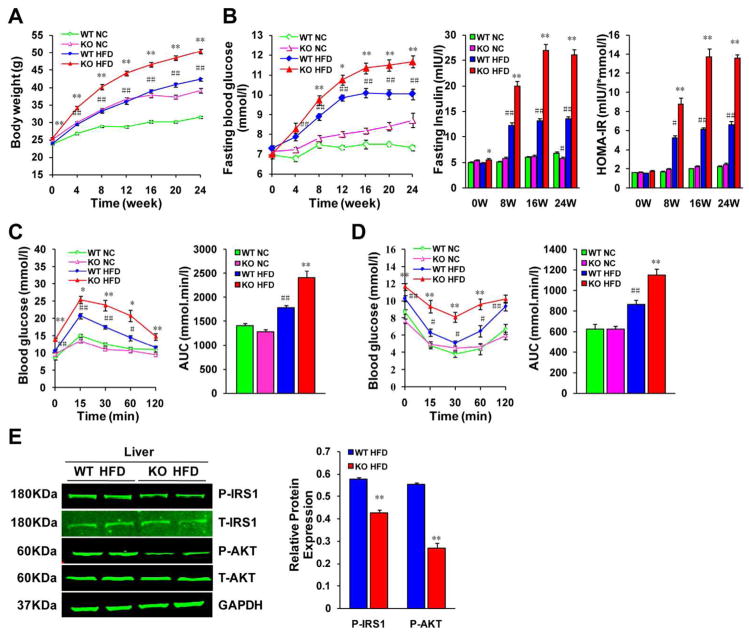
To full understand the effect of IRF9 on metabolism, we utilized IRF9 KO mice. After consuming an HFD, although there was no significant difference in the food consumption between the two genotypes (Supporting Fig. 1A), IRF9 KO mice were more obese (Fig. 2A) and displayed lower insulin sensitivity than the WT controls. IRF9 KO mice also had higher fasting blood glucose and insulin levels and a higher homeostasis model assessment of insulin resistance (HOMA-IR) index than the WT controls (Fig. 2B). During fasting, the liver generates glucose to stabilize serum glucose level; after feeding, insulin increases and gluconeogenesis slows down correspondingly. We found that although IRF9 KO mice had higher serum insulin level, gluconeogenic gene expression, such as phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase), was still higher in the IRF9 KO livers than in the WT ones (Supporting Fig. 1B). We also performed intraperitoneal glucose tolerance tests (IPGTT) and insulin tolerance tests (IPITT), both of which revealed compromised insulin sensitivity and glucose regulatory functions in the IRF9 KO mice as compared with the WT mice (Fig. 2C and 2D). Insulin regulates organ function in an endocrine manner. Upon insulin binding, insulin receptors (IRs) display increased kinase activity against intracellular adaptors, such as insulin receptor substrates (IRSs), which relay signals to downstream pathways (23). Western blot determined that the levels of tyrosine phosphorylation of IRS1 and serine phosphorylation of Akt were lower in the livers of the IRF9 KO mice than in the WT mice, indicating downregulation of the insulin signaling pathway (Fig. 2E).
Figure 2. IRF9 deficiency aggravates obesity and impairs glucose metabolism.
(A) The comparison of body weight gains in WT or IRF9 KO mice on the 26-week normal chow diet (NC) or high fat diet (HFD)-feeding, n=28–39 per group. (B) Fasting blood glucose levels were detected at an interval of 4 weeks from 0 to 24 weeks feeding duration in WT or KO mice with food deprivation for 6 hours. Serum fasting insulin levels were determined by ELISA every 8 weeks. Homeostasis model assessment of insulin resistance (HOMA-IR) index was calculated as [FBG (mmol/l) × FIns(mIU/l)]/22.5. n=4–8 per group at each time-point. (C) A glucose tolerance test (GTT) (1 g/kg body weight, glucose intraperitoneal injection) was performed on WT and KO mice in both the NC group and the HFD group with 24-week diet feeding. The corresponding area under the curve (AUC) of blood glucose levels in each group was calculated, n=12–16 per group. (D) An insulin tolerance test (ITT) (0.75 units/kg body weight, insulin intraperitoneal injection) was performed on WT and KO mice in both the NC and the HFD group at the 25th week of food administered. The corresponding area under the curve (AUC) of blood glucose levels in each group was calculated, n=13–17 per group. (E) Immunoblot analysis established systemic insulin resistance in KO mice and indicated marked inhibition of the phosphorylation of IRS1-tyr608 and Akt in insulin target organs. Quantification of the phosphorylated protein expression levels were normalized to GAPDH, n= 4 per group. For all the statistical significance is indicated and compared with the WT HFD group, *p < 0.05, **p < 0.01; compared with the WT NC group, #p < 0.05, ##p < 0.01. All values are expressed as the mean ± SEM.
Metabolic disorders involve a series of systemic changes. With continuous HFD feeding, metabolic dysfunction became increasingly significant in the IRF9 KO mice. Triglyceride (TG), total cholesterol (TC), low-density lipoprotein (LDL), free fatty acid (FFA) and β-hydroxybutyrate levels were higher in IRF9 KO mouse serum, whereas high-density lipoprotein (HDL) was lower (Table 1). All these data indicate catabolism insufficiency and energy overabundance in IRF9 KO mice compared with WT mice.
Table 1.
Serum lipid, hormone and cytokine levels in WT and KO mice with a 24-week-diet treatment.
| Parameters | WT NC | KO NC | WT HFD | KO HFD |
|---|---|---|---|---|
| Lipid Contents | ||||
| Triglyceride (mg/dl) | 39.96±1.71 | 43.50±1.75 | 49.82±2.28## | 69.67±2.46** |
| Cholesterol (mg/dl) | 130.79±5.67 | 144.82±5.52 | 251.55±11.69## | 322.62±12.05** |
| HDL (mg/dl) | 86.52±3.74 | 78.67±2.47 | 73.15±3.38## | 46.29±1.43** |
| LDL (mg/dl) | 17.33±0.72 | 19.34±0.60 | 24.18±1.09## | 39.62±1.26** |
| FFA (mmol/l) | 0.74±0.03 | 0.92±0.03# | 1.09±0.04## | 2.44±0.07** |
| β-hydroxybutyrate (mmol/l) | 0.23±0.03 | 0.29±0.03 | 0.89±0.07## | 1.28±0.09** |
| Hormones & Cytokines | ||||
| Leptin (pg/ml) | 5173.10±225.02 | 5520.90±223.06 | 12265.00±570.63## | 17324.00±433.83** |
| Resistin (pg/ml) | 891.67±38.50 | 924.62±31.87 | 1292.20±59.10## | 1663.50±55.35** |
| Adiponectin (μg/ml) | 1.10±0.04 | 1.03±0.04 | 0.57±0.02## | 0.22±0.01** |
| IL-1β (pg/ml) | 38.66±1.64 | 44.20±1.21 | 57.21±2.50## | 78.29±2.59** |
| IL-4 (pg/ml) | 21.73±0.90 | 21.61±0.69 | 24.34±1.04 | 45.64±1.46** |
| IL-6 (pg/ml) | 0.71±0.02 | 0.98±0.03# | 1.20±0.04## | 3.14±0.09** |
| TNF-α (pg/ml) | 2.79±0.12 | 3.11±0.11# | 3.63±0.14## | 4.98±0.16** |
| MCP-1 (pg/ml) | 23.40±0.99 | 27.58±0.93 | 29.40±1.31## | 47.28±1.60** |
| IL-10 (pg/ml) | 15.07±0.63 | 14.86±0.45 | 24.97±1.15## | 27.36±0.82** |
Data are expressed as mean ± SEM. n=5–8 per group.
P<0.05,
P<0.01 vs. WT NC;
P<0.01 vs. WT HFD.
IRF9 deficiency aggravates hepatic steatosis and inflammation
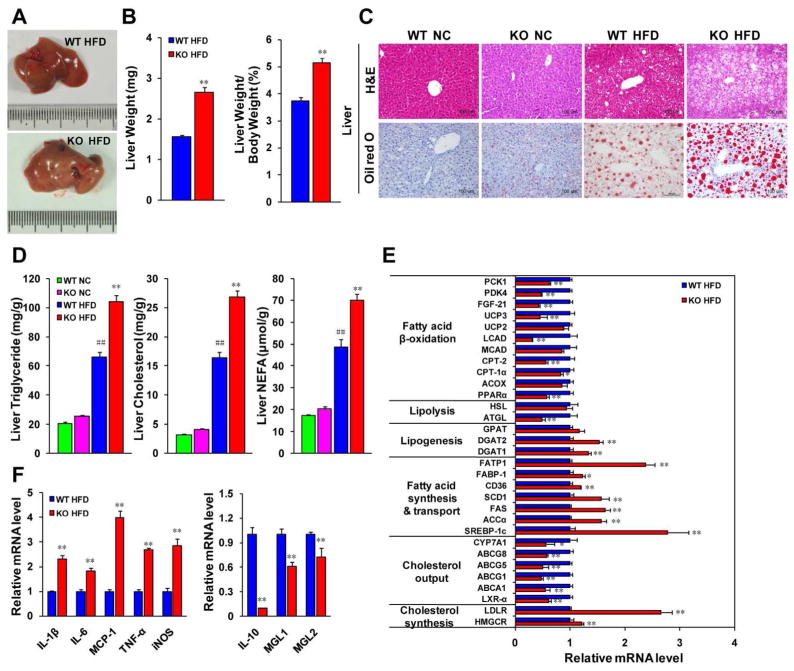
Hepatic steatosis is an important manifestation of metabolic dysfunction and insulin resistance. We found that the livers of the IRF9 KO mice were larger than those from the WT mice after 26 weeks of an HFD owing to cellular lipid accumulation, as determined by H&E and Oil red O staining (Fig. 3A–3C). Considering that steatohepatitis devastates liver integrity and function, we tested hepatic function in the mice. Alanine transaminase (ALT), aspartate transaminase (AST) and alkaline phosphatase (ALP) levels were all significantly higher in HFD-fed IRF9 KO mouse serum than in WT mouse serum, indicating poorer hepatic function in the IRF9 KO mice (Supporting Fig. 2A). The IRF9 KO mice also had higher hepatic TG, TC and FFA levels (Fig. 3D). Quantitative real-time PCR demonstrated that the expression levels of genes related to cholesterol synthesis (e.g., HMGCR and LDLR), lipogenesis (e.g., DGAT1 and DGAT2), fatty acid synthesis (e.g., SREBP-1c, ACC-α FAS and SCD1) and uptake (e.g., CD36, FABP-1 and FATP1) were higher, whereas the expression of genes regulating cholesterol output, lipolysis (e.g., ATGL) and fatty acid oxidation (e.g., PPARα LCAD and UCP3) were lower in the livers of the IRF9 KO mice than in the livers of WT mice (Fig. 3E). AMP-activated protein kinase (AMPK), a master regulator of cellular energy homeostasis, stimulates catabolism in response to low ATP levels (24). In the livers of the IRF9 KO mice, lower levels of phosphorylated AMPK and ACC2 indicated a compromised AMPK signaling pathway (Supporting Fig. 2B).
Figure 3. IRF9 deficiency aggravates the hepatic steatosis, exacerbates global and local inflammation levels in mice.
(A) The macroscopic pictures of livers in the WT and KO mice fed with the HFD. (B) Quantification of liver weight and the ratio to body weight, n=28–39 per group. (C) Representative liver sections stained with H&E (upper panel) or Oil red O (lower panel) in WT or KO mice with 26 weeks of NC or HFD-feeding. Scale bar indicates 100 μm. (D) The levels of triglyceride (TG), cholesterol, and non-esterified fatty acid (NEFA) were extracted from liver tissue in WT with KO mice upon the HFD, n=5–13 per group. (E) The mRNA levels of genes related to lipid metabolism were examined by Real-time PCR, n= 6–12 per group. (F) The mRNA levels of proinflammatory and anti-inflammatory markers in liver were measured by Real-time PCR, n=4 for each group. All values are expressed as the mean ± SEM. The statistical significance is indicated and compared with the WT HFD group, *p < 0.05, **p < 0.01; compared with the WT NC group, ##p < 0.01.
Considering that inflammation is intimately related to metabolic disorders, we further tested liver inflammation. Immunofluorescent staining of inflammatory markers (e.g., 7/4, CD45 and CD68) indicated more hepatic inflammatory cell infiltration in IRF9 KO mice (data not shown) than in the WT mice. Meanwhile, real-time PCR demonstrated Kupffer cell activation and M1 macrophage polarization in IRF9 KO livers. The levels of proinflammatory cytokines (e.g., TNF-α, IL-1β, IL-6 and MCP-1) were higher whereas those of anti-inflammatory markers (e.g., IL-10, MGL1 and MGL2) were lower in the livers of the IRF9 KO mice (Fig. 3F). Adipokines are important regulators of adipose inflammation and insulin sensitivity (25). Serum levels of leptin and resistin were higher and that of adiponectin was lower in IRF9 KO mice as compared with WT controls. Furthermore, levels of proinflammatory cytokines were higher whereas adiponectin was lower in the circulation of the IRF9 KO mice (Table 1). All these factors contribute to insulin resistance and metabolic dysfunction. In line with the results in liver, more proinflammatory factors and fewer anti-inflammatory factors were also detected in the serum of the IRF9 KO mice than in the WT mice. All the findings described above illustrate that IRF9 deficiency aggravates hepatic steatosis and enhances local and global inflammation in mice.
Hepatic IRF9 overexpression attenuates diet-induced hepatic steatosis, insulin resistance and inflammation
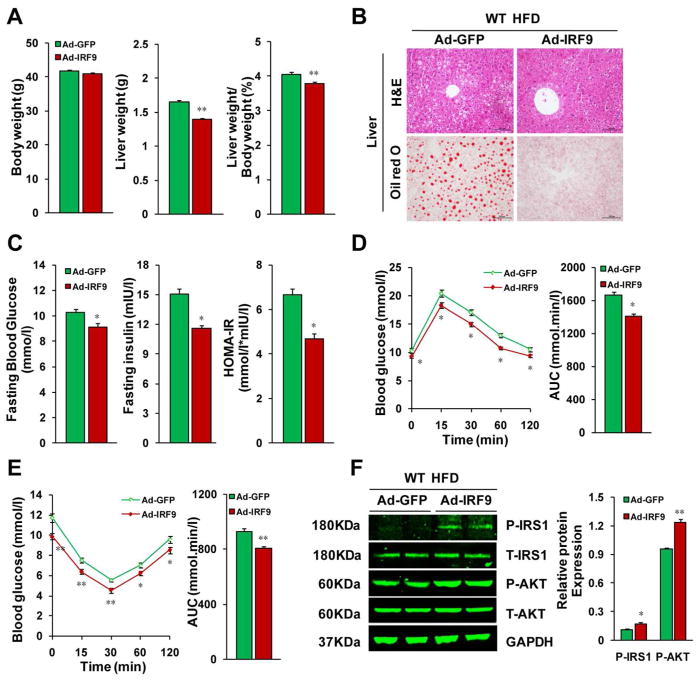
To determine the in vivo function of IRF9 on hepatic lipid metabolism and insulin sensitivity, we used adenovirus infection, a well-established method, to overexpress IRF9 in mouse liver. The adenovirus-mediated gene transfer approach acutely delivers genes to the liver without confounding developmental effects that commonly occur during chronic overexpression (26, 27). After 20 weeks of HFD feeding, the mice were injected with an IRF9-expressing adenovirus through the jugular vein. Four weeks after adenovirus injection, the protein expression level of IRF9 had a more than four-fold increase in the liver, but remained unchanged in WAT and skeletal muscle (Supporting Fig. 3A). Immunofluorescent staining of HNF4 and IRF9 confirmed the elevation of IRF9 expression in hepatocytes rather than in other types of cells (Supporting Fig. 3B). Four weeks after the adenovirus injection, the mice with IRF9 overexpression had lower liver weight than those of the WT mice injected with an adenovirus expressing green fluorescent protein (GFP) as a control (Fig. 4A). H&E and Oil red O staining revealed lower hepatic lipid accumulation in the livers with IRF9 overexpressed (Fig. 4B). Hepatic TG, TC and non-esterified fatty acid (NEFA) contents were also lower in IRF9-overexpressing mice than in the control mice (Supporting Fig. 3C). IRF9-injected mice displayed lower ALT, AST and ALP levels (Supporting Fig. 3D). All these factors indicate that IRF9 promotes hepatic lipid metabolism and protects liver function. The IRF9-overexpressing mice displayed lower fasting serum glucose and insulin levels when on an HFD than did the control animals (Fig. 4C). Both the IPGTT and IPITT showed improved glucose regulation in the IRF9-overexpressing mice (Fig. 4D and 4E). Consistent with these results, the insulin signaling pathway was upregulated in the IRF9-overexpressing livers compared with the control livers, as measured by immunoblot (Fig. 4F). Moreover, liver-specific IRF9 overexpression ameliorated obesity-induced inflammation in the liver. Decreased proinflammatory markers (e.g., F4/80, CD11c, TNF-α, IL-1β, IL-6 and MCP-1) and increased anti-inflammatory markers (e.g., IL-10, ARG1, MRC1, MGL1 and MGL2) were detected by real-time PCR and indicate a shift in the balance to M2-like macrophages (Supporting Fig. 3E).
Figure 4. Hepatic IRF9 overexpression improves metabolism in diet-induced obese mice.
(A) Body weight, liver weight and the liver to body weight ratio of WT mice fed with HFD were examined. These mice were either injected with IRF9 adenovirus or GFP adenovirus control, n=12–13 per group. (B) Representative liver sections stained with H&E (top panel), Oil-red O (lower panel) of IRF9 overexpressed and vector control mice fed with HFD. (C) Fasting blood glucose levels were detected with food deprivation for 6 hours. Serum fasting insulin levels were determined by ELISA. HOMA-IR index was also calculated. n=6 per group. (D) A glucose tolerance test (GTT) (1 g/kg body weight, glucose intraperitoneal injection) was performed on HFD fed WT mice injected with IRF9 adenovirus or vector control virus. The corresponding area under the curve (AUC) of blood glucose levels in each group was calculated, n=12–13 per group. (E) An insulin tolerance test (ITT) (0.75 units/kg body weight, insulin intraperitoneal injection) was also performed. The corresponding area under the curve (AUC) of blood glucose levels in each group was calculated, n=12–13 per group. (F) Immunoblot analysis indicated that the reverse insulin resistance in mice with IRF9 liver-specific overexpression and displayed marked upregulation of the phosphorylation of IRS1-tyr608 and Akt in liver. Quantification of the phosphorylated protein expression levels were normalized to GAPDH, n= 4 per group. All values are expressed as mean ± SEM. The statistical significance is indicated and compared with the GFP adenovirus injected group, *p < 0.05, **p < 0.01.
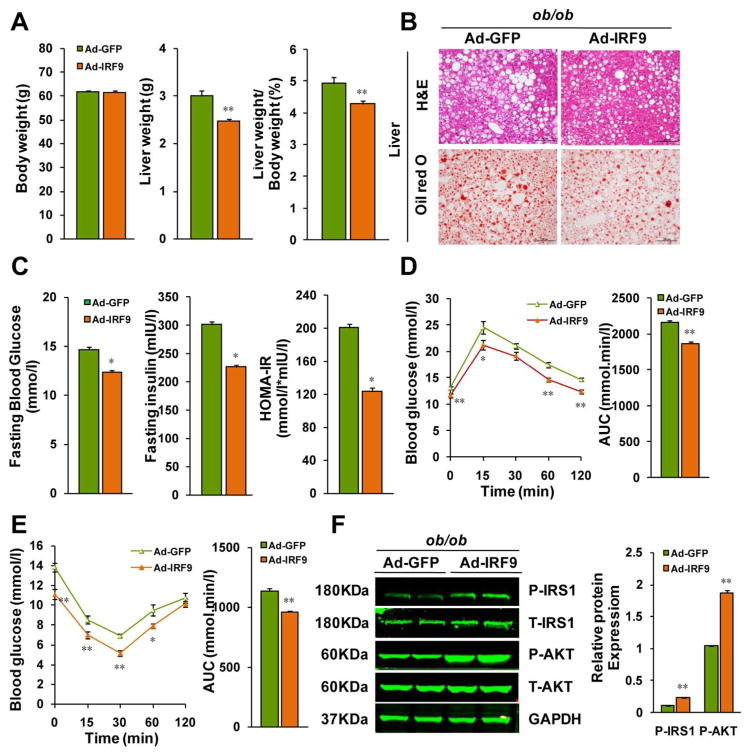
Hepatic IRF9 overexpression attenuates hepatic steatosis, insulin resistance and inflammation in ob/ob mice
To rule out any potential impact of unidentified components of the HFD on our results, we used a genetic obesity model to assess the metabolic role of IRF9. We fed normal chow to leptin-deficient (ob/ob) mice, which spontaneously develop obesity. As with the dietary model described above, we injected male ob/ob mice with IRF9 adenovirus through the jugular vein for liver-specific IRF9 overexpression (Supporting Fig. 4A and Supporting Fig. 4B). Four weeks later, hepatic lipid depots were greatly reduced in the IRF9-overexpressing mice compared with the GFP adenovirus-injected controls (Fig. 5A and 5B). IRF9-overepressing ob/ob mice had lower fasting serum glucose and insulin levels and lower hepatic TG, TC and NEFA content than did the control mice (Fig. 5C and Supporting Fig. 4C). Liver function was also protected from IRF9 overexpression (Supporting Fig. 4D). The IPGTT and IPITT results demonstrated improved glucose tolerance and reduced insulin resistance in IRF9-overexpressing mice compared with the control mice (Fig. 5D and 5E). The phosphorylation of key insulin signaling molecules, such as the IRS1 and Akt, was elevated after IRF9 overexpression (Fig. 5F). Downregulated proinflammatory factors and upregulated anti-inflammatory factors were also observed in the IRF9-overexpressing mice (Supporting Fig. 4E). Therefore, using dietary and genetic obesity models, we have now determined that IRF9 attenuates obesity-induced hepatic steatosis, insulin resistance and inflammation.
Figure 5. Hepatic IRF9 overexpression improves metabolism in ob/ob mice.
(A) Body weight, liver weight and the liver to body weight ratio of ob/ob mice injected IRF9 or GFP adenovirus were examined. n=12 per group. (B) Representative liver sections stained with H&E (top panel), Oil-red O (lower panel) of IRF9 overexpressed and GFP control ob/ob mice. (C) Fasting blood glucose levels were detected with food deprivation 6 hours. Serum fasting insulin levels were determined by ELISA. HOMA-IR index was also calculated. n=6 per group. (D) A glucose tolerance test (GTT) (1 g/kg body weight, glucose intraperitoneal injection) was performed on ob/ob mice. The corresponding area under the curve (AUC) of blood glucose levels in each group was calculated, n=12 per group. (E) An insulin tolerance test (ITT) (0.75 units/kg body weight, insulin intraperitoneal injection) was also performed. The corresponding area under the curve (AUC) of blood glucose levels in each group was calculated, n=12 per group. (F) Immunoblot analysis of the phosphorylation of IRS1-tyr608 and Akt in livers. Quantification of the phosphorylated protein expression levels were normalized to GAPDH, n= 4 per group. All values are expressed as mean ± SEM. The statistical significance is indicated and compared with the GFP adenovirus injected group, *p < 0.05, **p < 0.01.
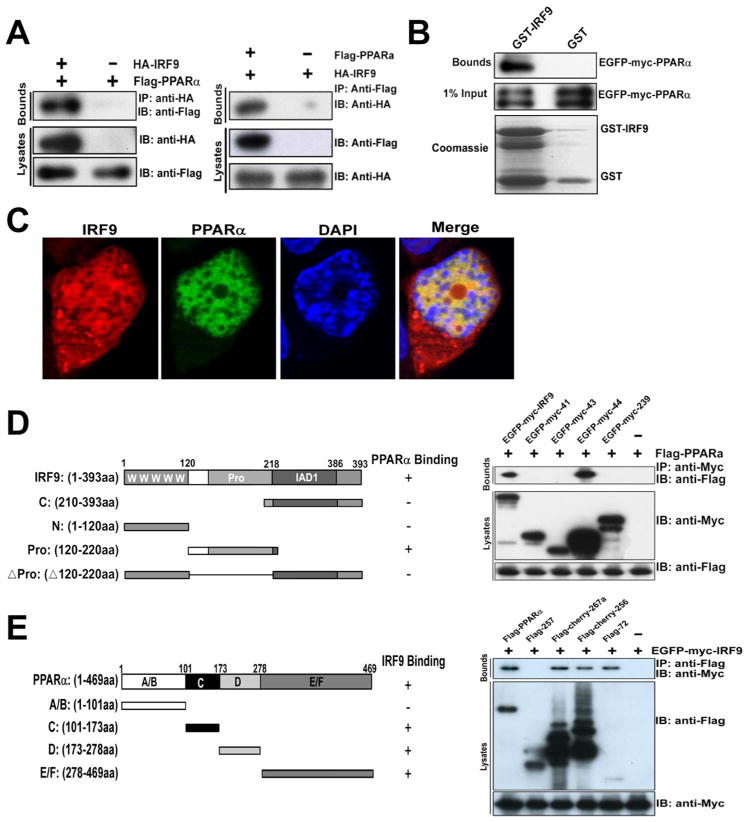
IRF9 interacts with PPARα to activate PPARα target genes
Transcription factors usually recruit cofactors to facilitate downstream gene expression. To investigate how IRF9 improves hepatic metabolism, we employed a yeast two-hybrid screening system and used IRF9 as bait to identify IRF9-interacting proteins in a human liver library. One of the candidate IRF9 interactors was PPARα; the prey clone encoded the N-terminal 254 residues of PPARα (data not shown). We confirmed the interaction between IRF9 and PPARα in HepG2 cells, a human hepatocellular carcinoma cell line, with co-immunoprecipitation (co-IP). We found that IRF9 co-immunoprecipitated with PPARα but not control IgG in HepG2 cells and vice versa (Fig. 6A). Additionally, a GST pulldown assay also confirmed the interaction between IRF9 and PPARα (Fig. 6B). To rule out the possibility that the interaction was newly formed during the co-IP or GST pulldown, we performed immunofluorescence to identify IRF9 and PPARα localization. We found that IRF9 and PPARα colocalized predominantly in the nucleus (Fig. 6C). To map the PPARα-interacting region of IRF9, a series of IRF9 deletion mutants were generated. Neither the IRF9 N-terminal DNA-binding domain (DBD) nor the C-terminal IRF association domain (IAD) associated with PPARα; only the less conserved IRF9 intermediate region interact with PPARα (Fig. 6D). We also generated a series of PPARα deletion mutants. The mapping demonstrated that the DNA-binding domain (DBD, C domain), the hinge region (D domain) and the ligand-binding domain (LBD, E/F domain) of PPARα were all able to interact with IRF9 (Fig. 6E), and only the N-terminal A/B domain was not.
Figure 6. IRF9 interacts with PPARα.
(A) co-immunoprecipitation (IP) of IRF9 and PPARα is shown. Cell lysates from HepG2 cells transfected with HA-tagged IRF9 and FLAG-tagged PPARα were prepared. These lysates were subjected to IP with HA antibody or IgG and analyzed by immunoblot using HA or FLAG antibodies. From the other side, these lysates were subjected to IP with FLAG antibody then went through immunoblot using HA or FLAG antibodies. (B) GST pulldown assays of EGFP-Myc-tagged PPARα with GST or GST-IRF9 are indicated. (C) Co-localization of IRF9 and PPARα in the nucleus is shown. pCherry-IRF9 and pEGFP-PPARα were transfected into primary mouse hepatocytes and indirect immunofluorescence analysis was performed. The cells were visualized by confocal microscopy, and nuclei were stained with DAPI. (D) Left panel: shown is a schematic representation of four kinds of IRF9 deletion mutants. Right panel: shown is mapping the PPARα binding region of IRF9. Cell lysates from HEK293T cells transfected with FLAG-tagged PPARα and EGFP-Myc-tagged IRF9 deletion mutants were immunoprecipitated with anti-FLAG followed by immunoblot with anti-FLAG and anti-Myc. (E) Left panel: schematic representation of four kinds of PPARα deletion mutants. Right panel: mapping the IRF9 binding region of PPARα. HEK293T cells were transfected with EGFP-Myc-tagged IRF9 and FLAG-tagged PPARα deletion mutants.
We next sought to determine why IRF9 binds to PPARα. As shown earlier, we found that the mRNA levels of PPARα target genes (e.g., ACOX, CPT-2, MCAD, LCAD, UCP2, UCP3, FGF-21, PDK4 and PCK1) were universally lower in the livers of the IRF9 KO mice than in the controls (Fig. 3E). We found that PPARα target genes were activated in primary mouse hepatocytes transfected with wild-type IRF9 plasmids (Supporting Fig. 5A). To confirm the activation of PPARα target genes through IRF9-PPARα interaction, we constructed a mutant IRF9 plasmid in which the PPARα-interaction domain was deleted. The expression of PPARα target genes didn’t change in cells transfected with mutant IRF9 plasmids (Supporting Fig. 5B). When we further overexpressed IRF9 specifically in liver, we saw the upregulation of PPARα target genes in the livers of both diet-induced and genetically obese mice (Supporting Fig. 5C and 5D). Taken together, these results suggest that IRF9 activates PPARα target gene expression by interacting with PPARα.
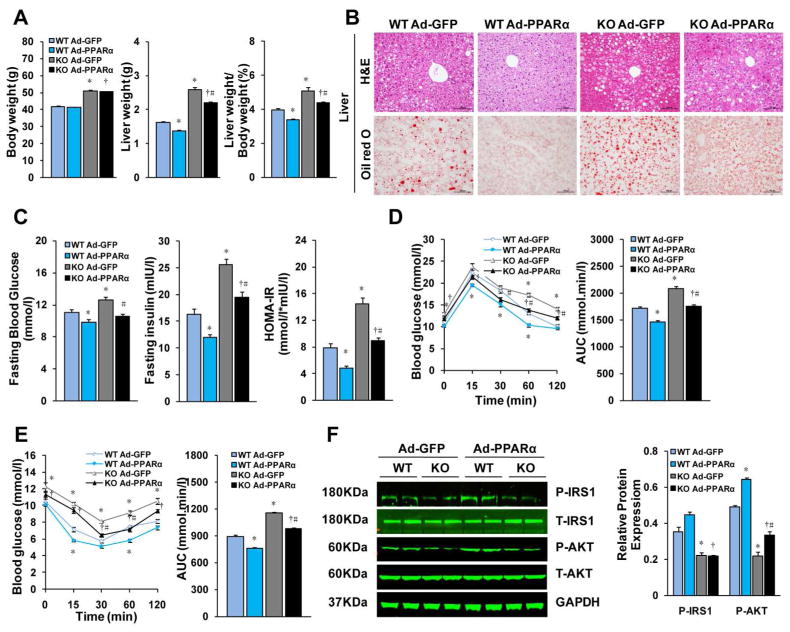
Hepatic PPARα overexpression rescues insulin sensitivity and ameliorates hepatic steatosis and inflammation in IRF9 KO mice
As expected, primary mouse hepatocytes trasfected with PPARα had markedly higher levels of its target genes than those of transfected with GFP controls (Supporting Fig. 6A). To determine the sufficiency of PPARα in mediating the metabolic functions of IRF9, we overexpressed PPARα specifically in the livers of WT mice and IRF9 KO mice. We injected the mice with PPARα adenovirus through the jugular vein. Four weeks later, PPARα and its target genes were significant increased in the liver (Supporting Fig. 6B and 6C). After 24 weeks of HFD feeding, the IRF9 KO mice displayed aggravated hepatic steatosis, insulin resistance and inflammation, as described earlier. However, after PPARα was overexpressed, the IRF9 KO mice displayed reduced body liver weight (Fig. 7A). H&E and Oil red O staining confirmed less hepatic lipid accumulation (Fig. 7B). Lower hepatic lipid content and preserved liver function indicated attenuated steatohepatitis (Supporting Fig. 6D and 6E). Fasting serum glucose and insulin levels and the HOMA-IR index in PPARα overexpressed IRF9 KO mice were similar to those of GFP adenovirus infected controls (Fig. 7C). Similar results were obtained with glucose and insulin tolerance tests (Fig. 7D and 7E). Insulin signaling was also upregulated upon IRF9 overexpression (Fig. 7F). Measurement of inflammation- related genes by real-time PCR indicated a shifting macrophage population from M1 to M2 (Supporting Fig. 6F and 6G). Thus, we demonstrated that liver-specific PPARα overexpression rescues insulin sensitivity and ameliorates hepatic steatosis and inflammation in IRF9 KO mice.
Figure 7. Hepatic PPARα overexpression rescues deregulated metabolism in IRF9 KO mice.
(A) Body weight, liver weight and the liver to body weight ratio of HFD fed mice injected PPARα or vector adenovirus were examined. n=12 per group. (B) Representative liver sections stained with H&E (top panel), Oil-red O (lower panel) of PPARα overexpressed and GFP control mice. (C) Fasting blood glucose levels were detected with food deprivation 6 hours. Serum fasting insulin levels were determined by ELISA. HOMA-IR index was also calculated. n=6–7 per group. (D) and (E) A glucose tolerance test (GTT) (1 g/kg body weight, glucose intraperitoneal injection) and an insulin tolerance test (ITT) (0.75 units/kg body weight, insulin intraperitoneal injection) were performed, n=12 per group. The corresponding area under the curve (AUC) of blood glucose levels in each group was calculated, n=12 per group. (F) Immunoblot analysis of the phosphorylation of IRS1-tyr608 and Akt signaling in livers. Quantification of the phosphorylated protein expression levels were normalized to GAPDH, n= 4 per group. All values are expressed as mean ± SEM. The statistical significance is indicated and compared with the GFP adenovirus injected WT group, *p < 0.05; compared with PPARα adenovirus injected WT group, +p<0.05; compared with the GFP adenovirus injected IRF9 KO group, #p< 0.05.
Discussion
IRF9 KO mice have a relatively normal physical appearance but are susceptible to virus infection because of the crucial role of IRF9 in mediating type I IFN responses (21, 28). Therefore, most studies on IRF9 have been focused on its involvement in innate immunity and oncogenesis (11). However, whether IRF9 is involved in the regulation of metabolism is unclear. In the present study, we for the first time demonstrated a critical role for IRF9 in hepatic lipid homeostasis. IRF9 expression was lower in the livers of both diet-induced and genetic obesity models. On an HFD, IRF9 KO mice exhibited more severe obesity, hepatic steatosis, insulin resistance and inflammation. When IRF9 was specifically overexpressed in the liver, diet-induced and genetically obese mice displayed attenuated hepatic steatosis, insulin resistance and inflammation, which indicate that IRF9 has an anti-diabetic role.
Eguchi et al. identified IRFs to have potential roles in adipogenesis and adipose biology via high-throughput DNase hypersensitivity (DHS) analysis (18). This group further reported that IRF4 expression was nutritionally regulated in adipocytes. After feeding, IRF4 was downregulated by insulin via effects of FoxO1 in WAT (19). In the present study, we investigated the metabolic effects of another IRF family member IRF9, which has ubiquitous distribution, rather than IRF4, the expression of which is highly restricted to adipose tissue and immune cells. In our study, obese mice displayed lower IRF9 expression in the liver than that of lean mice. Still, the mechanism by which IRF9 expression is downregulated during obesity remains to be elucidated.
IRF9 KO mice showed higher levels of hepatic cholesterol and FA synthesis, FA uptake and lipogenesis and lower levels of hepatic cholesterol output, lipolysis and FA oxidation, which all lead to hepatic lipid overload. All these indicate that IRF9 functions for hepatic lipid clearance and against hepatic steatosis. We further identified an interaction between IRF9 and PPARα and observed that PPARα target genes were significantly activated upon IRF9 overexpression. Because PPARα promotes lipid catabolism by increasing FA uptake and oxidation in the liver and other organs (29), PPARα mediates at least part of the anti-hepatic steatosis function of IRF9. PPARs are a family of nuclear receptors that initiate transactivation of target genes through ligand binding, corepressor removal and coactivator recruitment (29). Our results implicate IRF9 as a novel cofactor of PPARα, which is involved in the regulation of PPARα transactivation.
The present study demonstrated that hepatic insulin sensitivity in IRF9 KO mice was impaired but was rescued by liver-specific PPARα overexpression. It seems paradoxical given that PPARα-deficient mice were protected from HFD-induced insulin resistance, as reported by Guerre Millo et al. (30). Additionally, according to Koo et al., PPARα impairs liver insulin signaling by activating TRB3, which inhibits Akt activation (31). Therefore, PPARα-mediated enhancement of insulin signaling in the context of the current study might be attributed to its lipid-clearing functions and the associated prevention of inflammation (32).
Obesity-induced inflammation, as proposed by Gregor and Hotamisligil, originates from signals within metabolic cells, followed by metabolic tissue reconstruction to an inflammatory state (3). Activation of IKK-β/NF-κB and JNK1/AP1 pathways contributes to insulin resistance (33–36). Cytokines (e.g., TNF-α and IL-6) also induce hepatic lipogenesis and increase hepatic TG accumulation (37, 38). Thus, obesity and inflammation form a vicious cycle. Unlike the situation in adipose tissue, macrophage infiltration plays a secondary role in the liver during obesity; instead, liver-resident macrophage-like Kupffer cells become activated (39). On an HFD, IRF9 KO livers displayed increased obesity-induced inflammation and M1-type polarization of the Kupffer cells, both of which contribute to compromised insulin activities.
We employed IRF9 global KO mice to study the metabolic roles of IRF9 and found a poor hepatic metabolic phenotype. After overexpressing IRF9 specifically in the liver, nearly all the devastating metabolic effects of IRF9 deficiency were mitigated. This phenomenon reflects the importance of IRF9 in the liver to regulate glucose and lipid metabolism. Probably due to the short period of IRF9 overexpression using adenovirus injection method and the pre-existence of endogenous IRF9, the metabolic changes during IRF9 overexpression were, although statistically significant, not as drastic as those during IRF9 deficiency. Despite all these, IRF9 was vividly shown to relieve hepatic lipid overabundance and the development of hepatic steatosis in our obesity models.
In mammals, the IRF family consists of nine members that share similar structures. Different IRFs have overlapping targets and functions (12). Some may wonder whether other IRFs compensate for the loss of IRF9 in IRF9 KO mice. Through deletion mutant plasmid construction and immunoprecipitation mapping, we identified the less conserved intermediate region of IRF9, rather than the well-conserved DNA binding domain (DBD) or IRF association domain (IAD), interacts with PPARα. Therefore, the regulation of PPARα transactivation could be uniquely attributed to IRF9 rather than other IRF family members.
Our study reveals the versatility of IRF9 and broadens our view toward the IRF family, which as the name implies, was renowned for mediating immune responses. We now have successfully suggested a key role for IRF9 in metabolic function independent of its effect on immunity. However, uncovering the metabolic role of IRF9 in the liver is only the tip of the iceberg. There are many more unanswered questions, such as the tissue specificity of IRF function, interactions among IRFs and multiple cofactors, and influence of one IRF family member on the other family members. Investigating the mechanisms of IRF-mediated metabolic regulation will undoubtedly shed new light on treatment for obesity and diabetes.
Supplementary Material
Acknowledgments
We thank Dr. Tadatsugu Taniguchi (University of Tokyo, Japan) for providing the IRF9 knockout mice. This study was supported by grants from National Natural Science Foundation of China (grants 81170086 and 81000342) and the National Science and Technology Support Project (NO. 2011BAI15B02 and NO. 2012BAI39B05) and National Basic Research Program of China (2011CB503902).
Abbreviations
- ABCA1
ATP-binding cassette transporter A1
- ACC
acetyl-CoA carboxylase
- ACOX1
acyl-coenzyme A oxidase 1
- AMPK
AMP-activated protein kinase
- ARG1
arignase 1
- ATGL
adipose triacylglycerol lipase
- AUC
area under the curve
- CYP7A1
cytochrome P450 7A1
- DGAT
diglyceride acyltra nsferase
- FABP
fatty-acid-binding protein
- FAS
fatty acid synthase
- FATP
fatty-acid- transporting protein
- G6Pase
glucose-6-phosphatase
- GPAT
glycerol-3-phosphate acyltransferase
- HMGCR
3-hydroxy-3-methyl-glutaryl-CoA reductase
- HOMA-IR
homeostasis model assessment-insulin resistance
- HSL
hormone sensitive lipase
- IRF
interferon regulatory factor
- LCAD
long-chain acyl-CoA dehydrogenase
- LDL
low-density lipoprotein
- LXR-α liver X receptor-α MCAD
medium-chainacyl-CoA dehydrogenase
- MCP-1
monocyte chemoattractant protein-1
- MGL
macrophage galactose-type C-type lectin
- MRC1
mannose receptor, C type 1
- PCK1
phosphoenolpyruvate carboxykinase 1
- PEPCK
Phosphoenolpyruvate carboxykinase
- PDK4
pyruvate dehydrogenase lipoamide kinase isozyme 4
- PPAR
peroxisome proliferator activated receptor
- SREBP1c
sterol response element binding protein 1c
- SCD1
stearoyl-CoA desaturase 1
- UCP
uncoupling protein
Footnotes
All authors have declared that no potential conflict of interest relevant to this article.
References
- 1.Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 2.Chen L, Magliano DJ, Zimmet PZ. The worldwide epidemiology of type 2 diabetes mellitus--present and future perspectives. Nat Rev Endocrinol. 2012;8:228–236. doi: 10.1038/nrendo.2011.183. [DOI] [PubMed] [Google Scholar]
- 3.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 4.Glass CK, Olefsky JM. Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab. 2012;15:635–645. doi: 10.1016/j.cmet.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 6.Donath MY, Shoelson SE. Type 2 diabetes as an inflammatory disease. Nat Rev Immunol. 2011;11:98–107. doi: 10.1038/nri2925. [DOI] [PubMed] [Google Scholar]
- 7.Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harding HP, Ron D. Endoplasmic reticulum stress and the development of diabetes: a review. Diabetes. 2002;51 (Suppl 3):S455–461. doi: 10.2337/diabetes.51.2007.s455. [DOI] [PubMed] [Google Scholar]
- 9.Tyagi S, Gupta P, Saini AS, Kaushal C, Sharma S. The peroxisome proliferator-activated receptor: A family of nuclear receptors role in various diseases. J Adv Pharm Technol Res. 2011;2:236–240. doi: 10.4103/2231-4040.90879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 11.Savitsky D, Tamura T, Yanai H, Taniguchi T. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol Immunother. 2010;59:489–510. doi: 10.1007/s00262-009-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 13.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 14.Takaoka A, Tamura T, Taniguchi T. Interferon regulatory factor family of transcription factors and regulation of oncogenesis. Cancer Sci. 2008;99:467–478. doi: 10.1111/j.1349-7006.2007.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu J, Bian ZY, Zhang R, Zhang Y, Liu C, Yan L, Zhang SM, et al. Interferon regulatory factor 3 is a negative regulator of pathological cardiac hypertrophy. Basic Res Cardiol. 2013;108:326. doi: 10.1007/s00395-012-0326-9. [DOI] [PubMed] [Google Scholar]
- 16.Chow EK, Castrillo A, Shahangian A, Pei L, O’Connell RM, Modlin RL, Tontonoz P, et al. A role for IRF3-dependent RXRalpha repression in hepatotoxicity associated with viral infections. J Exp Med. 2006;203:2589–2602. doi: 10.1084/jem.20060929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castrillo A, Joseph SB, Vaidya SA, Haberland M, Fogelman AM, Cheng G, Tontonoz P. Crosstalk between LXR and toll-like receptor signaling mediates bacterial and viral antagonism of cholesterol metabolism. Mol Cell. 2003;12:805–816. doi: 10.1016/s1097-2765(03)00384-8. [DOI] [PubMed] [Google Scholar]
- 18.Eguchi J, Yan QW, Schones DE, Kamal M, Hsu CH, Zhang MQ, Crawford GE, et al. Interferon regulatory factors are transcriptional regulators of adipogenesis. Cell Metab. 2008;7:86–94. doi: 10.1016/j.cmet.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, et al. Transcriptional control of adipose lipid handling by IRF4. Cell Metab. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kraus TA, Lau JF, Parisien JP, Horvath CM. A hybrid IRF9-STAT2 protein recapitulates interferon-stimulated gene expression and antiviral response. J Biol Chem. 2003;278:13033–13038. doi: 10.1074/jbc.M212972200. [DOI] [PubMed] [Google Scholar]
- 21.Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Tarutani M, et al. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells. 1996;1:115–124. doi: 10.1046/j.1365-2443.1996.08008.x. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto M, Tanaka N, Harada H, Kimura T, Yokochi T, Kitagawa M, Schindler C, et al. Activation of the transcription factor ISGF3 by interferon-gamma. Biol Chem. 1999;380:699–703. doi: 10.1515/BC.1999.087. [DOI] [PubMed] [Google Scholar]
- 23.Saltiel AR, Pessin JE. Insulin signaling pathways in time and space. Trends Cell Biol. 2002;12:65–71. doi: 10.1016/s0962-8924(01)02207-3. [DOI] [PubMed] [Google Scholar]
- 24.Steinberg GR, Kemp BE. AMPK in Health and Disease. Physiol Rev. 2009;89:1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 25.Maury E, Brichard SM. Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Mol Cell Endocrinol. 2010;314:1–16. doi: 10.1016/j.mce.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Xu S, Giles A, Nakamura K, Lee JW, Hou X, Donmez G, et al. Hepatic overexpression of SIRT1 in mice attenuates endoplasmic reticulum stress and insulin resistance in the liver. FASEB J. 2011;25:1664–1679. doi: 10.1096/fj.10-173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaffe HA, Danel C, Longenecker G, Metzger M, Setoguchi Y, Rosenfeld MA, Gant TW, et al. Adenovirus-mediated in vivo gene transfer and expression in normal rat liver. Nat Genet. 1992;1:372–378. doi: 10.1038/ng0892-372. [DOI] [PubMed] [Google Scholar]
- 28.Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Kimura T, Kitagawa M, Yokochi T, et al. Regulation of IFN-alpha/beta genes: evidence for a dual function of the transcription factor complex ISGF3 in the production and action of IFN-alpha/beta. Genes Cells. 1996;1:995–1005. doi: 10.1046/j.1365-2443.1996.870287.x. [DOI] [PubMed] [Google Scholar]
- 29.Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J Clin Invest. 2006;116:571–580. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guerre-Millo M, Rouault C, Poulain P, Andre J, Poitout V, Peters JM, Gonzalez FJ, et al. PPAR-alpha-null mice are protected from high-fat diet-induced insulin resistance. Diabetes. 2001;50:2809–2814. doi: 10.2337/diabetes.50.12.2809. [DOI] [PubMed] [Google Scholar]
- 31.Koo SH, Satoh H, Herzig S, Lee CH, Hedrick S, Kulkarni R, Evans RM, et al. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat Med. 2004;10:530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- 32.Stienstra R, Mandard S, Patsouris D, Maass C, Kersten S, Muller M. Peroxisome proliferator-activated receptor alpha protects against obesity-induced hepatic inflammation. Endocrinology. 2007;148:2753–2763. doi: 10.1210/en.2007-0014. [DOI] [PubMed] [Google Scholar]
- 33.Solinas G, Karin M. JNK1 and IKKbeta: molecular links between obesity and metabolic dysfunction. FASEB J. 2010;24:2596–2611. doi: 10.1096/fj.09-151340. [DOI] [PubMed] [Google Scholar]
- 34.Cai D, Yuan M, Frantz DF, Melendez PA, Hansen L, Lee J, Shoelson SE. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat Med. 2005;11:183–190. doi: 10.1038/nm1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arkan MC, Hevener AL, Greten FR, Maeda S, Li ZW, Long JM, Wynshaw-Boris A, et al. IKK-beta links inflammation to obesity-induced insulin resistance. Nat Med. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- 36.Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, et al. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–336. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 37.Feingold KR, Grunfeld C. Tumor necrosis factor-alpha stimulates hepatic lipogenesis in the rat in vivo. J Clin Invest. 1987;80:184–190. doi: 10.1172/JCI113046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grunfeld C, Adi S, Soued M, Moser A, Fiers W, Feingold KR. Search for mediators of the lipogenic effects of tumor necrosis factor: potential role for interleukin 6. Cancer Res. 1990;50:4233–4238. [PubMed] [Google Scholar]
- 39.Baffy G. Kupffer cells in non-alcoholic fatty liver disease: the emerging view. J Hepatol. 2009;51:212–223. doi: 10.1016/j.jhep.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.