Abstract
Chromosome replication is performed by numerous proteins that function together as a “replisome”. The replisome machinery duplicates both strands of the parental DNA simultaneously. Upon DNA damage to the cell, replisome action produces single-strand DNA to which RecA binds, enabling its activity in cleaving the LexA repressor and thus inducing the SOS response. How single-strand DNA is produced by a replisome acting on damaged DNA is not clear. For many years it has been assumed the single-strand DNA is generated by the replicative helicase, which continues unwinding DNA even after DNA polymerase stalls at a template lesion. Recent studies indicate another source of the single-strand DNA, resulting from an inherently dynamic replisome that may hop over template lesions on both leading and lagging strands, thereby leaving single-strand gaps in the wake of the replication fork. These single-strand gaps are proposed to be the origin of the single-strand DNA that triggers the SOS response after DNA damage.
Keywords: SOS response, Translesion Synthesis, DNA repair, lesion bypass, DNA polymerase, Sliding Clamp, Review
2. INTRODUCTION
DNA damage in E. coli can induce the SOS response, which expresses over 40 proteins, some of which may aid cell survival (1–4). The SOS response is initiated by accumulation of single-strand (ss) DNA during replication of DNA containing lesions, which block the replicase, preventing conversion of ssDNA to dsDNA. RecA binds the ssDNA to form RecA* which acquires a coprotease activity that facilitates self-cleavage of the LexA repressor and de-represses SOS-regulated genes (2, 4, 5). There are several different routes by which SOS induced proteins may remove, circumvent or bypass DNA lesions and we refer the reader to other sources for details of these processes (6–10). This review is centered on recent studies of the replisome that suggest a new mechanism by which the replisome may generate ssDNA upon encountering DNA lesions, thereby enabling RecA* to trigger the SOS response.
The E. coli replication machinery, referred to as the “replisome”, consists of about a dozen different proteins present in stoichiometries that range from one to six (11–14) (Figure 1A). At the head of the fork is the homohexameric DnaB helicase that encircles the lagging strand. DnaB unwinds the parental duplex by using rNTPs and translocates along the lagging strand while excluding the leading strand, acting as a wedge to peel the strands apart. The replicative polymerase is polymerase III (Pol III) holoenzyme consisting of three components: Pol III core, beta clamp and clamp loader. DNA Pol III core is a heterotrimer that contains alpha (the catalytic polymerase), epsilon (the proofreading 3’–5’ exonuclease) and theta subunits. Pol III core is slow and distributive when acting alone, but at the replication fork Pol III core associates with the beta sliding clamp, a ring shaped protein that encircles DNA and tethers Pol III to the template for rapid (500–1000 nucleotides/s) and processive (>80 kb) synthesis. The beta clamp is assembled onto DNA by a multisubunit clamp loader in an ATP driven reaction. Each of the three tau subunits in the clamp loader can bind a Pol III core, providing three DNA polymerases at the replication fork (15). The tau subunits also bind the DnaB helicase and enhance its rate of unwinding from 35 bp/s to nearly 1 kb/s (16). One Pol III core-beta extends the leading strand in the direction of fork movement, while the other two Pol III cores function on the lagging strand (17). Due to the antiparallel structure of DNA and the ability of DNA polymerases to only synthesize DNA in the 5’–3’ direction, the lagging strand must be extended in the opposite direction of fork movement. The lagging strand is made discontinuously as numerous 1–2 kb Okazaki fragments. Each Okazaki fragment is initiated by RNA primase, which must first bind to DnaB to produce RNA primers of about 10–12 nucletotides in length. The ssDNA on the lagging strand is rapidly coated by ssDNA binding protein (SSB), which protects the template from nucleases and also enhances the rate of DNA synthesis by Pol III-beta.
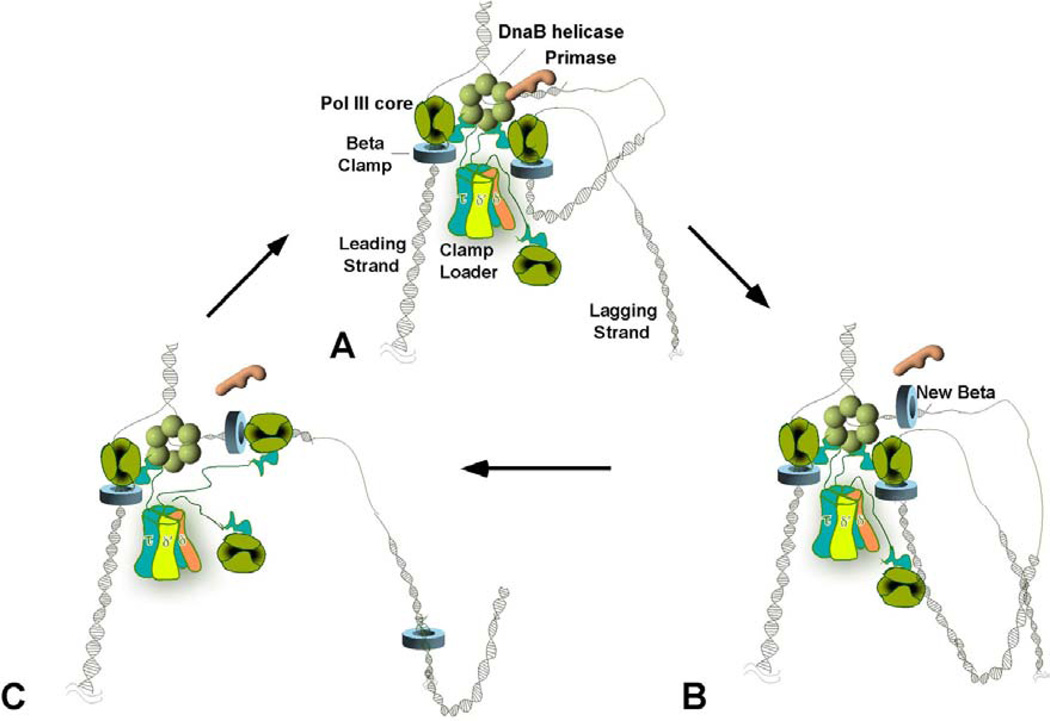
Figure 1.
Organization of a Trimeric Replicase at the E. coli Replication Fork. The figures show a replication fork containing three Pol III core subunits bound to the same clamp loader during leading and lagging strand synthesis on an undamaged template. In (A), two Pol III cores (dark green) function on the lagging strand, one of which is illustrated off DNA. The tau subunits (blue) of the clamp loader are represented with a flexible linker that connects the clamp loader to the DnaB helicase (light green hexamer) and Pol III core. (B) After synthesis of the RNA primer, the clamp loader displaces the primase (orange) and loads the clamp onto the new primer/template junction. The two lagging-strand Pol III cores are depicted here as extending two Okazaki fragments at the same time, producing two lagging-strand loops. (C) After an Okazaki fragment is fully extended the lagging strand polymerase recycles to the newly loaded clamp and starts elongation of a new fragment, leaving the old clamp behind. This completes a full cycle of lagging strand synthesis. Fork unwinding and leading strand synthesis continue throughout the cycle.
The high processivity of Pol III-beta must be overcome on the lagging strand in order for the replisome to repeatedly dissociate from the end of each Okazaki fragment and recycle to a new primer. This is accomplished by the ability of Pol III core to disengage from the beta clamp at the end of each Okazaki fragment, leaving the beta clamp on the DNA (see Figure 1B and C). The Pol III core can then re-associate at a new primed site for extension of the next Okazaki fragment. The clamp loader repeatedly assembles beta clamps onto new primed sites, enabling processive synthesis during the polymerase hopping process. The three Pol III cores at the fork increase the efficiency of the replisome in filling Okazaki fragments, since two Pol III cores can work on the lagging strand (17). RNA primers are removed by the 5’–3’ exonuclease in Pol I, but can also be removed by RNaseH. Okazaki fragments are then sealed together by ligase.
The above description of the replisome enables simultaneous and uninterrupted duplication of both parental stands of DNA under normal growth conditions in which there are no blocks to forward progression. However, even in the best of the circumstances the replisome will encounter obstacles, including DNA bound proteins such as repressors and RNA polymerase transcription complexes (18–22). Many of these obstacles can block the replisome, and it is often assumed that the fate of a stalled replisome is to collapse and require reassembly. Although use of the term “collapsed fork” may sometimes refer to stalled forks, this review defines a collapsed fork as a fork that has lost the helicase. The slow step of origin-independent replication fork assembly is the re-loading of DnaB helicase, and the cell contains factors that perform this task (23). It is also unavoidable that replication machinery occasionally stalls at DNA lesions formed by metabolic byproducts (24). In general, these DNA lesions are produced at a low frequency and are fixed by repair enzymes before the replisome encounters them. Under conditions of heavy DNA damage, DNA lesions are more likely to be encountered considering the rapid rate of E. coli replication fork progression. Experimentally the rate of DNA synthesis measured by 3H-TTP incorporation decreases immediately after UV irradiation (25–29). The decrease in 3H-TTP incorporation has been interpreted as due to replication fork collapse upon encounter a lesion. After removal of the DNA damaging agent, replication eventually restarts. Repair of DNA lesions and reassembly of the replication machinery is presumed to explain “replication restart”.
It is generally believed that accumulation of ssDNA is the trigger for activation of the SOS response by RecA (5, 30). However, the exact mechanism by which ssDNA is produced after DNA damage is not clearly understood. In the last few years several studies indicate that the replication machinery is quite stabile on DNA, and that a replication fork may not simply collapse upon encountering a lesion. Furthermore, the replisome has been demonstrated to be highly dynamic, capable of exchanging protein subunits during its movement and of hopping from one primed site to another. Based on recent biochemical studies of replisome dynamics, this review proposes a new process by which ssDNA may be generated in response to DNA damage.
3. THE “RUNAWAY HELICASE” MODEL OF ssDNA PRODUCTION
A long-standing model suggests that a lesion in the leading strand template blocks fork progression whereas a lesion in the lagging strand template does not and proposes how ssDNA is generated during DNA damage. (see Figure 2) (31–34). According to this model, DnaB, which binds to the lagging strand, uncouples from Pol III core and continues unwinding DNA ahead of the stalled Pol III, thereby producing ssDNA. Primase binds DnaB for activity on the lagging strand, and thus it is assumed that the unwound lagging strand is continuously primed and converted to duplex DNA. Indeed, recent studies of a fork with a stalled leading strand polymerase indicate that the lagging strand is converted to duplex DNA (35). As a result the lagging strand may not provide the ssDNA needed to induce the SOS response. However, if the helicase continues unwinding upon fork stalling, the unwound leading strand may persist as ssDNA. The ssDNA binding protein, SSB, rapidly binds ssDNA in the cell, protecting it from nucleases. Progressive accumulation of ssDNA enables the RecF,O,R proteins to eventually displace SSB and load RecA onto ssDNA, allowing RecA* to initiate the SOS response (4, 5, 36). DnaB, acting in the context of a stalled leading stand Pol III, is thought to be unstable and soon dissociates from DNA, resulting in replication fork collapse. Replication restart requires reassembly of the DnaB helicase on DNA, a time consuming process performed by helicase loading factors (23, 37).
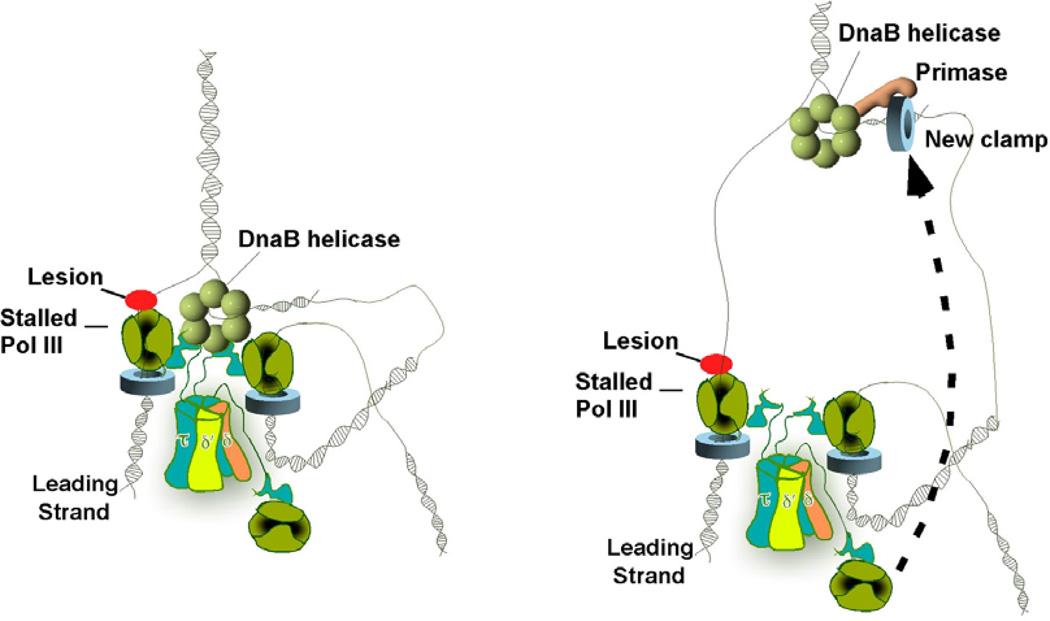
Figure 2.
Runaway helicase model of how ssDNA is generated during DNA damage. The leading strand polymerase stalls upon encountering a lesion (red circle) (Left). Despite a block in the leading strand, lagging-strand synthesis proceeds, implying transient uncoupling of concurrent leading/lagging strand synthesis. This creates a long ssDNA gap on the leading strand template (Right).
The above-mentioned model seems quite plausible, but to our knowledge there is no direct evidence for the formation of leading strand ssDNA upon DNA damage. This review proposes a different mechanism to generate ssDNA, which, however, does not exclude the runaway helicase model. It is interesting to note that in an early model based on studies of the DNA synthesized in UV-irradiated E. coli cells, Rupp and Howard-Flanders proposed that replication forks simply proceed past the damage and resume synthesis downstream, leaving gaps on both the leading and lagging strands, not just one gap on the leading strand, that are then filled in by RecA-mediated recombination (38). These observations seem to have been largely ignored. Indeed, it was not easy to conceive how ssDNA gaps could be generated on both leading and lagging strands considering the prevailing semi-discontinuous model of DNA replication. The semi-discontinuous model is currently being brought into question by several studies (12, 35, 39). In addition, biochemical findings provide new insights into the dynamic behavior of the replisome, and offer an explanation for how ssDNA gaps may be produced on both strands of damaged DNA (40–42). Formation of ssDNA gaps on both leading and lagging strands requires that the replisome continues to move on damaged DNA, skipping over lesions. This implies that replication forks do not collapse, but they have ways of circumventing lesions while continuing forward progression. The following sections briefly describe some of the findings that support these actions, and propose a model for production of ssDNA gaps on both leading and lagging strands. We suggest that these ssDNA gaps may persist and enable RecA to bind. The accumulation of a sufficient number of gaps could lead to the induction of the SOS response.
4. STALLED REPLICATION FORKS DO NOT SIMPLY COLLAPSE
During chromosomal duplication forks can stall at “replication fork barriers” (RFBs), where particular proteins bind tightly to DNA. In E. coli, examples of RFBs include Lac or Tet repressors bound to their cognate operators and efficient stalling requires tandem arrays of such complexes (43–45). Several studies have shown that the pausing of forks at Tus-Ter and Lac repressor-operator complexes can stimulate recombination at these sites (46–48). Since these sites are hotspots for recombination, it has been proposed that paused forks collapse (i.e. disassemble), enabling access of recombination proteins to the DNA (49, 50). However, it now seems that paused forks are surprisingly stable, and DNA synthesis can resume if the barrier protein is removed (43–45). Furthermore, there is evidence, in several cases, to indicate that hotspots of recombination are dependent on other factors, in addition to the pausing of a DNA replication fork (51). This brings into question the relative frequency of fork collapse/recombination compared to stalled forks and resumption of synthesis without collapse.
Clashes between forks and the transcriptional machinery can also represent an impediment to fork progression. An in vitro study utilized RNA polymerase, among the tightest of DNA binding proteins, to test the stability of the replication fork upon encounter with a transcribing RNA polymerase (21). RNA polymerase, transcribing in the opposite direction of fork movement, utilizes the lagging strand as template, the same strand that DnaB helicase encircles. Thus the replisome and RNA polymerase will collide head-on. Given the tight binding of RNA polymerase, and the presumed collapse of a stalled replication fork, the expected outcome of this collision is that DnaB helicase will dissociate, the fork will collapse and transcription will continue. However, the study showed quite the opposite. Even though fork progression was blocked by the tightly bound RNA polymerase, the helicase remained stably attached to DNA and halted transcription. In fact, the helicase finally displaced the RNA polymerase from DNA, with a half-time of about a half-hour. Hence, the fork does not spontaneously collapse upon encountering a block.
These studies indicate that replication forks are highly stabile. However, it still remains possible that the helicase continues to unwind DNA even though the leading stand polymerase is stalled at the lesion, thereby producing ssDNA for RecA to induce the SOS response.
5. TRANSLESION DNA POLYMERASES CAN REMODEL THE FORK, FUNCTION WITH HELICASE, AND MOVE SLOWLY
As described earlier, the observation that DNA damage halts replication, as measured by 3H-TTP incorporation, was explained as replication fork collapse upon encounter with a DNA lesion (25, 26, 52, 53). However, a low rate of 3H-TTP incorporation could also be due to forks that slow down, but do not collapse (i.e. the replisome could remain on DNA). Maybe this possibility was not previously proposed for lack of precedent that replication fork speed could be modulated.
Recent in vitro studies provide precedent for SOS induced factors that slow replication fork progression (54, 55). These factors are translesion synthesis DNA polymerases (TLS Pols), which are induced during the SOS response (some TLS Pols were only discovered in the late 1990s) (8–10, 56–58) TLS Pols are a class of DNA polymerases capable of extending DNA across a template lesion. TLS Pols, especially those in the Y-family, lack a 3’–5’ exonuclease activity, and thus have low fidelity compared to enzymes that contain a proofreading nuclease. This facilitates elongation over a damaged DNA as enzymes with a proofreading exonuclease often remove the misincorporated base and prevent the advance of a DNA chain over a template lesion. In addition, the structures of Y-family polymerases reveal a more open active site, making them more capable of accepting distortions in DNA, and thus more readily traverse template lesions compared to high fidelity DNA polymerases (59). Most cells contain several different TLS Pols, presumably to synthesize DNA across different types of damage (9, 56, 60–62).
The majority of DNA damage is repaired before fork encounter by excision repair, which is a high fidelity mechanism since it functions in the context of duplex DNA. In fact, excision repair enables correction of the excised damaged base using the complimentary undamaged strand as template. However, when the replication fork encounters a damaged DNA base, excision repair system cannot correct this lesion because it is located at a primed ds/ss junction. These lesions can be repaired by fork reversal followed by high fidelity excision repair, or by TLS Pol assisted bypass which can result in mutations (63). It is thought that TLS Pols act at a stalled replication fork by switching with the replicase and moving the replication fork past the leading strand lesion. Once the lesion is traversed the high fidelity replicase rebinds for continued chromosome replication (9, 64–67).
E. coli possesses two classic TLS Pols, Pols IV and V, which belong to the Y-family of DNA polymerases and lack a proofreading exonuclease (10, 58, 68). These TLS Pols are highly induced during the SOS response triggered by DNA damage, as one may expect from their function in traversing lesions. E. coli Pol II is also considered to be a TLS polymerase because it is induced in the SOS response. Pol II is a B-family polymerase and contains a proofreading exonuclease, but its proficiency in template slippage at a block can enable bypass of a lesion at the expense of a single nucleotide deletion (69, 70). Both Pols II and IV are induced in the very early stage (within the first 5 minutes) of the SOS response (71–74).
Mutagenesis is most closely associated with Pol V (70), which is induced quite late in the SOS response (after 45 minutes) (72, 75). Pol V is composed of a polymerase (UmuC) and two copies of UmuD’ (the RecA self-cleaved form of UmuD) and requires the RecA-ssDNA nucleoprotein filament, RecA*, for activity (reviewed in (76)). The 3’ terminus of a RecA* filament in trans transfers a RecA monomer to Pol V to generate the active form of the enzyme (UmuC-UmuD’2-RecA-ATP), referred to as the Pol V mutasome (76–78).
The beta clamp interacts with each of the 5 E. coli Pols, as well as several other proteins involved in DNA replication and repair (79). Many models propose that the clamp binds simultaneously to different partners and regulates their access to the DNA primer during switching. One such model, referred to as the “toolbelt” model, suggests that 2 different Pols bind to the dimeric beta clamp at the same time, with each Pol contacting a separate protomer (65, 80). Another model indicates that a single protomer is sufficient for Pol III* and Pol IV switching, and that the second protomer may be available to accommodate yet additional partners (81). A similar situation is true for many eukaryotic Pols (9, 82). The eukaryotic sliding clamp, PCNA, is a homotrimer and thus may bind up to three different proteins at once (83). Support for the toolbelt model came from studies in the T4 system, which indicated that wild-type and mutant T4 gp43 DNA polymerases trade places with one another through interaction with the same clamp (84). This was first demonstrated in the E. coli system using Pol III and TLS Pol IV (65). The results showed that when E. coli Pol III stalls on DNA, the TLS polymerase rapidly switches with Pol III on the clamp, and after the stall is relieved (e.g. as it would occur upon lesion bypass) the high fidelity Pol III regains the clamp from Pol IV for continued replication. Further in vitro studies using SOS induced levels of Pols II and Pol IV showed that both TLS Pols can take over the clamp and DnaB helicase without Pol III stalling (Figure 3) (54). Simply adding TLS Pols II or IV at levels comparable to those that exist during the SOS response is sufficient to displace Pol III from a moving replication fork, yet retain the clamp and helicase to form a “TLS replisome” (i.e. replisome containing DnaB, beta clamp and either Pol II or Pol IV) (Figure 3). Pol II and Pol IV are 200–1000 times slower than Pol III respectively, thus Pol III/TLS Pol switching results in a slow moving TLS replisome. The rates of the TLS replisomes (10 ntd/s for Pol II and 1 ntd/s for Pol IV) are also significantly lower than the intrinsic rate of DnaB helicase (35 ntd/s), implying that TLS Pols slow the helicase. These observations suggest that upon SOS induction, TLS Pols take over the replication fork from Pol III even prior to encountering a template lesion. As partial evidence that these events occur inside the cell, controlled expression of Pol II and Pol IV to levels similar to those in a SOS induced cell result in a lower 3H-TTP incorporation, even in the absence of DNA damage (54, 55).
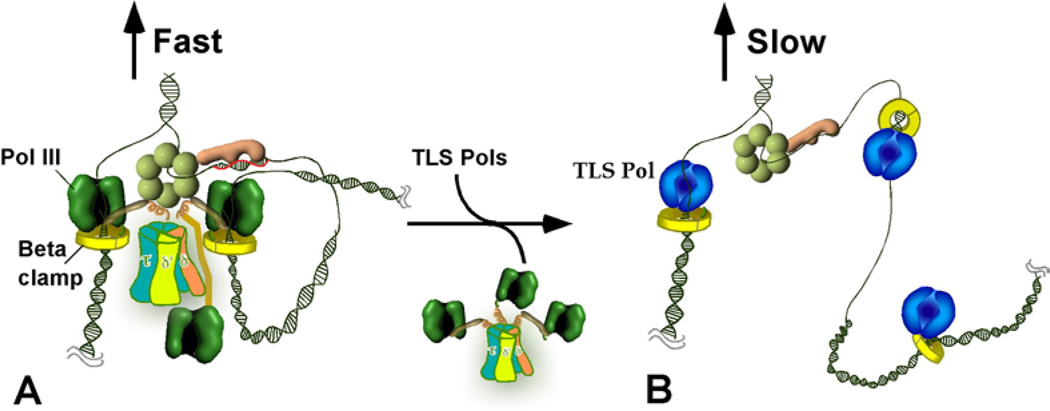
Figure 3.
Pol III and TLS replisomes. Conversion of the coupled Pol III replisome to an uncoupled alternative TLS replisome. A) The coupled trimeric Pol III holoenzyme-DnaB replisome. B) Take-over of beta clamps by a TLS Pol displaces Pol III form the fork, resulting in a TLS Pol-replisome in which TLS Pols act distributively on beta clamps.
A somewhat perplexing question that arises from these in vitro experiments is: “Why would Pol III be replaced at a replication fork by a low fidelity enzyme even before it encounters a lesion?”. One may expect this to increase mutation frequency. As unreasonable as it may seem at first, a slow, albeit lower fidelity replisome has its advantages. First, a rapid 1kb/s replisome would almost surely encounter a lesion whereas a much slower replisome (1–10 ntds/s) would allow more time for excision repair to correct it preventing fork stalling . Second, even though TLS Pols are low fidelity polymerases, the amount of DNA made by a slow replisome will minimize the number of mutations. For example, Pol IV generates about one misincorporation every 1 kb (85), but the Pol IV replisome travels at a speed of 1 ntd/s and thus would require over 15 minutes to synthesize 1 kb and insert a single mutation. Furthermore, UmuD binds to Pol IV during the early stage of the SOS response in a way that modulates its mutagenic effect (86). Finally, a mistake introduced by Pol IV acting on undamaged DNA would be a mismatch, not a base incorporated opposite a template lesion. Mismatches can be corrected by the mismatch repair system.
6. DISCONTINUOUS LAGGING STRAND SYNTHESIS GENERATES ssDNA GAPS IN RESPONSE TO DNA DAMAGE ON THE LAGGING STRAND
According to the “trombone model” of replication discussed in the introduction, the leading strand is extended continuously in the direction of fork movement, while the lagging strand is made discontinuously as a series of Okazaki fragments(87). Okazaki fragments are only about 1–2 kb, while Pol III-beta has an intrinsic processivity far greater than this, and in the context of a replisome it has been shown to extend DNA about 86 kb per binding event(88). This high processivity contrasts with the action of the lagging strand Pol III, which must repeatedly disengage from DNA at the end of each Okazaki fragment in order to extend a new fragment. This “processivity barrier” is circumvented by specific mechanisms that trigger release of Pol III from the clamp upon completing an Okazaki fragment, enabling Pol III to hop to a new beta clamp loaded on an upstream RNA primer (89, 90).
Polymerase hopping among clamps provides a means to get around a lesion on the lagging strand. Specifically, upon encountering a lesion the polymerase could dissociate from the clamp and bind a new clamp at the next available RNA primer, thus leaving the lesion behind in a ssDNA gap. However, simple model studies show that Pol III remains stably attached to its clamp upon stalling at a lesion (91). Furthermore, it was assumed for many years that the leading and lagging strand polymerases were strictly coordinated, and if one polymerase were blocked, it would stop the other polymerase (92–95). Thus a lesion on the lagging strand would stop both the lagging and leading stand polymerases, thereby halting the replisome until the lesion was repaired or bypassed.
In vitro studies put this long-standing belief of leading/lagging strand coordination to rest (32–34, 96). A block to the lagging strand polymerase does not stop the leading strand polymerase, and leading strand extension continues at the same rate. Hence, the two polymerases are functionally uncoupled, even though they are physically linked together. In fact, the stalled lagging strand Pol III dissociates from its clamp and then continues lagging strand synthesis as new RNA primers are synthesized, leaving the block behind in a ssDNA gap (see Figure 4). Hence, even though a stalled Pol III-beta is stably bound to DNA at a lesion in a simple model system (i.e. a singly primed M13 ssDNA), it acts differently in the context of a moving replication fork. Presumably continued fork progression generates a DNA loop that eventually builds up enough “drag” on the moving replisome to pull the stalled lagging strand Pol III from its clamp. These events explain how DNA damage can result in ssDNA gaps on the lagging strand, but do not explain how ssDNA gaps are created on the leading strand, the topic of the next section.
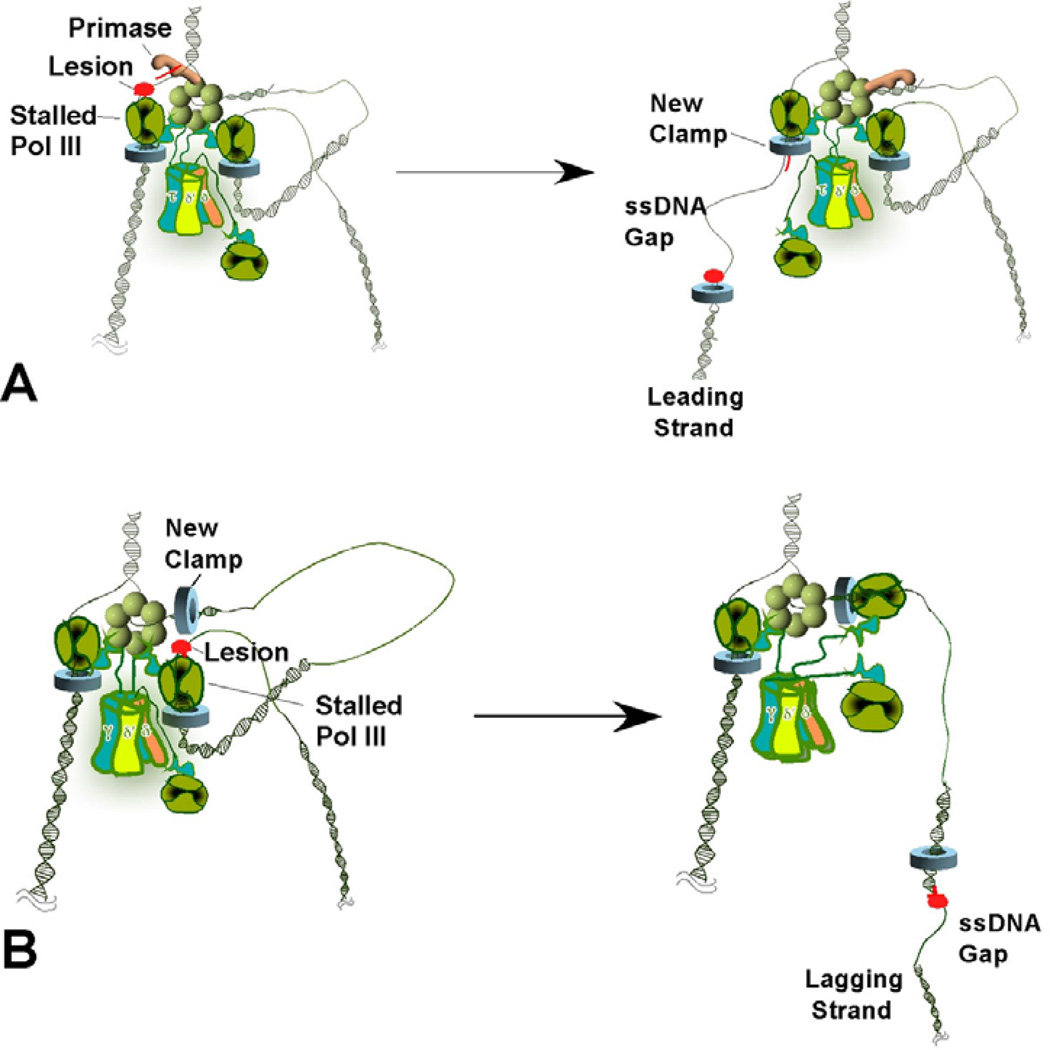
Figure 4.
Lesion skipping model of ssDNA generation during DNA damage. (A) Leading strand lesion. The leading strand polymerase stalls upon encountering a lesion (left). The helicase recruits primase to reinitiate leading strand synthesis ahead of the lesion, leaving a single-strand gap (right). If stalling causes the replication fork to collapse, additional factors (e.g., PriA or PriC) are required to reload the helicase at the collapsed fork. (B) Lagging strand lesion. Upon encountering a lesion on the lagging strand template (Left), leading strand synthesis continues and the stalled lagging strand polymerase recycles to a new primer/template junction, leaving a single-strand gap with a template lesion (right).
7. THE LEADING STRAND CAN BE DISCONTINUOUS, AND PRODUCES ssDNA GAPS IN RESPONSE TO DNA DAMAGE
Early work by Okazaki suggested that replication might be discontinuous on both the leading and lagging strands (97). Yet continuous leading strand synthesis dominates current models, largely based on in vitro studies, which did not include blocks to replisome progression. However, sources of discontinuities on the leading strand have been demonstrated by two recent in vitro studies (35, 98). In one study, a replication fork was allowed to collide in-line with a transcribing RNA polymerase (i.e. moving in the same direction of fork progression while transcribing the leading strand) (98). Replication forks advance at 12–30 times the rate of transcription, and considering the abundance of closely spaced genes in bacterial genomes, collisions of the replisome with RNA polymerase during replication are likely quite frequent. The study showed that the replisome dislodges the RNA polymerase, but retains the RNA transcript and utilizes it as a primer to continue elongation of the leading strand. The end result is a discontinuity between the leading strand DNA and the RNA transcript. This action further supports a view of the replisome as a highly dynamic entity, capable of circumventing obstacles in unexpected ways. Presumably, the clamp loader places a new beta clamp on the transcript, enabling the leading strand polymerase to hop from the site of encounter, to the 3’ terminus of the transcript, possibly using the third Pol III core to do so. The RNA transcript is likely removed and filled in with DNA by the same process that removes RNA primers and seals Okazaki fragments on lagging strand. The only difference is that the leading strand RNA is produced by RNA polymerase rather than primase.
A second example of discontinuous synthesis on the leading strand utilized a cyclobutane pyrimidine dimer lesion on the leading stand template to block the Pol III replisome (35). This study demonstrated that a stalled fork does not collapse, and that the DnaB helicase on the lagging strand enables primase to prime the leading strand. After priming downstream of the lesion, the replisome continued, leaving the lesion behind in a ssDNA gap on the leading strand (Figure 4A). In cells growing without extensive DNA damage, this source of leading strand discontinuity is not expected to be as frequent as collisions with RNA polymerase. However, priming ahead of leading strand lesions likely explains early observations of ssDNA gaps on both leading and lagging strands following DNA damage (38, 99). The lesion, left in a ssDNA gap behind the fork, is probably repaired by either recombination/excision repair, using the sister chromosome, or it may be bypassed using a TLS Pol.
In overview, the replisome has evolved various means to overcome and circumvent barriers of all kinds, allowing replication to continue and leaving problem areas behind to be sorted out by other enzymes. However, lesion hopping is only one scenario that occurs upon encounter of a fork with a leading strand lesion. One may expect that a high density of DNA damage will finally impair fork progression and bring it to a griding halt. Moreover, some lesions generate strand breaks, which will result in fork collapse, and require replication restart. The observation that DnaC is needed after DNA damage is consistent with fork collapse in some cells, or upon a certain level of damage (100). Regardless, the idea that ssDNA gaps can be generated by replisome hopping (prior to fork collapse) suggests a mechanism that may produce ssDNA for the SOS response, as discussed in the section below.
8. LESION SKIPPING MODEL FOR GENERATION OF ssDNA DURING DNA DAMAGE
In light of the recent work that illustrates the unexpected dynamic flexibility of the replisome, we propose another source of ssDNA, besides the runaway helicase model, that may initiate the SOS response. Under normal growth conditions, the fork will occasionally encounter a DNA obstacle (e.g. DNA damage and/or DNA-protein complex). Regardless of the strand the obstacle is on, the block will be skipped and left behind in the wake of the fork (as illustrated in Figure 4A, B). When there is a low density of lesions the ssDNA gaps will not persist for long before being repaired. Hence, the SOS response will not be elicited. On the contrary, in the face of heavy DNA damage, the replisome will encounter many lesions and thereby produce more ssDNA gaps on both daughter strands. At some tipping point, the concentration of ssDNA gaps will exceed the capability of the cell to repair them, enabling them to persist long enough for the RecFOR pathway to displace SSB and load RecA onto the ssDNA gaps. The RecA* (i.e. RecA nucleoprotein filament on ssDNA) will then trigger the SOS response. Once initiated, the SOS response will rapidly induce TLS Pols II and IV which can perform double duty by: 1) slowing the fork, preventing most future encounters with lesions, and 2) traversing lesions in ssDNA gaps that are not dealt with by the recombination repair pathway.
9. RESOLVING INCONSISTENCIES WITH GENETIC STUDIES
Admittedly, not all the genetic evidence is seamlessly consistent with the model proposed here. For example, E. coli mutants in which the SOS response is constitutive appear healthy and do not grow poorly (101, 102). This is clearly inconsistent with the view that slow TLS Pols take over the replisome. A reasonable proposition that may resolve this apparent discrepancy is that SOS constitutive cells have a second site suppressor mutation(s) that prevents TLS Pol takeover of the replisome, such as lower expression of TLS Pols, or higher expression of Pol III. Second site suppressors of this sort would be likely considering that the defective cells are under selection for rapid growth (i.e. suppressors would outcompete the unsuppressed constitutive cells). It is also possible that unidentified factors control access of TLS Pols to the replisome, thereby enabling continued rapid replication. Another case in which the proposed hypothesis for ssDNA at first appears inconsistent with genetic studies is the finding that fork slowdown still occurs in Rec F,O,R mutants (52, 53, 103–105). One may imagine that without Rec FOR, RecA would not assemble onto SSB coated ssDNA, thus the SOS response would not be induced and increased levels of TLS Pols would not be produced for fork slowdown. However, recent biochemical studies may provide an explanation for this observation. Specifically, Pol IV is efficient at taking over a clamp once Pol III has stalled (65). Considering the constitutive high levels of Pol IV (250/cell) without SOS induction relative to Pol III (10– 20/cell), Pol IV may takeover a fork that has encountered a lesion. The advantage conferred by SOS response is that SOS-induced Pol IV (2500/cell) can also takeover the fork from a moving Pol III, prior to Pol III stalling at a lesion, and thus mutagenic TLS synthesis may prevent fork blocking.
In any case, the central point of this review is that the runaway helicase at a stalled fork is not the only imaginable solution to generating ssDNA upon DNA damage. This proposal is highlighted by the in vivo observation of ssDNA gaps on both daughter strands after DNA damage, the slow down of helicase when Pol III is blocked in vitro, and the demonstrated ability of the replisome to circumvent barriers on both leading and lagging strands that leaves ssDNA gaps in the wake of the replication fork.
ACKNOWLEDGEMENTS
The authors are grateful to a grant from the NIH (GM38839) for this report.
REFERENCES
- 1.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158(1):41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker GC. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernandez De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol. 2000;35(6):1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 4.Janion C. Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int J Biol Sci. 2008;4(6):338–344. doi: 10.7150/ijbs.4.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Little JW. Autodigestion of lexA and phage lambda repressors. Proc Natl Acad Sci U S A. 1984;81(5):1375–1379. doi: 10.1073/pnas.81.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courcelle J, Hanawalt PC. RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet. 2003;37:611–646. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- 7.Courcelle J, Belle JJ, Courcelle CT. When replication travels on damaged templates: bumps and blocks in the road. Res Microbiol. 2004;155(4):231–237. doi: 10.1016/j.resmic.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 8.Bichara M, Meier M, Wagner J, Cordonnier A, Lambert IB. Postreplication repair mechanisms in the presence of DNA adducts in Escherichia coli. Mutat Res. 2011;727(3):104–122. doi: 10.1016/j.mrrev.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13(3):141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh JM, Hawver LA, Beuning PJ. Escherichia coli Y family DNA polymerases. Front Biosci. 2011;17:3164–3182. doi: 10.2741/3904. [DOI] [PubMed] [Google Scholar]
- 11.Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- 12.Langston LD, Indiani C, O'Donnell M. Whither the replisome: emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle. 2009;8(17):2686–2691. doi: 10.4161/cc.8.17.9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McHenry CS. DNA replicases from a bacterial perspective. Annu Rev Biochem. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. [DOI] [PubMed] [Google Scholar]
- 14.Yao NY, O'Donnell M. Replisome structure and conformational dynamics underlie fork progression past obstacles. Curr Opin Cell Biol. 2009;21(3):336–343. doi: 10.1016/j.ceb.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McInerney P, Johnson A, Katz F, O'Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27(4):527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 16.Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell. 1996;84(4):643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 17.Georgescu RE, Kurth I, O'Donnell ME. Single-molecule studies reveal the function of a third polymerase in the replisome. Nat Struct Mol Biol. 2009 doi: 10.1038/nsmb.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirkin EV, Mirkin SM. Replication fork stalling at natural impediments. Microbiol Mol Biol Rev. 2007;71(1):13–35. doi: 10.1128/MMBR.00030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol. 2002;3(11):859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 20.Merrikh H, Machon C, Grainger WH, Grossman AD, Soultanas P. Co-directional replication-transcription conflicts lead to replication restart. Nature. 2011;470(7335):554–557. doi: 10.1038/nature09758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pomerantz RT, O'Donnell M. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science. 2010;327(5965):590–592. doi: 10.1126/science.1179595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guy CP, Atkinson J, Gupta MK, Mahdi AA, Gwynn EJ, Rudolph CJ, Moon PB, van Knippenberg IC, Cadman CJ, Dillingham MS, Lloyd RG, McGlynn P. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol Cell. 2009;36(4):654–666. doi: 10.1016/j.molcel.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7(12):932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- 24.Lindahl T. The Croonian Lecture 1996: endogenous damage to DNA. Philos Trans R Soc Lond B Biol Sci. 1996;351(1347):1529–1538. doi: 10.1098/rstb.1996.0139. [DOI] [PubMed] [Google Scholar]
- 25.Witkin EM. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Setlow RB, Swenson PA, Carrier WL. Thymine Dimers and Inhibition of DNA Synthesis by Ultraviolet Irradiation of Cells. Science. 1963;142(3598):1464–1466. doi: 10.1126/science.142.3598.1464. [DOI] [PubMed] [Google Scholar]
- 27.Courcelle CT, Belle JJ, Courcelle J. Nucleotide excision repair or polymerase V-mediated lesion bypass can act to restore UV-arrested replication forks in Escherichia coli. J Bacteriol. 2005;187(20):6953–6961. doi: 10.1128/JB.187.20.6953-6961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Courcelle CT, Chow KH, Casey A, Courcelle J. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc Natl Acad Sci U S A. 2006;103(24):9154–9159. doi: 10.1073/pnas.0600785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khidhir MA, Casaregola S, Holland IB. Mechanism of transient inhibition of DNA synthesis in ultraviolet-irradiated E. coli: inhibition is independent of recA whilst recovery requires RecA protein itself and an additional, inducible SOS function. Mol Gen Genet. 1985;199(1):133–140. doi: 10.1007/BF00327522. [DOI] [PubMed] [Google Scholar]
- 30.Bridges BA. Error-prone DNA repair and translesion DNA synthesis. II: The inducible SOS hypothesis. DNA Repair (Amst) 2005;4(6):725–736. 739. doi: 10.1016/j.dnarep.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Meneghini R, Hanawalt PC. Postreplication repair in human cells: on the presence of gaps opposite dimers and recombination. Basic Life Sci. 1975;5B:639–642. doi: 10.1007/978-1-4684-2898-8_36. [DOI] [PubMed] [Google Scholar]
- 32.Pages V, Fuchs RP. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science. 2003;300(5623):1300–1303. doi: 10.1126/science.1083964. [DOI] [PubMed] [Google Scholar]
- 33.McInerney P, O'Donnell M. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J Biol Chem. 2004;279(20):21543–21551. doi: 10.1074/jbc.M401649200. [DOI] [PubMed] [Google Scholar]
- 34.Higuchi K, Katayama T, Iwai S, Hidaka M, Horiuchi T, Maki H. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes to Cells. 2003;8(5):437–449. doi: 10.1046/j.1365-2443.2003.00646.x. [DOI] [PubMed] [Google Scholar]
- 35.Yeeles JT, Marians KJ. The Escherichia coli replisome is inherently DNA damage tolerant. Science. 2011;334(6053):235–238. doi: 10.1126/science.1209111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rangarajan S, Woodgate R, Goodman MF. Replication restart in UV-irradiated Escherichia coli involving pols II, III, V, PriA, RecA and RecFOR proteins. Mol Microbiol. 2002;43(3):617–628. doi: 10.1046/j.1365-2958.2002.02747.x. [DOI] [PubMed] [Google Scholar]
- 37.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439(7076):557–562. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 38.Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol. 1968;31(2):291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 39.Wang TC, Smith KC. Discontinuous DNA replication in a lig-7 strain of Escherichia coli is not the result of mismatch repair, nucleotide-excision repair, or the base-excision repair of DNA uracil. Biochem Biophys Res Commun. 1989;165(2):685–688. doi: 10.1016/s0006-291x(89)80020-8. [DOI] [PubMed] [Google Scholar]
- 40.Amado L, Kuzminov A. The replication intermediates in Escherichia coli are not the product of DNA processing or uracil excision. J Biol Chem. 2006;281(32):22635–22646. doi: 10.1074/jbc.M602320200. [DOI] [PubMed] [Google Scholar]
- 41.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21(1):15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 42.Wang TC. Discontinuous or semi-discontinuous DNA replication in Escherichia coli? Bioessays. 2005;27(6):633–636. doi: 10.1002/bies.20233. [DOI] [PubMed] [Google Scholar]
- 43.Payne BT, van Knippenberg IC, Bell H, Filipe SR, Sherratt DJ, McGlynn P. Replication fork blockage by transcription factor-DNA complexes in Escherichia coli. Nucleic Acids Res. 2006;34(18):5194–5202. doi: 10.1093/nar/gkl682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Possoz C, Filipe SR, Grainge I, Sherratt DJ. Tracking of controlled Escherichia coli replication fork stalling and restart at repressor-bound DNA in vivo. EMBO J. 2006;25(11):2596–2604. doi: 10.1038/sj.emboj.7601155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGlynn P, Guy CP. Replication forks blocked by protein-DNA complexes have limited stability in vitro. J Mol Biol. 2008;381(2):249–255. doi: 10.1016/j.jmb.2008.05.053. [DOI] [PubMed] [Google Scholar]
- 46.Horiuchi T, Fujimura Y. Recombinational rescue of the stalled DNA replication fork: a model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J Bacteriol. 1995;177(3):783–791. doi: 10.1128/jb.177.3.783-791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma B, Hill TM. Insertion of inverted Ter sites into the terminus region of the Escherichia coli chromosome delays completion of DNA replication and disrupts the cell cycle. Mol Microbiol. 1995;18(1):45–61. doi: 10.1111/j.1365-2958.1995.mmi_18010045.x. [DOI] [PubMed] [Google Scholar]
- 48.Vilette D, Uzest M, Ehrlich SD, Michel B. DNA transcription and repressor binding affect deletion formation in Escherichia coli plasmids. EMBO J. 1992;11(10):3629–3634. doi: 10.1002/j.1460-2075.1992.tb05447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lambert S, Carr AM. Checkpoint responses to replication fork barriers. Biochimie. 2005;87(7):591–602. doi: 10.1016/j.biochi.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 50.Admire A, Shanks L, Danzl N, Wang M, Weier U, Stevens W, Hunt E, Weinert T. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes Dev. 2006;20(2):159–173. doi: 10.1101/gad.1392506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Labib K, Hodgson B. Replication fork barriers: pausing for a break or stalling for time? EMBO Rep. 2007;8(4):346–353. doi: 10.1038/sj.embor.7400940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Courcelle J, Crowley DJ, Hanawalt PC. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and recF protein function. J Bacteriol. 1999;181(3):916–922. doi: 10.1128/jb.181.3.916-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Courcelle J, Carswell-Crumpton C, Hanawalt PC. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci U S A. 1997;94(8):3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Indiani C, Langston LD, Yurieva O, Goodman MF, O'Donnell M. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. Proc Natl Acad Sci U S A. 2009;106(15):6031–6038. doi: 10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uchida K, Furukohri A, Shinozaki Y, Mori T, Ogawara D, Kanaya S, Nohmi T, Maki H, Akiyama M. Overproduction of Escherichia coli DNA polymerase DinB (Pol IV) inhibits replication fork progression and is lethal. Mol Microbiol. 2008;70(3):608–622. doi: 10.1111/j.1365-2958.2008.06423.x. [DOI] [PubMed] [Google Scholar]
- 56.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases (Pol II Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19(22):6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, Prakash L, Prakash S, Todo T, Walker GC, Wang Z, Woodgate R. The Y-family of DNA polymerases. Mol Cell. 2001;8(1):7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 58.Jarosz DF, Beuning PJ, Cohen SE, Walker GC. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 2007;15(2):70–77. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 59.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107(1):91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 60.Wagner J, Etienne H, Janel-Bintz R, Fuchs RP. Genetics of mutagenesis in E. coli: various combinations of translesion polymerases (Pol II, IV and V) deal with lesion/sequence context diversity. DNA Repair (Amst) 2002;1(2):159–167. doi: 10.1016/s1568-7864(01)00012-x. [DOI] [PubMed] [Google Scholar]
- 61.Jarosz DF, Godoy VG, Delaney JC, Essigmann JM, Walker GC. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature. 2006;439(7073):225–228. doi: 10.1038/nature04318. [DOI] [PubMed] [Google Scholar]
- 62.Fujii S, Fuchs RP. Interplay among replicative and specialized DNA polymerases determines failure or success of translesion synthesis pathways. J Mol Biol. 2007;372(4):883–893. doi: 10.1016/j.jmb.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 63.Berdichevsky A, Izhar L, Livneh Z. Error-free recombinational repair predominates over mutagenic translesion replication in E. coli. Mol Cell. 2002;10(4):917–924. doi: 10.1016/s1097-2765(02)00679-2. [DOI] [PubMed] [Google Scholar]
- 64.Furukohri A, Goodman MF, Maki H. A dynamic polymerase exchange with Escherichia coli DNA polymerase IV replacing DNA polymerase III on the sliding clamp. J Biol Chem. 2008;283(17):11260–11269. doi: 10.1074/jbc.M709689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Indiani C, McInerney P, Georgescu R, Goodman MF, O'Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol Cell. 2005;19(6):805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 66.Fujii S, Fuchs RP. Defining the position of the switches between replicative and bypass DNA polymerases. EMBO J. 2004;23(21):4342–4352. doi: 10.1038/sj.emboj.7600438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 68.Fuchs RP, Fujii S, Wagner J. Properties and functions of Escherichia coli: Pol IV and Pol V. Adv Protein Chem. 2004;69:229–264. doi: 10.1016/S0065-3233(04)69008-5. [DOI] [PubMed] [Google Scholar]
- 69.Fuchs RP, Fujii S. Translesion synthesis in Escherichia coli: lessons from the NarI mutation hot spot. DNA Repair (Amst) 2007;6(7):1032–1041. doi: 10.1016/j.dnarep.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 70.Pham P, Rangarajan S, Woodgate R, Goodman MF. Roles of DNA polymerases V and II in SOS-induced error-prone and error-free repair in Escherichia coli. Proc Natl Acad Sci U S A. 2001;98(15):8350–8354. doi: 10.1073/pnas.111007198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bonner CA, Stukenberg PT, Rajagopalan M, Eritja R, O’Donnell M, McEntee K, Echols H, Goodman MF. Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins. J Biol Chem. 1992;267(16):11431–11438. [PubMed] [Google Scholar]
- 72.Kim SR, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Multiple pathways for SOS-induced mutagenesis in Escherichia coli: an overexpression of dinB/dinP results in strongly enhancing mutagenesis in the absence of any exogenous treatment to damage DNA. Proc Natl Acad Sci U S A. 1997;94(25):13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim SR, Matsui K, Yamada M, Gruz P, Nohmi T. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol Genet Genomics. 2001;266(2):207–215. doi: 10.1007/s004380100541. [DOI] [PubMed] [Google Scholar]
- 74.Qiu Z, Goodman MF. The Escherichia coli polB locus is identical to dinA, the structural gene for DNA polymerase II. Characterization of Pol II purified from a polB mutant. J Biol Chem. 1997;272(13):8611–8617. doi: 10.1074/jbc.272.13.8611. [DOI] [PubMed] [Google Scholar]
- 75.Woodgate R, Ennis DG. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol Gen Genet. 1991;229(1):10–16. doi: 10.1007/BF00264207. [DOI] [PubMed] [Google Scholar]
- 76.Patel M, Jiang Q, Woodgate R, Cox MM, Goodman MF. A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V. Crit Rev Biochem Mol Biol. 2010;45(3):171–184. doi: 10.3109/10409238.2010.480968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jiang Q, Karata K, Woodgate R, Cox MM, Goodman MF. The active form of DNA polymerase V is UmuD'(2)C-RecA-ATP. Nature. 2009;460(7253):359–363. doi: 10.1038/nature08178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schlacher K, Cox MM, Woodgate R, Goodman MF. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature. 2006;442(7105):883–887. doi: 10.1038/nature05042. [DOI] [PubMed] [Google Scholar]
- 79.Lopez de Saro FJ, O'Donnell M. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase. Proceedings of the National Academy of Sciences. 2001;98(15):8376. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pages V, Fuchs R. How DNA lesions are turned into mutations within cells? Oncogene. 2002;21(58):8957–8966. doi: 10.1038/sj.onc.1206006. [DOI] [PubMed] [Google Scholar]
- 81.Heltzel JM, Maul RW, Scouten Ponticelli SK, Sutton MD. A model for DNA polymerase switching involving a single cleft and the rim of the sliding clamp. Proc Natl Acad Sci U S A. 2009;106(31):12664–12669. doi: 10.1073/pnas.0903460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moldovan GL, Pfander B, Jentsch S. PCNA the maestro of the replication fork. Cell. 2007;129(4):665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 83.Warbrick E. The puzzle of PCNA's many partners. Bioessays. 2000;22(11):997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 84.Yang J, Zhuang Z, Roccasecca RM, Trakselis MA, Benkovic SJ. The dynamic processivity of the T4 DNA polymerase during replication. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(22):8289. doi: 10.1073/pnas.0402625101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kobayashi S, Valentine MR, Pham P, O’Donnell M, Goodman MF. Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment. J Biol Chem. 2002;277(37):34198–34207. doi: 10.1074/jbc.M204826200. [DOI] [PubMed] [Google Scholar]
- 86.Godoy VG, Jarosz DF, Simon SM, Abyzov A, Ilyin V, Walker GC. UmuD and RecA directly modulate the mutagenic potential of the Y family DNA polymerase DinB. Mol Cell. 2007;28(6):1058–1070. doi: 10.1016/j.molcel.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sinha N, Morris C, Alberts B. Efficient in vitro replication of double-stranded DNA templates by a purified T4 bacteriophage replication system. Journal of Biological Chemistry. 1980;255(9):4290–4293. [PubMed] [Google Scholar]
- 88.Yao NY, Georgescu RE, Finkelstein J, O'Donnell ME. Single-molecule analysis reveals that the lagging strand increases replisome processivity but slows replication fork progression. Proceedings of the National Academy of Sciences. 2009;106(32):13236–13241. doi: 10.1073/pnas.0906157106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, Van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439(7076):621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 90.Georgescu RE, Kurth I, Yao NY, Stewart J, Yurieva O, O'Donnell M. Mechanism of polymerase collision release from sliding clamps on the lagging strand. The EMBO journal. 2009;28(19):2981–2991. doi: 10.1038/emboj.2009.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McInerney P, O'Donnell M. Replisome fate upon encountering a leading strand block and clearance from DNA by recombination proteins. Journal of Biological Chemistry. 2007;282(35):25903. doi: 10.1074/jbc.M703777200. [DOI] [PubMed] [Google Scholar]
- 92.Salinas F, Benkovic SJ. Characterization of bacteriophage T4-coordinated leading-and lagging-strand synthesis on a minicircle substrate. Proceedings of the National Academy of Sciences. 2000;97(13):7196. doi: 10.1073/pnas.97.13.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee J, Chastain PD, Kusakabe T, Griffith JD, Richardson CC. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Molecular cell. 1998;1(7):1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 94.Falkenberg M, Lehman I, Elias P. Leading and lagging strand DNA synthesis in vitro by a reconstituted herpes simplex virus type 1 replisome. Proceedings of the National Academy of Sciences. 2000;97(8):3896. doi: 10.1073/pnas.97.8.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Benkovic SJ, Valentine AM, Salinas F. Replisome-mediated DNA replication. Annual review of biochemistry. 2001;70(1):181–208. doi: 10.1146/annurev.biochem.70.1.181. [DOI] [PubMed] [Google Scholar]
- 96.Nelson SW, Benkovic SJ. Response of the bacteriophage T4 replisome to noncoding lesions and regression of a stalled replication fork. Journal of molecular biology. 2010;401(5):743–756. doi: 10.1016/j.jmb.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sakabe K, Okazaki R. A unique property of the replicating region of chromosomal DNA. Biochimica et biophysica acta. 1966;129(3):651. doi: 10.1016/0005-2787(66)90088-8. [DOI] [PubMed] [Google Scholar]
- 98.Pomerantz RT, O'Donnell M. The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature. 2008;456(7223):762–766. doi: 10.1038/nature07527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Iyer VN, Rupp WD. Usefulness of benzoylated naphthoylated DEAE-cellulose to distinguish and fractionate double-stranded DNA bearing different extents of single-stranded regions. Biochim Biophys Acta. 1971;228(1):117–126. doi: 10.1016/0005-2787(71)90551-x. [DOI] [PubMed] [Google Scholar]
- 100.Rudolph CJ, Upton AL, Lloyd RG. Replication fork stalling and cell cycle arrest in UV-irradiated Escherichia coli. Genes Dev. 2007;21(6):668–681. doi: 10.1101/gad.417607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krueger JH, Elledge SJ, Walker GC. Isolation and characterization of Tn5 insertion mutations in the lexA gene of Escherichia coli. Journal of Bacteriology. 1983;153(3):1368. doi: 10.1128/jb.153.3.1368-1378.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mount DW. A mutant of Escherichia coli showing constitutive expression of the lysogenic induction and error-prone DNA repair pathways. Proceedings of the National Academy of Sciences. 1977;74(1):300. doi: 10.1073/pnas.74.1.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rudolph CJ, Upton AL, Lloyd RG. Maintaining replication fork integrity in UV-irradiated Escherichia coli cells. DNA Repair (Amst) 2008;7(9):1589–1602. doi: 10.1016/j.dnarep.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 104.Courcelle J. Recs preventing wrecks. Mutat Res. 2005;577(1–2):217–227. doi: 10.1016/j.mrfmmm.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 105.Courcelle J, Donaldson JR, Chow KH, Courcelle CT. DNA damage-induced replication fork regression and processing in Escherichia coli. Science. 2003;299(5609):1064–1067. doi: 10.1126/science.1081328. [DOI] [PubMed] [Google Scholar]