Abstract
Mitotic defects leading to aneuploidy have been recognized as a hallmark of tumor cells for over 100 years. Current data indicate that ~85% of human cancers have missegregated chromosomes to become aneuploid. Some maintain a stable, aneuploid karyotype while others consistently missegregate chromosomes over multiple divisions due to Chromosomal INstability (CIN). Both aneuploidy and CIN serve as markers of poor prognosis in diverse human cancers. Despite this, aneuploidy is generally incompatible with viability during development, and some aneuploid karyotypes cause a proliferative disadvantage in somatic cells. In vivo, the intentional introduction of aneuploidy can promote tumors, suppress them, or do neither. Here, we summarize current knowledge of the effects of aneuploidy and CIN on proliferation and cell death in nontransformed cells, as well as on tumor promotion, suppression, and prognosis.
1. Introduction
Cancer is the leading cause of death worldwide [1]. It was recognized as early as the late 1800s that mitotic defects that give rise to aneuploidy, an abnormal chromosome complement that differs from a multiple of the haploid, are a common characteristic of tumor cells. Based on this, Boveri proposed in the early 1900s that aneuploid cells may initiate tumors [2]. With the discovery of oncogenes and tumor suppressors, the role of aneuploidy has become hotly debated. This review summarizes the effects of abnormal chromosome contents on cell growth and tumor progression, suppression, and prognosis.
2. Aneuploidy, polyploidy and CIN
Aneuploidy can be described as numerical or structural, depending on whether whole chromosomes or portions of chromosomes are gained or lost. Both of these are distinct from polyploidy, in which cells contain more than two complete sets of chromosomes, but always contain an exact multiple of the haploid number, so the chromosomes remain balanced.
Aneuploid cells, including near-polyploid cells, can maintain stable karyotypes or exhibit Chromosomal INstability (CIN), in which the genome continuously evolves over the course of multiple cell cycles. Like aneuploidy, CIN can involve gains and losses of whole chromosomes or of chromosomal segments, sometimes abbreviated as W-CIN and S-CIN, respectively.
2.1. State versus rate
CIN is sometimes inferred from the presence of aneuploidy. However, the observance of the state of aneuploidy at a given moment in time does not necessarily indicate the presence of CIN, which is an ongoing rate of instability. Even a tumor cell with a complex karyotype such as 86, XXXX, add(1)(p22), der(3)(p14),+der(11)t(11;20)(p12; p13),−14, −15,+16, −18, −21, −22, which could be described as “highly aneuploid,” is not necessary chromosomally unstable. The presence of these genomic alterations does not provide information about the rate of karyotypic change.
2.2. CIN causes aneuploidy; aneuploidy can cause CIN
CIN produces aneuploid progeny. However, whether or not aneuploidy causes CIN is debated. Aneuploidy can occur due to a single insult that is not repeated, resulting in chromosomal imbalances that can be stably propagated. However, it has been proposed since the late 1990s that aneuploidy destabilizes the genome by unbalancing genes required for mitosis, resulting in the autocatalytic formation of randomly generated karyotypes due to recurrent segregation errors [3]. Evidence for this comes from studies showing that transformed CHO cells with increased ploidy have an increased rate of CIN [3] and that polyploidy increases the rate of chromosome loss in yeast [4]. More recently, it has been shown that 9 of 13 Saccharomyces cerevisiae strains exhibiting aneuploidy due to the presence of a single additional chromosome missegregate yeast artificial chromosomes at rates that are 1.7–3.3 fold higher than a haploid strain. Conversely, the other 31% of strains containing an additional chromosome did not exhibit CIN [5]. Additionally, 12.5% of strains generated through meiosis of 3n or 5n yeast produced stably aneuploid offspring [6] (also see review by Rancati and Pavelka in this issue).
There are also examples of stable and unstable aneuploidy in vertebrate cells. The chromosomally stable colorectal cancer cell lines HCT116 and SW48 contain 45 chromosomes (-Y) and 47 chromosomes (+7), respectively. Gain or loss of a single chromosome did not induce CIN in these cell lines. Similarly, addition of a single extra copy of chromosome 3 into HCT116 cells does not cause chromosomal instability, as assessed by FISH of five different chromosomes. Nor does polyploidy due to fusion of two chromosomally stable cells (two HCT116 or two DLD1) result in CIN [7]. Thus, while aneuploidy can induce CIN, it does not necessarily do so.
3. Causes of aneuploidy and CIN
3.1 Mitotic checkpoint defects
Deficits in the mitotic checkpoint, also known as the spindle assembly checkpoint, result in numerical aneuploidy and W-CIN. The mitotic checkpoint is the major regulator of chromosome segregation during mitosis (reviewed in [8]). It delays separation of the replicated sister chromatids until each pair has made stable attachments to both poles of the mitotic spindle, which is necessary for accurate chromosome segregation. Each sister chromatid assembles a kinetochore, a proteinaceous structure that serves as the binding site between the chromosome and spindle microtubules, at its centromere. Mitotic checkpoint components, including Mad1, Mad2, Bub1, BubR1, Bub3 and CENP-E, are recruited to kinetochores on chromosomes that are not yet properly attached, and would be likely to missegregate if the cells entered anaphase. At unattached kinetochores, mitotic checkpoint components are converted into active inhibitors of the Anaphase Promoting Complex (APC), an E3 ubiquitin ligase that, in the context of its specificity factor Cdc20, is necessary for anaphase onset and mitotic exit. Once all the kinetochores have become stably attached to spindle microtubules, the mitotic checkpoint is satisfied and APC-Cdc20 becomes active. It then ubiquitinates Securin, which frees its binding partner, the protease Separase. Separase cleaves the cohesins that link sister chromatids, resulting in anaphase onset. In this fashion, the mitotic checkpoint ensures accurate chromosome segregation during mitosis.
Defects in the mitotic checkpoint, caused by reduction, or in some cases, overexpression of mitotic checkpoint proteins, lead to numerical aneuploidy and W-CIN. While heterozygous deletion of mitotic checkpoint genes results in viable progeny in mice, homozygous deletion is uniformly lethal [9–13]. Similarly, while partial depletion of these components in cell culture results in missegregation of small numbers of chromosomes per division (low CIN), complete depletion of Mad2 or BubR1 results in massive chromosome missegregation (high CIN) and rapid cell death, even in cancer cell lines in which p53 function is impaired (further discussed in section 4.3) [14, 15].
3.2. Merotelic attachments and abnormal spindles
Another mechanism that causes chromosome missegregation is inappropriate connections between kinetochores and spindle microtubules. Merotelic attachments, in which a single kinetochore is attached to microtubules originating from both poles, can generate chromosomes that lag behind the segregating masses of DNA during anaphase and telophase (lagging chromosomes). These frequently occur in cells that missegregate chromosomes [16]. Merotelic attachments can be caused by defects in the Aurora B dependent error correction mechanism that destabilizes improper attachments, by a reduction in microtubule dynamics, or by the focusing of multipolar spindles [17, 18]. Importantly, since the kinetochores are attached to microtubules from opposite poles, neither merotely nor multipolar spindles are detected by the mitotic checkpoint. Interestingly, recent evidence indicates that lagging chromosomes can also cause S-CIN either because they are damaged by the cytokinetic furrow [19], or because they are localized to micronuclei which are not completely replicated by the start of the next mitosis [20].
Both merotely and deficits in mitotic checkpoint signaling are thought to contribute to numerical aneuploidy and CIN in human cancers. Lagging chromosomes have been detected in the CIN cancer cell lines HT29, Caco2, MCF-7, SW480 and SW837 [21, 22], and in diffuse large B-cell lymphomas [23], while MDA-MB-231 cells enter anaphase in the presence of misaligned chromosomes, indicative of a weakened mitotic checkpoint [24].
4. Effects of aneuploidy and CIN on cellular fitness
Recently, aneuploidy has been described as uniformly causing a proliferative disadvantage. A portion of the evidence for this comes from budding yeast containing stably aneuploid karyotypes. Most aneuploid yeast strains grew more slowly than haploid yeast under conditions optimized for growth of the euploid controls [6, 25]. Interestingly, however, some aneuploid karyotypes conferred a proliferative advantage under growth conditions suboptimal for euploid yeast (reduced temperature, addition of chemotherapeutic or antifungal drugs, etc) [6] (also see review by Rancati and Pavelka in this issue).
Evidence for proliferative defects caused by chromosome gains, at least under standard growth conditions, can also be found in mammalian cells. Mouse Embryonic Fibroblasts (MEFs) containing a single extra chromosome were generated by breeding mice bearing Robertsonian translocations, resulting in MEFs containing an extra copy of the long arm of chromosomes 1, 13, 16 or 19. MEFs with an additional chromosome arm showed a decrease in growth rate. Effects on growth rate were more pronounced and less variable in cells that had gained larger chromosome arms [26]. In contrast to the situation in yeast, in which 16 of 20 aneuploid strains had a delay in G1 [25], no delay in any specific stage of the cell cycle was identified in MEFs [26].
Additionally, human skin fibroblasts containing an extra copy of chromosome 21 have long been known to proliferate more slowly than diploid skin fibroblasts under standard culture conditions (35.6 versus 23.0 hour doubling times, respectively) [27]. Consistent with this, individuals with trisomy 21/Down’s syndrome show decreased stature and head circumference when compared with the general population [28]. Thus, stable aneuploidy often, although not always, confers a growth disadvantage under standard conditions.
CIN, on the other hand, usually does not inhibit population growth, at least in mouse cells. Heterozygous deletion of the mitotic checkpoint components BubR1, Bub3, or CENP-E or the mRNA export factor Rae1 in mammalian cells all cause significant aneuploidy due to CIN without a proliferative disadvantage [9, 10, 29–31], as does overexpression of the E2 ubiquitin conjugating enzyme UbcH10 [32].
While CIN does not adversely affect population growth rates, it does appear to inhibit the colony forming ability of individual cells. CIN induced in HCT116 cells, or immortalized but non-transformed RPE1 cells, produced only transient increases in aneuploid cells in the colonies that formed [22], suggesting that the aneuploid cells were out-proliferated by the diploids. In a follow up study, cells that missegregated a marked chromosome were less able to grow into colonies than those that did not [33]. Similarly, MEFs that exhibit CIN due to haploinsufficiency of CENP-E form fewer colonies than wild type cells when plated under identical conditions [31]. Reduced colony forming ability of individual aneuploid cells could be a result of a lengthened cell cycle time or activation of checkpoint proteins such as p53 (discussed in section 4.3). Together with the population growth rates, these data suggest that some aneuploid karyotypes confer a proliferative disadvantage, while others do not.
Evidence in mammalian cells suggests that chromosome gains and losses are not equivalent with respect to their consequences on cellular survival and proliferation. This relationship does not hold true in diploid yeast [6]. However, in many examples of CIN in mammals, chromosome loss is more prevalent than chromosome gains when assaying dividing cells. A skewing towards chromosome loss has been observed in murine neurons, BRCA1 11−/− embryonic cells, MEFs from APCMin/+, Bub1 overexpressing, and one of two BubR1+/− animals, splenocytes from CENP-E+/−, Bub1−/H;p53+/−, Cdc20+/AAA and Cdc20+/AAA;p53−/− mice, and in Mad2+/− HCT116 cells [10, 29, 31, 34–38]. Although this could occur because of loss of chromosomes during analysis, it suggests the intriguing possibility that chromosome loss is less likely to cause a proliferative disadvantage than chromosome gain. This could be due to an increase in cell cycle duration or because of activation of cell cycle checkpoints, perhaps as a result of damage to the gained chromosome(s), as recently shown for lagging chromosomes [19, 20].
4.1. Aneuploidy often causes embryonic lethality
It is estimated that only 30–40% of human conceptions result in live births [39]. The majority of early pregnancy losses are attributed to chromosomal abnormalities. A significant portion of these are likely due to aneuploid gametes. Reported rates of numerical aneuploidy vary from 5–7% in sperm from healthy men but average around 26% in oocytes [40]. In addition, 29–89% of developing embryos have mosaic aneuploidy and/or polyploidy [39].
The most viable autosomal trisomies, those for chromosomes 13, 18, and 21, can produce live births, but are estimated to have prenatal survival rates of only 3%, 5% and 20%, respectively [39]. Aneuploidy affecting sex chromosomes is more common and tends to result in fewer phenotypic effects, with the exception of monosomy X, which is embryonic lethal in 98% of cases [39]. Thus, aneuploidy is usually deleterious during embryogenesis.
4.2. Aneuploidy is common in normal somatic tissues
Although aneuploidy often has significant detrimental effects during development, aneuploidy in somatic cells is surprisingly common. Remarkably, 33% of developing neuronal precursors in wild type mice exhibit numerical aneuploidy [41]. Although a portion of these cells may be eliminated, there is evidence that neuronal progenitor cells that have lost a copy of chromosome 15 bearing an eGFP transgene divide and survive at normal rates, [36] and that aneuploid neurons are functional, at least in retrograde transport [42] (also see the review by Bushman and Chun in this issue).
There is also evidence for aneuploidy in asymptomatic humans in cells of the blood, liver and brain. In peripheral blood lymphocytes, it has been recognized since the 1960s that aneuploidy occurs and increases with age, with chromosome losses seen more often than gains [43, 44]. In healthy livers, ≥ 25% of hepatocytes were found to exhibit numerical aneuploidy, again with losses more common than gains. This has been proposed to be beneficial, allowing hepatocytes with karyotype(s) that confer resistance to specific insults to clonally expand in response to liver injury [45] (also see the review by Duncan in this issue). In the brain, approximately 4% of normal postmortem cells were aneuploid for chromosome 21, as assessed by FISH. Aneuploidy was found in both neurons and glia, and monosomy was more common than trisomy in each cell type [46]. Thus, despite the negative effects of aneuploidy during development, it occurs quite commonly in asymptomatic adults.
4.3. The role of p53 in eliminating aneuploid cells
Why do some aneuploid cells have a proliferative disadvantage? Insight into this question comes from chromosomally stable HCT116 cells containing a fluorescently tagged chromosome. When induced to undergo chromosome missegregation, ≥ 75% of cells missegregating the marked chromosome accumulated nuclear p53 and its downstream activator, the CDK inhibitor p21. Importantly, p53 and p21 accumulated in the cell that had lost the marked chromosome as well as the one that had gained it. As discussed previously, aneuploidy levels transiently increase in HCT116 cells induced to missegregate chromosomes, but return to baseline after extended treatment, suggesting that the diploid cells out-proliferate the aneuploid ones. However, in p53−/− HCT116 cells, aneuploidy levels remain high, suggesting that p53 is responsible for the proliferative disadvantage [33].
The long tumor latency and low penetrance in most CIN mice (see section 5.1), despite significant percentages of aneuploid cells, suggests that only specific, complex aneuploid karyotypes result in tumors, and/or that there are factors that restrain the transforming potential of aneuploidy. Consistent with the hypothesis that p53 is one of those factors, introducing a mutation that causes chromosome missegregation into a p53 deficient background accelerates tumor onset in most cases. To date, mice deficient in Bub1, Bub3, Mad1, Mad2, Separase and Cdc20 have been crossed into p53 heterozygous or null backgrounds. Significantly shorter tumor latencies occurred in p53+/− and p53−/− animals expressing approximately 30% (Bub1H/H) or 20% (Bub1−/H) of normal levels of Bub1, [47, 48], a hypomorphic allele of Separase [49], or an allele of Cdc20 that cannot be inhibited by the mitotic checkpoint (Cdc20+/AAA) [48] (Table 1).
Table 1.
Aneuploidy and CIN can promote tumors, suppress them, or do neither.
| mutation | Effect on tumors: | Aneuploidy: mutant vs wt, % | Effect on tumors in genetic backgrounds: | ref | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| spontaneous | carcinogen-induced | splenocytes, 5mo | MEFs | p53 +/− | p53−/− | Apc Min/+ | Rb +/− | p19 ARF−/− | Pten +/− | ||
| Bub1+/H | = | 6 vs 1 | 11 vs 7 | = | = | = | [11, 47, 48] | ||||
| Bub1+/− | = | DMBA: ↑ | 16 vs 1 | 14 vs 7 | = | SI =; LI ↑ | |||||
| Bub1H/H | ↑ | 35 vs 1 | 35 vs 7 | ↑ | |||||||
| Bub1−/H | ↑a | 39 vs 1 | 36 vs 7 | ↑ | SI =; LI ↑ | = | ↓ | ||||
| Bub1Δ2–3/+ | ↑ | 23 vs 9 | [85] | ||||||||
| Bub1Δ2–3/Δ2–3 | ↑ | 76 vs 9 | |||||||||
| Bub1 aa1-331b | = | = | = | [62] | |||||||
| Bub1 OE (T85)c | ↑ | 35 vs 4d | 21 vs 11 | [37] | |||||||
| Bub1 OE (T264) | ↑ | 28 vs 4d | 25 vs 11 | ||||||||
| BubR1+/− | AOM: ↑ | SI ↓; LI ↑ | [29, 71, 72] | ||||||||
| BubR1+/− | = | 15 vs 0 | 14 vs 9 | [10, 30, 84] | |||||||
| BubR1H/H e | = | DMBA: ↑ | 36 vs 9 | = | |||||||
| Bub3+/− | = | 42 vs 35 | = | = | = | [63] | |||||
| Bub3+/− | = | DMBA: ↑ | 9 vs 0 | 19 vs 9 | [9, 30] | ||||||
| Rae1+/− | = | DMBA: ↑ | 9 vs 0 | 19 vs 9 | |||||||
| Bub3+/−; Rae1+/− | = | DMBA: ↑ | 37 vs 0 | 41 vs 9 | |||||||
| Mad1+/− | ↑ | vincristine: ↑ | ↑ | [50, 64] | |||||||
| Mad2+/− | ↑ | ↑f | [35, 50, 73] | ||||||||
| Mad1+/−;Mad2+/− | ↑g | [50, 64] | |||||||||
| Mad2 OEh | ↑ | [65, 86] | |||||||||
| CENP-E+/− | ↑ spleen & lung; ↓ liver | DMBA: ↓ | 35 vs 10 | 18 vs 8 | ↓ | [31] | |||||
| Separase+/hyp | = | ↑ | ↑ | [49] | |||||||
| Securin (Pttg)−/− | = | ↓ | [67, 68] | ||||||||
| Cdc20+/AAA | ↑ | 35 vs 6 | 28 vs 5 | ↑ | [38, 48] | ||||||
| Cdc20+/− | = | DMBA: = | 15 vs 0 | 17 vs 12 | [12] | ||||||
| Cdc20H/H | = | DMBA: = | 17 vs 0 | 23 vs 12 | |||||||
| Cdc20−/H | = | DMBA: = | 21 vs 0 | 27 vs 12 | |||||||
| Ts65Dn (Down’s model) | ↓i | ↓ | [69, 70] | ||||||||
| UbcH10 OE (T2) | ↑ | DMBA: ↑ | 6 vs 0 | 29 vs 13 | [32] | ||||||
| UbcH10 OE (T2/T2)j | ↑ | DMBA: ↑ | 19 vs 0 | 33 vs 13 | |||||||
| Hec1 OE | ↑ | 35 vs 16 | [66] | ||||||||
= indicates that the mutation has no effect on tumor formation; ↑ indicates tumor promotion; ↓ indicates tumor suppression; blank space=not reported; OE=overexpression; SI=small intestine; LI=large intestine.
Bub1−/H animals have a higher rate of CIN, but develop fewer liver tumors than Bub1H/H animals.
Bub1aa1-331 does not cause a mitotic checkpoint defect in vivo.
Bub1 overexpression promotes tumors in the Eμ-myc model.
hepatic lymphocytes.
BubR1 haploinsufficiency promotes tumors in p16INK4a−/− mice.
Mad2 heterozygosity suppresses tumors in Wap121/+ and suppresses lymphomas in p53c/c mice.
Mad1+/−;Mad2+/−;p53+/− animals develop fewer lymphomas than Mad2+/−;p53+/− mice.
Mad2 overexpression promotes tumors in Eμ-myc and Kras G12D models.
This p53+/− model is also NF1+/−.
UbcH10 T2/T2 animals express higher levels of UbcH10, but develop fewer spontaneous tumors than T2 mice.
Though reduction of Mad1 and/or Mad2 in p53+/− animals was not reported to have an effect on tumor latency, tumor incidence increased from 67% in p53+/− to 76% in Mad1+/−;p53+/− to 88% in Mad2+/−;p53+/− and to 95% in Mad1+/−;Mad2+/−;p53+/− animals. Decreasing Mad2 or both Mad1 and Mad2 in p53+/− animals also resulted in a higher number of tumors per mouse [50].
The published examples of mutations which did not affect tumor onset in animals with deficits in p53 caused relatively subtle effects on chromosome missegregation (Table 1). Thus, to date, the introduction of substantial levels of chromosome missegregation into p53+/− and p53−/− backgrounds accelerates tumor initiation and/or progression.
These data clearly implicate p53 in restricting the growth and transforming potential of aneuploid cells. However, as mentioned above, primary MEFs that exhibit aneuploidy and CIN due to a variety of genetic manipulations do not exhibit a proliferative disadvantage despite having an intact p53 pathway [9, 10, 29–31]. Similarly, the frequent occurrence of aneuploid cells in vivo suggests that p53 is not stabilized in all aneuploid cells. Support for this comes from experiments using HCT116 cells depleted of Mad2, which have elevated levels of active, phosphorylated p53 [48]. However, while 28% of HCT116 cells exhibited numerical aneuploidy after Mad2 depletion, only 5.6% of similarly depleted cells were positive for active p53 [48]. Thus, p53 is activated in some, but not all, aneuploid cells. Further work will be required to elucidate what types of aneuploidy result in a p53-mediated response.
5. Aneuploidy, polyploidy and CIN in cancer
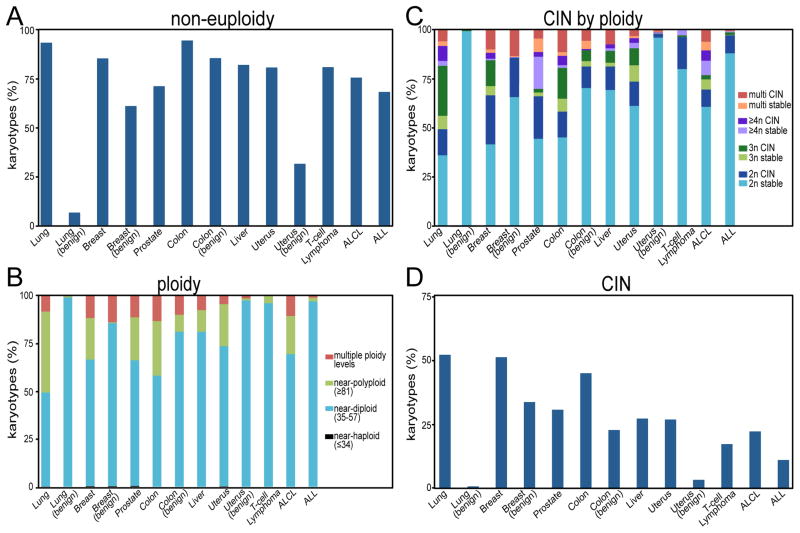
Karyotyping data available in the Mitelman database of chromosomal aberrations in cancer [51] indicate that approximately 86% of solid tumors and 72% of hematopoietic cancers have gained or lost whole chromosomes to become numerically aneuploid (Fig 1A). 26% of solid tumors and 6% of hematopoietic cancers exhibit near-polyploidy (≥58 chromosomes; Fig 1B). Most near-polyploid tumors (74%) are near triploid (58–80 chromosomes). Only 11% of solid tumors and 3% of hematopoietic cancers have a near-tetraploid or greater number of chromosomes (≥81; Fig. 1C).
Figure 1. Ploidy and CIN in human tumors.
A–D) Compiled from the Mitelman database [49]. n = 485 (lung), 172 (benign lung), 896 (breast), 228 (benign breast), 208 (prostate), 399 (colon), 145 (benign colon), 150 (liver), 290 (uterine), 506 (benign uterine), 110 (angioimmunoblastic T-cell lymphoma), 135 (anaplastic large cell lymphoma; ALCL), 400 (acute lymphoblastic leukemia/lymphoblastic lymphoma; ALL). A) Non-euploidy in human tumors, defined as any numerical deviation from a multiple of 23. Non-euploidy also includes pseudoeuploidy (i.e. 46, XY,−5,+9). B) Ploidy in human tumors, according to ISCN definitions. Multiple ploidy indicates a tumor with multiple clones from more than one ploidy level. C-D) CIN, defined as tumors having a range of chromosome numbers ≥ 2 (i.e. 47–49 chromosomes). C) ClN and chromosomally stable tumors classified by ploidy level. D) CIN by tumor site.
To estimate CIN in human tumors, we used intercellular variability in chromosome number in the Mitelman database. For this analysis, tumors with a range of chromosome numbers ≥ 2 were considered CIN (i.e. 47–49 chromosomes), while tumors with a single chromosome number or a range of 1 were considered stable (i.e. 48–49 chromosomes). Based on this, 56% of solid tumors had a stable karyotype, while 44% exhibited CIN. 86% of hematopoietic tumors were stable, and only 14% were CIN (Fig. 1C-D). These values are consistent with the range of CIN detected by other methods [52–54].
Although genomic instability is commonly regarded as an enabling characteristic in tumor development, and aneuploidy can lead to CIN in some circumstances, at least half of human tumors are karyotypically stable. Since microsatellite instability occurs in only a subset of tumors (e.g. ~15% of colorectal cancers), this suggests that a significant fraction of cancers have relatively stable genomes. This is consistent with what has been shown for cancer cell lines. Despite ongoing structural and numerical chromosomal instability, HCT116 and other established cancer cell lines maintain a stable consensus karyotype [22, 33, 55]. This suggests these genomes have been optimized for cell growth under specific culture conditions. A similar genomic optimization for the tumor site and microenvironment may explain the relative stability of a large number of cancers.
5.1. Tumor initiation, progression and suppression
Many attempts have been made to test Boveri’s hypothesis that aneuploidy drives tumorigenesis, first with aneuploidy-inducing drugs and later with genetically engineered mouse models. The challenge lies in generating aneuploidy without additional, confounding, defects. The majority of aneuploidy-inducing drugs are also mutagens. Most genes required for chromosome segregation are expressed throughout the cell cycle and participate in additional cellular functions, including some that would be expected to influence tumor outcomes. For instance, Bub1, BubR1 and Bub3 have been implicated in cell death [11, 56, 57]. Bub1 and BubR1 participate in the DNA damage response [58, 59]. Mad2, Bub3 and the budding yeast homolog of BubR1, Mad3, contribute to gross chromosomal rearrangements [60]. A large number of animal models that exhibit aneuploidy and/or CIN have now been tested for their tumor forming ability. The results have shown that aneuploidy and CIN can promote tumors, suppress them, or do neither, depending on the context and the specific genes involved.
Mutations that cause aneuploidy fall into five categories with respect to their tumor phenotypes. Tumor incidence can be: 1) unaffected; 2) increased; 3) decreased; 4) unaffected for spontaneous tumors but increased in response to carcinogens and/or in a tumor prone background; and 5) increased in some contexts and decreased in others (Table 1). Mutations which cause aneuploidy and CIN but do not affect tumor incidence under any conditions tested include those that cause inactivation of Bub1 kinase activity [61], Cdc20 hypomorphism [12], a 25% reduction in protein levels of Bub1 (Bub1+/H) [11, 47, 48], a dominant negative fragment of Bub1 [62], and one of two models of Bub3+/− mice [63]. Animals that reliably promote tumors include Mad1+/− [50, 64] and Cdc20+/AAA mice [38, 48] as well as those that overexpress Bub1 [37], Mad2 [65], UbcH10 [32] or Hec1 [66]. Mutations that suppress tumors when they have an effect include Securin−/− [67, 68] and Ts65Dn animals, a model of Down’s syndrome [69, 70]. Animals that promote induced but not spontaneous tumors include Bub1+/− [11, 47, 48], BubR1H/H [10, 30], Rae1+/−, Bub3+/−;Rae1+/−, the second model of Bub3+/− [9, 30], and Separase+/H animals [49]. Mutations that both promote and suppress tumors depending on the context include Bub1−/H [11, 47, 48], BubR1+/− [29, 71, 72], Mad2+/− [35, 50, 73] and CENP-E+/− animals [31]. Thus, the mere presence of aneuploidy and/or CIN is insufficient to predict the effect on tumor phenotype.
5.2. Patient prognosis
Since the 1970s, efforts have been made to link aneuploidy with patient prognosis. Early attempts relied primarily on flow cytometry of DNA content. Subsequently, DNA content has been measured using image cytometry. SNP microarrays and comparative genomic hybridization are now also used to measure genome wide copy number changes.
Recently, estimates of CIN have been made based on cell-to-cell variability using interphase FISH, nuclear diameter, or nuclear area. In rare cases of tumors with high mitotic indices, CIN can be inferred from the number of abnormal anaphase and telophase figures, as performed in diffuse large B-cell lymphoma [23]. This is one of the most direct measures of CIN available but, unfortunately, is not possible in most tumors due to a low mitotic index. Recently, gene expression data from near diploid/low CIN tumors or tumor cell lines has been compared with data from aneuploid/high CIN tumors to develop gene expression signatures indicative of aneuploidy and/or CIN.
It should be noted that most measures of aneuploidy and CIN in human tumors also reflect proliferative index. For instance, DNA content analysis frequently classifies tumors as aneuploid due to the presence of anything more than a single G0/G1 peak of DNA, including peaks that correspond to S and G2/M cells [74]. Similarly, a Stemline Scatter Index (SSI), derived from image cytometry data, has been used as a measure of ploidy and genomic stability for prognostic testing, and for the generation of gene expression signatures. SSI accounts for the percentage of cells in S phase, the percentage of cells with DNA content exceeding the G2 value, and the coefficient of variation. However, S phase percentage contributes most to SSI [75]. Gene expression signatures also frequently reflect proliferation as well as aneuploidy and CIN. The CIN70 and CIN25 signatures that have been found to be prognostic indicators in a variety of human tumors (Table 2) reflect a measure known as “total functional aneuploidy,” which reflects structural aneuploidy and DNA copy number profiles in the NCI60 cell lines [76]. ≥ 50% of CIN70 and CIN25 genes were identified as cell cycle regulated in a gene expression study in synchronized HeLa cells [77], although this gene signature remained prognostic even after removal of the cell cycle regulated cohort [76].
Table 2.
Prognostic value of aneuploidy, polyploidy and CIN.
| method of determining ploidy/CIN | method actually reflects | tumor type | n | type of survival | __ year survival | survival rate for: | ref | |
|---|---|---|---|---|---|---|---|---|
| stable diploid | altered ploidy/ CIN | |||||||
| DNA content (aneuploidy and polyploidy) | ||||||||
| DNA image cytometry- SSIa | proliferative index + heterogeneity | breast | 154 | overall | 2–3 | 98% | 74% | [75] |
| 890 | overall | 10 | ~90% | ~65% | [54] | |||
| flow cytometry | # and location of all DNA peaks | breast | 1391 | relapse-free | 10 | ~80–95% | ~60–75% | [87] |
| not specified | STSb | 102 | metastasis-free | 5 | 77% | 48% | [88] | |
| multiple stemlinesc | 46 | overall | 15 | ~55% | ~20% | [89] | ||
| nuclear grading | nuclear aread | lunge | 133 | recurrence-free | 5 | 90% | 58% | [90] |
| largest nuclear diameterd | 133 | recurrence-free | 5 | 89% | 62% | |||
| genomic index | segmental gains & losses by CHG/# chromosomes involved | STSb (GIST)f | 60 | metastasis-free | 5 | 93% | 16% | [78] |
| FISH (cellular heterogeneity or CIN) | ||||||||
| FISH (centromeres for Chr 3, 10, 11 & 17) | heterogeneity of 4 chromosomes | lunge | 50 | overall | 4 | ~75% | ~35% | [91] |
| FISH (EGFR-7p12, MYC-8q24, 5p15, 6cen, p16–9p21 | heterogeneity of ≥ 4 chromosomes | 47 | overall | 5 | 77% | 33% | [92] | |
| heterogeneity of ≥ 3 chromosomes | 63 | overall | 5 | 94% | 69% | [53] | ||
| abnormal anaphases (lagging chromosomes and chromatin bridges) | ||||||||
| lagging/bridge chromosomes | chromosome missegregation | lymphomag | 54 | overall | 10 | ~65 | ~45 | [23] |
| gene signatures | ||||||||
| CIN70 and CIN25 gene signaturesh | structural aneuploidy and proliferation | 6 typesi | 1944 | overall (breast cohort) | 10 | ~70 | ~55 | [76] |
| 12 gene genomic instability signaturej | proliferative index/SSIa | breast | 469 | overall | 5 | ~70–90% | ~40–60% | [52] |
| metastasis-free | ~60–80% | ~30–50% | ||||||
| lunge | 637 | overall | 5 | ~50–60% | ~10–40% | [93] | ||
| ovarian | 124 | relapse-free | 10 | ~65% | ~15% | |||
| 67 gene CINSARC signaturek | combination of structural aneuploidy, proliferation and tumor grade | STSb | 127 | metastasis-free | 5 | 75–84% | 35–48% | [94] |
| STSb (GIST)f | 32 | metastasis-free | 5 | 100% | 61% | |||
| 10 | 100% | 30% | ||||||
| breast | 373 | metastasis-free | 10 | ~75–80% | ~35–60% | |||
| lymphomag | 278 | overall | 10 | ~65–75% | ~35–40% | |||
| Aurora A expression | AURKA mRNA level | STSb (GIST)f | 60 | metastasis-free | 5 | 100% | 38% | [78] |
| 100% | 12% | |||||||
| 112 CIN gene signaturel | Loss of heterozygosity | colon | 548 | disease-free | 5 | ~95% | ~75% | [95] |
Stemline Scatter Index (SSI) is the percentage of cells in S phase + the percentage of cells > 5c + coefficient of variation (standard deviation/mean). S
phase percentage contributes most to SSI.
Soft tissue sarcoma.
”Tumors were considered aneuploid when a distinct separate second or more G0/G1
peak(s) was present”.
based on H&E staining.
Non-Small Cell Lung Cancer.
Gastrointestinal Stromal Tumor.
Diffuse large B-cell lymphoma.
To identify the CIN70 and CIN25 gene signatures, the authors inferred aneuploidy based on gene expression data. They then confirmed that in NCI60 cell lines, this correlates with structural aneuploidy as measured by SKY and SNPchip DNA copy number. Tumors with low versus high aneuploidy were compared to identify the 25 or 70 genes with the highest CIN score (designated CIN25 and CIN70). 43/70=61% of CIN70 genes are cell cycle regulated [77].
The CIN25 signature is prognostic in 1 lung, 5 breast, 1 mesothelioma, 3 glioma, 1 medulloblastoma and 1 lymphoma cohort, but not in 3 lung, 1 ovarian, 1 lymphoma and 1 prostate cohort.
The 12 gene signature was identified by analyzing gene expression data from 48 primary breast carcinomas stratified into 17 diploid genomically stable, 15 aneuploid genomically stable, and 16 aneuploid genomically unstable tumors by SSI. Only Aurora A overlaps with the CIN70 genes.
CINSARC (Complexity INdex in SARComa) genes were identified by comparing expression differences between 1) tumors with CGH imbalances of <20 with >35; 2) grade 3 vs grade 2 tumors; 3) CIN70 signature.
The 112 CIN gene signature was identified by determining the LOH status in 745 tumors using 9 microsatellite markers representing 4 chromosome arms. An LOH ratio of 33% was used to separate CIN high from CIN low tumors. 25 CIN high and 10 CIN low tumors were used to identify the 112 gene signature. These 112 genes are non-overlapping with the CIN70.
These analyses have been tested for prognostic value on a wide range of tumors (Table 2). Though there is variability, each type of assay has shown differences in outcome for stable diploid versus aneuploid, polyploid, or CIN tumors. In some cases, the difference is quite stark. For instance, the 5 year metastasis-free survival rate of soft tissue sarcoma patients was 16% for tumors with altered ploidy versus 96% for stable diploid tumors [78]. In others, the difference is less pronounced. Overall, aneuploidy and CIN are markers of poor prognosis in numerous tumor types, including lung, breast, and colon cancers, as well as lymphomas and soft tissue sarcomas (Table 2).
5.3. High CIN and tumor suppression
Interestingly, patients with tumors having the highest rate of CIN, defined as the highest quartile of CIN70 scores, have improved outcome relative to patients whose tumors have CIN70 scores in the third quartile. High CIN was associated with improved prognosis in lung, breast, ovarian, and gastric cancers [79]. Similarly, when FISH analysis was used to measure CIN, ER negative breast cancer patients with the highest CIN score had improved outcomes compared to all other groups. A similar relationship was found for patients with lung, ovarian and gastric tumors [80].
These findings in human cancers are reminiscent of experiments in murine models, in which an intermediate rate of CIN promotes tumors, but further increasing CIN leads to tumor suppression. For instance, ~28% of Bub1H/H mice develop hepatocellular carcinomas, but this drops to ~18%, near wild type levels, in Bub1−/H mice, which express lower levels of Bub1 and have a higher rate of CIN [11]. Similarly, while an intermediate level of overexpression of UbcH10 causes an increase in spontaneous tumors, further overexpression leads to increased CIN and decreased tumor formation [32]. Likewise, reducing Mad1 in Mad2+/−;p53+/− animals decreases the incidence of lymphomas [50]. Since high rates of chromosome missegregation lead to rapid cell death [14, 15], these data suggest that there is an optimal rate of CIN for promoting tumors and that further elevating this rate leads to cell death and tumor suppression.
5.4. Response to chemotherapy
Aneuploidy and CIN are generally associated with resistance to chemotherapy, since specific combinations of altered gene dosages can often rescue growth in the presence of drug concentrations that are cytotoxic to diploid cells. There is evidence for both spontaneous and acquired drug resistance in aneuploid cells. CIN cells can acquire drug resistance, even in the absence of multidrug efflux pumps, presumably due to chromosomal reassortments. MEFs lacking the multidrug resistance genes Mdr1a, Mdr1b and Mrp1 acquired resistance to puromycin after exposure to increasing concentrations of the drug. Puromycin-selected cells showed a multidrug resistance phenotype, as they also had reduced sensitivity to cytosine arabinoside and colcemid [81]. Spontaneous drug resistance has been observed in a panel of 18 CIN colorectal cancer cell lines that were more resistant to a library of kinase inhibitors and cytotoxic compounds than 9 near-diploid colorectal cancer lines. Similarly, induction of CIN in HCT116 cells by heterozygous loss of the mitotic checkpoint gene Mad2 results in decreased sensitivity to the cytotoxic anticancer library [82]. The available data support the hypothesis that abnormal karyotypes can confer growth advantages in the presence of a variety of chemical insults, and that low CIN offers cells the opportunity to select for karyotypes that are optimal under specific growth conditions. Resistance to therapy is likely to explain, at least in part, why aneuploidy and CIN serve as markers of poor prognosis.
The effect of aneuploidy and CIN on response to chemotherapy that specifically targets mitotic cells remains controversial. Evidence in cultured cells has indicated that CIN cells are more, less, and equally sensitive to anti-mitotic drugs (reviewed in [83]). As with tumorigenesis, the effects of a given CIN-inducing mutation may depend on the specific gene involved and the role(s), if any, it plays outside of chromosome segregation. The limited amount of in vivo data in humans suggests that CIN confers resistance to anti-mitotic drugs. This is based on gene expression data, which indicates that most CIN70 genes are overexpressed in taxol-resistant, carboplatin-sensitive ovarian tumors, while a low median CIN70 score was associated with taxol sensitivity [77]. Consistent with this, we have recently found that overexpression of Mad1, which causes a low rate of numerical CIN and is common in breast cancer, also confers resistance to concentrations of the chemotherapeutic microtubule poisons taxol and vinblastine that cause mitotic arrest [24]. Together, these data suggest that CIN and/or genes that generate CIN have the potential to be used as predictive biomarkers for response to chemotherapy.
6. Conclusions
The term aneuploidy describes a nearly infinite set of karyotypes. Unsurprisingly, these karyotypic alterations have differing effects. During development, most aneuploidy results in embryonic lethality. However, in somatic cells, aneuploidy is remarkably common in asymptomatic individuals. Intentionally introducing aneuploidy, polyploidy, and/or CIN into experimental models results in a p53-dependent proliferative disadvantage in some, but not all, cases. In mouse models, aneuploidy and CIN can promote or suppress tumors depending on the context and the specific mutations involved. Interestingly, data from mouse models and human patients suggests that while low and intermediate rates of CIN can promote tumors, high CIN causes tumor suppression and correlates with improved patient outcomes. Additional studies will be required to fully understand the ramifications of aneuploidy and CIN in tumors in order to exploit these conditions therapeutically.
Highlights.
Aneuploidy is detrimental during development, but is common in asymptomatic adults
Aneuploidy and polyploidy occur frequently in tumors
Aneuploidy and CIN can promote or suppress tumors, depending on the context
Aneuploidy and CIN are used as markers of poor prognosis in a variety of cancers
Acknowledgments
We would like to thank our colleagues for helpful discussions and suggestions. Work in the Weaver lab is supported in part by National Institutes of Health Grant 1R01CA140458. Support also provided from T32 GM008688 (E.B. and L.Z.) and T32 CA009135 (E.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.JF, HRS, FB, DF, CM, DMP . IARC CancerBase No 10. Lyon, France: GLOBOCAN; 2008. Cancer Incidence and Mortality Worldwide. [Google Scholar]
- 2.Boveri T. Gustav Fischer, Jena; English translation The Origin of Malignant Tumors by Boveri. M Williams and Wilkins; Baltimore: 1929. Zur Frage der Entstehung maligner Tumoren. 1914. [Google Scholar]
- 3.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci U S A. 1998;95:13692–7. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer VW, Aguilera A. High levels of chromosome instability in polyploids of Saccharomyces cerevisiae. Mutat Res. 1990;231:177–86. doi: 10.1016/0027-5107(90)90024-x. [DOI] [PubMed] [Google Scholar]
- 5.Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, et al. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–30. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–5. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 8.Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–85. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- 9.Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–53. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–9. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 11.Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–67. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malureanu L, Jeganathan KB, Jin F, Baker DJ, van Ree JH, Gullon O, et al. Cdc20 hypomorphic mice fail to counteract de novo synthesis of cyclin B1 in mitosis. J Cell Biol. 2010;191:313–29. doi: 10.1083/jcb.201003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwanaga Y, Kasai T, Kibler K, Jeang KT. Characterization of regions in hsMAD1 needed for binding hsMAD2. A polymorphic change in an hsMAD1 leucine zipper affects MAD1-MAD2 interaction and spindle checkpoint function. J Biol Chem. 2002;277:31005–13. doi: 10.1074/jbc.M110666200. [DOI] [PubMed] [Google Scholar]
- 14.Michel L, Diaz-Rodriguez E, Narayan G, Hernando E, Murty VV, Benezra R. Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc Natl Acad Sci U S A. 2004;101:4459–64. doi: 10.1073/pnas.0306069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci U S A. 2004;101:8699–704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson SL, Compton DA. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc Natl Acad Sci U S A. 2011;108:17974–8. doi: 10.1073/pnas.1109720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009 doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–8. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 20.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–8. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–22. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–72. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakhoum SF, Danilova OV, Kaur P, Levy NB, Compton DA. Chromosomal instability substantiates poor prognosis in patients with diffuse large B-cell lymphoma. Clin Cancer Res. 2011;17:7704–11. doi: 10.1158/1078-0432.CCR-11-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan SD, Britigan EM, Zasadil LM, Witte K, Audhya A, Roopra A, et al. Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons. Proc Natl Acad Sci U S A. 2012;109:E2205–14. doi: 10.1073/pnas.1201911109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–24. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 26.Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–9. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segal DJ, McCoy EE. Studies on Down’s syndrome in tissue culture. I. Growth rates and protein contents of fibroblast cultures. J Cell Physiol. 1974;83:85–90. doi: 10.1002/jcp.1040830112. [DOI] [PubMed] [Google Scholar]
- 28.Van Gameren-Oosterom HB, Van Dommelen P, Oudesluys-Murphy AM, Buitendijk SE, Van Buuren S, Van Wouwe JP. Healthy growth in children with Down syndrome. PLoS One. 2012;7:e31079. doi: 10.1371/journal.pone.0031079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao CV, Yang YM, Swamy MV, Liu T, Fang Y, Mahmood R, et al. Colonic tumorigenesis in BubR1+/−ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci U S A. 2005;102:4365–70. doi: 10.1073/pnas.0407822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker DJ, Jeganathan KB, Malureanu L, Perez-Terzic C, Terzic A, van Deursen JM. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172:529–40. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 32.van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188:83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–81. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen SX, Weaver Z, Xu X, Li C, Weinstein M, Chen L, et al. A targeted disruption of the murine Brca1 gene causes gamma-irradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115–24. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- 35.Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–9. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 36.Kaushal D, Contos JJ, Treuner K, Yang AH, Kingsbury MA, Rehen SK, et al. Alteration of gene expression by chromosome loss in the postnatal mouse brain. J Neurosci. 2003;23:5599–606. doi: 10.1523/JNEUROSCI.23-13-05599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ricke RM, Jeganathan KB, van Deursen JM. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol. 2011;193:1049–64. doi: 10.1083/jcb.201012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li M, Fang X, Wei Z, York JP, Zhang P. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol. 2009;185:983–94. doi: 10.1083/jcb.200904020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macklon NS, Geraedts JP, Fauser BC. Conception to ongoing pregnancy: the ‘black box’ of early pregnancy loss. Hum Reprod Update. 2002;8:333–43. doi: 10.1093/humupd/8.4.333. [DOI] [PubMed] [Google Scholar]
- 40.Plachot M. Chromosomal abnormalities in oocytes. Mol Cell Endocrinol. 2001;183 (Suppl 1):S59–63. doi: 10.1016/s0303-7207(01)00577-9. [DOI] [PubMed] [Google Scholar]
- 41.Rehen SK, McConnell MJ, Kaushal D, Kingsbury MA, Yang AH, Chun J. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc Natl Acad Sci U S A. 2001;98:13361–6. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kingsbury MA, Friedman B, McConnell MJ, Rehen SK, Yang AH, Kaushal D, et al. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc Natl Acad Sci U S A. 2005;102:6143–7. doi: 10.1073/pnas.0408171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobs PA, Brunton M, Court Brown WM, Doll R, Goldstein H. Change of human chromosome count distribution with age: evidence for a sex differences. Nature. 1963;197:1080–1. doi: 10.1038/1971080a0. [DOI] [PubMed] [Google Scholar]
- 44.Nowinski GP, Van Dyke DL, Tilley BC, Jacobsen G, Babu VR, Worsham MJ, et al. The frequency of aneuploidy in cultured lymphocytes is correlated with age and gender but not with reproductive history. Am J Hum Genet. 1990;46:1101–11. [PMC free article] [PubMed] [Google Scholar]
- 45.Duncan AW, Hanlon Newell AE, Smith L, Wilson EM, Olson SB, Thayer MJ, et al. Frequent aneuploidy among normal human hepatocytes. Gastroenterology. 2012;142:25–8. doi: 10.1053/j.gastro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–80. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–86. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li M, Fang X, Baker DJ, Guo L, Gao X, Wei Z, et al. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:14188–93. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukherjee M, Ge G, Zhang N, Huang E, Nakamura LV, Minor M, et al. Separase loss of function cooperates with the loss of p53 in the initiation and progression of T- and B-cell lymphoma, leukemia and aneuploidy in mice. PLoS One. 2011;6:e22167. doi: 10.1371/journal.pone.0022167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chi YH, Ward JM, Cheng LI, Yasunaga J, Jeang KT. Spindle assembly checkpoint and p53 deficiencies cooperate for tumorigenesis in mice. Int J Cancer. 2009;124:1483–9. doi: 10.1002/ijc.24094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitelman F, Johansson B, Mertens F. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2010 http://cgapncinihgov/Chromosomes/Mitelman.
- 52.Habermann JK, Doering J, Hautaniemi S, Roblick UJ, Bündgen NK, Nicorici D, et al. The gene expression signature of genomic instability in breast cancer is an independent predictor of clinical outcome. Int J Cancer. 2009;124:1552–64. doi: 10.1002/ijc.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi CM, Seo KW, Jang SJ, Oh YM, Shim TS, Kim WS, et al. Chromosomal instability is a risk factor for poor prognosis of adenocarcinoma of the lung: Fluorescence in situ hybridization analysis of paraffin-embedded tissue from Korean patients. Lung Cancer. 2009;64:66–70. doi: 10.1016/j.lungcan.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 54.Kronenwett U, Ploner A, Zetterberg A, Bergh J, Hall P, Auer G, et al. Genomic instability and prognosis in breast carcinomas. Cancer Epidemiol Biomarkers Prev. 2006;15:1630–5. doi: 10.1158/1055-9965.EPI-06-0080. [DOI] [PubMed] [Google Scholar]
- 55.Roschke AV, Stover K, Tonon G, Schäffer AA, Kirsch IR. Stable karyotypes in epithelial cancer cell lines despite high rates of ongoing structural and numerical chromosomal instability. Neoplasia. 2002;4:19–31. doi: 10.1038/sj.neo.7900197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niikura Y, Dixit A, Scott R, Perkins G, Kitagawa K. BUB1 mediation of caspase-independent mitotic death determines cell fate. J Cell Biol. 2007;178:283–96. doi: 10.1083/jcb.200702134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niikura Y, Ogi H, Kikuchi K, Kitagawa K. BUB3 that dissociates from BUB1 activates caspase-independent mitotic death (CIMD) Cell Death Differ. 2010;17:1011–24. doi: 10.1038/cdd.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang Y, Liu T, Wang X, Yang YM, Deng H, Kunicki J, et al. BubR1 is involved in regulation of DNA damage responses. Oncogene. 2006 doi: 10.1038/sj.onc.1209392. [DOI] [PubMed] [Google Scholar]
- 59.Yang C, Wang H, Xu Y, Brinkman KL, Ishiyama H, Wong ST, et al. The kinetochore protein Bub1 participates in the DNA damage response. DNA Repair (Amst) 2012;11:185–91. doi: 10.1016/j.dnarep.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Myung K, Smith S, Kolodner RD. Mitotic checkpoint function in the formation of gross chromosomal rearrangements in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:15980–5. doi: 10.1073/pnas.0407010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ricke RM, Jeganathan KB, Malureanu L, Harrison AM, van Deursen JM. Bub1 kinase activity drives error correction and mitotic checkpoint control but not tumor suppression. J Cell Biol. 2012;199:931–49. doi: 10.1083/jcb.201205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cowley DO, Muse GW, Van Dyke T. A dominant interfering Bub1 mutant is insufficient to induce or alter thymic tumorigenesis in vivo, even in a sensitized genetic background. Mol Cell Biol. 2005;25:7796–802. doi: 10.1128/MCB.25.17.7796-7802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalitsis P, Fowler KJ, Griffiths B, Earle E, Chow CW, Jamsen K, et al. Increased chromosome instability but not cancer predisposition in haploinsufficient Bub3 mice. Genes Chromosomes Cancer. 2005 doi: 10.1002/gcc.20215. [DOI] [PubMed] [Google Scholar]
- 64.Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, Peloponese JM, Jr, et al. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–6. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- 65.Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–40. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diaz-Rodríguez E, Sotillo R, Schvartzman JM, Benezra R. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci U S A. 2008;105:16719–24. doi: 10.1073/pnas.0803504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chesnokova V, Kovacs K, Castro AV, Zonis S, Melmed S. Pituitary hypoplasia in Pttg−/− mice is protective for Rb+/− pituitary tumorigenesis. Mol Endocrinol. 2005;19:2371–9. doi: 10.1210/me.2005-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z, Yu R, Melmed S. Mice lacking pituitary tumor transforming gene show testicular and splenic hypoplasia, thymic hyperplasia, thrombocytopenia, aberrant cell cycle progression, and premature centromere division. Mol Endocrinol. 2001;15:1870–9. doi: 10.1210/mend.15.11.0729. [DOI] [PubMed] [Google Scholar]
- 69.Sussan TE, Yang A, Li F, Ostrowski MC, Reeves RH. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down’s syndrome. Nature. 2008;451:73–5. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- 70.Yang A, Reeves RH. Increased survival following tumorigenesis in Ts65Dn mice that model Down syndrome. Cancer Res. 2011;71:3573–81. doi: 10.1158/0008-5472.CAN-10-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dai W, Wang Q, Liu T, Swamy M, Fang Y, Xie S, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–5. doi: 10.1158/0008-5472.can-03-3119. [DOI] [PubMed] [Google Scholar]
- 72.Wang Q, Liu T, Fang Y, Xie S, Huang X, Mahmood R, et al. BUBR1 deficiency results in abnormal megakaryopoiesis. Blood. 2004;103:1278–85. doi: 10.1182/blood-2003-06-2158. [DOI] [PubMed] [Google Scholar]
- 73.Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell. 2011;19:701–14. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut. 2008;57:941–50. doi: 10.1136/gut.2007.135004. [DOI] [PubMed] [Google Scholar]
- 75.Kronenwett U, Huwendiek S, Ostring C, Portwood N, Roblick UJ, Pawitan Y, et al. Improved grading of breast adenocarcinomas based on genomic instability. Cancer Res. 2004;64:904–9. doi: 10.1158/0008-5472.can-03-2451. [DOI] [PubMed] [Google Scholar]
- 76.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 77.Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci U S A. 2009;106:8671–6. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lagarde P, Pérot G, Kauffmann A, Brulard C, Dapremont V, Hostein I, et al. Mitotic checkpoints and chromosome instability are strong predictors of clinical outcome in gastrointestinal stromal tumors. Clin Cancer Res. 2012;18:826–38. doi: 10.1158/1078-0432.CCR-11-1610. [DOI] [PubMed] [Google Scholar]
- 79.Birkbak NJ, Eklund AC, Li Q, McClelland SE, Endesfelder D, Tan P, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71:3447–52. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roylance R, Endesfelder D, Gorman P, Burrell RA, Sander J, Tomlinson I, et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:2183–94. doi: 10.1158/1055-9965.EPI-11-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Duesberg P, Stindl R, Hehlmann R. Origin of multidrug resistance in cells with and without multidrug resistance genes: chromosome reassortments catalyzed by aneuploidy. Proc Natl Acad Sci U S A. 2001;98:11283–8. doi: 10.1073/pnas.201398998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee AJ, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 2011;71:1858–70. doi: 10.1158/0008-5472.CAN-10-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamada HY, Gorbsky GJ. Spindle checkpoint function and cellular sensitivity to antimitotic drugs. Mol Cancer Ther. 2006;5:2963–9. doi: 10.1158/1535-7163.MCT-06-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baker DJ, Perez-Terzic C, Jin F, Pitel KS, Pitel K, Niederländer NJ, et al. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat Cell Biol. 2008;10:825–36. doi: 10.1038/ncb1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schliekelman M, Cowley DO, O’Quinn R, Oliver TG, Lu L, Salmon ED, et al. Impaired Bub1 function in vivo compromises tension-dependent checkpoint function leading to aneuploidy and tumorigenesis. Cancer Res. 2009;69:45–54. doi: 10.1158/0008-5472.CAN-07-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bagwell CB, Clark GM, Spyratos F, Chassevent A, Bendahl PO, Stål O, et al. Optimizing flow cytometric DNA ploidy and S-phase fraction as independent prognostic markers for node-negative breast cancer specimens. Cytometry. 2001;46:121–35. doi: 10.1002/cyto.1097. [DOI] [PubMed] [Google Scholar]
- 88.Bauer HC, Kreicbergs A, Tribukait B. DNA content prognostic in soft tissue sarcoma. 102 patients followed for 1–10 years. Acta Orthop Scand. 1991;62:187–94. doi: 10.3109/17453679108993591. [DOI] [PubMed] [Google Scholar]
- 89.el-Naggar AK, Ayala AG, Abdul-Karim FW, McLemore D, Ballance WW, Garnsey L, et al. Synovial sarcoma. A DNA flow cytometric study. Cancer. 1990;65:2295–300. doi: 10.1002/1097-0142(19900515)65:10<2295::aid-cncr2820651022>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 90.Nakazato Y, Minami Y, Kobayashi H, Satomi K, Anami Y, Tsuta K, et al. Nuclear grading of primary pulmonary adenocarcinomas: correlation between nuclear size and prognosis. Cancer. 2010;116:2011–9. doi: 10.1002/cncr.24948. [DOI] [PubMed] [Google Scholar]
- 91.Nakamura H, Saji H, Idiris A, Kawasaki N, Hosaka M, Ogata A, et al. Chromosomal instability detected by fluorescence in situ hybridization in surgical specimens of non-small cell lung cancer is associated with poor survival. Clin Cancer Res. 2003;9:2294–9. [PubMed] [Google Scholar]
- 92.Yoo JW, Seo KW, Jang SJ, Oh YM, Shim TS, Kim WS, et al. The relationship between the presence of chromosomal instability and prognosis of squamous cell carcinoma of the lung: fluorescence in situ hybridization analysis of paraffin-embedded tissue from 47 Korean patients. J Korean Med Sci. 2010;25:863–7. doi: 10.3346/jkms.2010.25.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mettu RK, Wan YW, Habermann JK, Ried T, Guo NL. A 12-gene genomic instability signature predicts clinical outcomes in multiple cancer types. Int J Biol Markers. 2010;25:219–28. doi: 10.5301/jbm.2010.6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chibon F, Lagarde P, Salas S, Pérot G, Brouste V, Tirode F, et al. Validated prediction of clinical outcome in sarcomas and multiple types of cancer on the basis of a gene expression signature related to genome complexity. Nat Med. 2010;16:781–7. doi: 10.1038/nm.2174. [DOI] [PubMed] [Google Scholar]
- 95.Watanabe T, Kobunai T, Yamamoto Y, Matsuda K, Ishihara S, Nozawa K, et al. Chromosomal instability (CIN) phenotype, CIN high or CIN low, predicts survival for colorectal cancer. J Clin Oncol. 2012;30:2256–64. doi: 10.1200/JCO.2011.38.6490. [DOI] [PubMed] [Google Scholar]