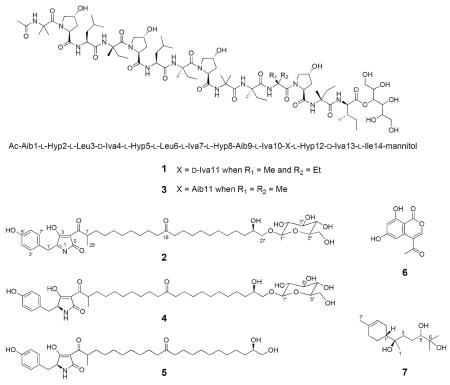
Abstract
An extract of the filamentous fungus Bionectria sp. (MSX 47401) showed both promising cytotoxic activity (>90% inhibition of H460 cell growth at 20 μg/mL) and antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA). A bioactivity-directed fractionation study yielded one new peptaibol (1) and one new tetramic acid derivative (2), and the fungus biosynthesized diverse secondary metabolites with mannose derived units. Five known compounds were also isolated: clonostachin (3), virgineone (4), virgineone aglycone (5), AGI-7 (6), and 5,6-dihydroxybisabolol (7). Compounds 5 and 7 have not been described previously from natural sources. Compound 1 represents the second member of the peptaibol structural class that contains an ester-linked sugar alcohol (mannitol) instead of an amide-linked amino alcohol, and peptaibols and tetramic acid derivatives have not been isolated previously from the same fungus. The structures of the new compounds were elucidated primarily by high field NMR (950 and 700 MHz), HRESIMS/MS, and chemical degradations (Marfey's analysis). All compounds (except 6) were examined for antibacterial and antifungal activities. Compounds 2, 4, and 5 showed antimicrobial activity against S. aureus and several MRSA isolates.
Fungi are a rich source of biologically active secondary metabolites that often possess unique structures. Estimates on the number of fungal species range between 1.5 M1 to 5 M,2 but fewer than 100,000 have been described taxonomically, and an even smaller number have been studied for pharmaceutical leads. The Dictionary of Natural Products3 contains over 246,000 compounds, of which approximately 14,260 (or 5.7%) have been isolated from fungi.3,4 Despite these and other statistics that demonstrate that fungi are prolific at the production of bioactive secondary metabolites,5–7 many pharmaceutical companies have minimized or eliminated natural products research programs.8 In this context we have established a collaborative program between university researchers and biotechnology entrepreneurs to probe a library of over 55,000 filamentous fungi for new anticancer and antimicrobial leads.9
An extract of the filamentous fungus Bionectria sp. (MSX 47401) showed both cytotoxic activity (>90% inhibition of H460 cell growth at 20 μg/mL) and antibacterial activity against methicillin-resistant Staphylococcus aureus (MRSA). While this fungus was first isolated from twigs collected in 1990, to date neither cytotoxic nor antibacterial compounds had been isolated, even though it has been part of several different screening campaigns.10 In this study, a suite of structurally diverse compounds was isolated, including peptaibols containing an ester-linked sugar alcohol (mannitol) instead of an amide-linked amino alcohol [a new peptaibol (1) and clonostachin (3)], a series of tetramic acid derivatives with mannosyl moieties [a new tetramic acid derivative (2) and virgineone (4)], the aglycone of 4 [virgineone aglycone (5)], one isocoumarin [AGI-7 (6)], and the bisabolane sesquiterpene 5,6-dihydroxybisabolol (7)]. Of the later, compounds 5 and 7 have not been described previously from natural sources. The structures of the new compounds were elucidated primarily by high field NMR (950 MHz), HRESIMS/MS, and chemical degradations (Marfey's analysis). Finally, while potent cytotoxicity was not observed, compounds 2, 4, and 5 were antimicrobial, even showing activity against several MRSA isolates.
RESULTS AND DISCUSSION
As part of ongoing studies to discover new anticancer and antimicrobial leads from filamentous fungi,9,11 an extract of the solid phase culture of Bionectria sp. (MSX 47401) was examined for cytotoxic and antimicrobial properties. The 1:1 CHCl3/MeOH extract of the scaled up fermentation of MSX 47401 was subjected to bioactivity-directed fractionation as monitored by inhibition of NCI-H460 (large cell lung carcinoma), MCF-7 (breast carcinoma), and SF-268 (astrocytoma) cells lines.12,13 Interestingly, most of the fractions were inactive against the three cell lines, with the exception of F-4, which had a precipitate that concentrated the activity (>75% inhibition of the three cell lines at 20 μg/mL). The UV, 1H and 13C NMR, and HRESIMS data of the white precipitate matched favorably with those of the isocoumarin AGI-7 (6), reported by Lee et al.14 from an unidentified fungus. Natural (cytogenin, sescandelin and compound 6) and synthetic (NM-3 and other derivatives) isocoumarins have been studied extensively for their antitumor activity and cytotoxicity, possibly due to antiangiogenic properties.14–16 In our assays, 6 was inactive, with an IC50 value >10 μM.
Fraction F-6 accounted for most of the mass (450 mg) after fractionation of the extract, and it was subjected to further purification using preparative RP-HPLC. This resulted in the isolation and characterization of a new peptaibol (1) and a new tetramic acid derivative (2) along with four known compounds, specifically clonostachin (3), virgineone (4), virgineone aglycone (5), and 5,6-dihydroxybisabolol (7).
Peptaibols 1 and 3 were isolated as white powders, and the molecular formulas were established as C79H136N14O25 and C78H134N14O25, respectively, on the basis of the HRESIMS (m/z 1, 1681.9863 [M + H]+; 3, 1667.9700 [M + H]+) and NMR data (Table 1), indicating 19 degrees of unsaturation for both compounds. The 1H NMR spectra in methanol-d3 of both 1 and 3 each displayed ten resonances between δH 7.30 and 8.70, which were assigned to the NH protons of the amide linkages. Similarly, 15 carbonyl signals were observed in the 13C NMR data between δC 170.0 and 176.0, confirming 1 and 3 to be peptides. Signals indicating a sugar alcohol were observed in the 1H (δH 3.40–5.30) and 13C (δC 60.0–73.0) NMR spectra for each compound. By analogy to compound 3,17 the sugar moiety in 1 was linked to the peptide core via an ester bond to the carboxy terminus (CO, δC 172.0). Finally, the presence of a sharp singlet at δH 2.04 in the 1H NMR spectrum of each compound suggested an acetylated N-terminal residue. Coupling these data with searches of the literature based on molecular formula, exact mass, and UV maxima in the Dictionary of Natural Products and SciFinder databases suggested that compound 3 was the known peptaibol clonostachin,17 isolated from Clonostachys sp., which is an anamorph of Bionectria, while 1, a larger analogue, showed no match. From the 317 peptaibol sequences reported in the Peptaibol Database (http://peptaibol.cryst.bbk.ac.uk),18,19 clonostachin (3) is the only peptaibol containing an ester-linked sugar alcohol (mannitol) instead of an amide-linked amino alcohol. The structure elucidation of 3 was based on Edman and chemical degradation, FABMS, EIMS and NMR analysis; however, a complete NMR characterization was not performed.17
Table 1.
NMR Spectroscopic Data for Compounds 1 and 3 (950, 150, and 700 MHz for 1H, 13C, and HMBC, respectively) in Methanol-d3a
| Clonostachin B (1) |
Clonostachin (3) |
|||||||
|---|---|---|---|---|---|---|---|---|
| δC type. | δH mult. (J in Hz) | HMBC (H → C) | δC type. | δH mult. (J in Hz) | ||||
| Ac | Ac | |||||||
| CO | 170.6 | C | CO | 170.7 | C | |||
| Me | 22.0 | CH3 | 2.04, s | CO | Me | 22.0 | CH3 | 2.04, s |
| Aib1 | Aib1 | |||||||
| CO | 175.1 | C | CO | 175.1 | C | |||
| NH | 8.69, s | CO (Ac), CO, α, β1,2 | NH | 8.69, s | ||||
| α | 57.1 | C | α | 57.1 | C | |||
| β 1 | 25.0 | CH3 | 1.58, s | CO, α, β2 | βMe1 | 25.0 | CH3 | 1.58, s |
| β 2 | 24.2 | CH3 | 1.51, s | CO, α, β1 | βMe2 | 24.2 | CH3 | 1.51, s |
| Hyp2 * | Hyp2 | |||||||
| CO | 172.9 | C | CO | 172.8 | C | |||
| α | 63.3 | CH | 4.61, dd (7.6, 10.5) | CO, β, γ | α | 63.3 | CH | 4.61, dd (7.6, 10.5) |
| β | 35.7 | CH2 | 1.88, m;1.82, m | CO, γ | β | 35.7 | CH2 | 1.88, m;1.82, m |
| γ | 69.1 | CH | 4.48, brs | δ, β | γ | 69.3 | CH | 4.48, brs |
| δ | 56.4 | CH2 | 3.96, d (14.2); 3.63, d (14.2) | γ | δ | 56.4 | CH2 | 3.96, d (14.2); 3.63, d (14.2) |
| Leu3 | Leu3 | |||||||
| CO | 173.0 | C | CO | 173.1 | C | |||
| NH | 8.24, d (8.5) | CO (O2), CO, α, β | NH | 8.24, d (8.5) | ||||
| α | 51.7 | CH | 4.30, m | CO, β | α | 51.7 | CH | 4.30, m |
| β | 38.9 | CH2 | 1.90, m, 1.59, m | α, γ | β | 38.9 | CH2 | 1.90, m, 1.59, m |
| γ | 24.1 | CH | 1.92, m | β, δ, δ' | γ | 24.1 | CH | 1.92, m |
| δ | 19.2 | CH3 | 0.94, d (5.7) | γ, δ' | δ | 19.2 | CH3 | 0.94, d (5.7) |
| δ' | 19.2 | CH3 | 0.98, d (5.7) | γ, δ | δ' | 19.2 | CH3 | 0.98, d (5.7) |
| Iva4 | Iva4 | |||||||
| CO | 174.4 | C | CO | 174.4 | C | |||
| NH | 7.76, s | CO (L3), CO, α, β2 | NH | 7.76, s | ||||
| α | 58.5 | C | α | 58.7 | C | |||
| β 1 | 18.5 | CH3 | 1.59, s | α, CO, β2 | β 1 | 18.6 | CH3 | 1.60, s |
| β 2 | 28.3 | CH2 | 2.40, m; 1.66, m | α, γ, β1 | β 2 | 28.4 | CH2 | 2.40, m; 1.66, m |
| γ | 6.7 | CH3 | 0.89, m | β2, α | α | 6.7 | CH3 | 0.89, m |
| Hyp5 * | Hyp5 | |||||||
| CO | 173.2 | C | CO | 173.4 | C | |||
| α | 61.9 | CH | 4.61, dd (7.6, 10.5) | CO, β, γ | α | 61.9 | CH | 4.61, dd (7.6, 10.5) |
| β | 35.9 | CH2 | 1.88, m;1.82, m | CO, γ | β | 36.1 | CH2 | 1.88, m;1.82, m |
| γ | 69.2 | CH | 4.48, brs | δ, β | γ | 69.3 | CH | 4.48, brs |
| δ | 56.2 | CH2 | 3.96, d (14.2); 3.63, d (14.2) | γ | δ | 56.2 | CH2 | 3.96, d (14.2); 3.63, d (14.2) |
| Leu6 | Leu6 | |||||||
| CO | 173.3 | C | CO | 173.3 | C | |||
| NH | 8.14, d (8.5) | CO (O5), CO, α, β | NH | 8.14, d (8.5) | ||||
| α | 51.5 | CH | 4.27, m | CO, β | α | 51.5 | CH | 4.27, m |
| β | 38.8 | CH2 | 1.90, m, 1.59, m | α, γ | β | 38.8 | CH2 | 1.90, m, 1.59, m |
| γ | 24.1 | CH | 1.92, m | β, δ, δ' | γ | 24.1 | CH | 1.92, m |
| δ | 19.2 | CH3 | 0.94, d (5.7) | γ, δ' | δ | 19.2 | CH3 | 0.94, d (5.7) |
| δ' | 19.2 | CH3 | 0.98, d (5.7) | γ, δ | δ' | 19.2 | CH3 | 0.98, d (5.7) |
| Iva7 | Iva7 | |||||||
| CO | 174.3 | C | CO | 174.4 | C | |||
| NH | 7.75, s | CO (L6), CO, α, β2 | NH | 7.75, s | ||||
| α | 58.5 | C | α | 58.6 | C | |||
| β 1 | 18.5 | CH3 | 1.54, s | α, CO, β2 | β 1 | 18.5 | CH3 | 1.55, s |
| β 2 | 27.8 | CH2 | 2.40, m; 1.66, m | α, γ, β1 | β 2 | 27.8 | CH2 | 2.40, m; 1.66, m |
| γ | 5.9 | CH3 | 0.88, m | β2, α | γ | 5.9 | CH3 | 0.88, m |
| Hyp8 * | Hyp8 | |||||||
| CO | 170.1 | C | CO | 170.1 | C | |||
| α | 62.8 | CH | 4.61, dd (7.6, 10.5) | CO, β, γ | α | 62.8 | CH | 4.61, dd (7.6, 10.5) |
| β | 36.2 | CH2 | 1.88, m;1.82, m | CO, γ | β | 36.2 | CH2 | 1.88, m;1.82, m |
| γ | 69.3 | CH | 4.45, brs | δ, β | γ | 69.4 | CH | 4.45, brs |
| δ | 55.9 | CH2 | 3.94, d (14.2); 3.87, d (14.2) | γ | δ | 56.1 | CH2 | 3.94, d (14.2); 3.87, d (14.2) |
| Aib9 | Aib9 | |||||||
| CO | 173.2 | C | CO | 173.1 | C | |||
| NH | 7.95, s | CO (O8), CO, α, β1,2 | NH | 7.95, s | ||||
| α | 56.1 | C | α | 56.1 | C | |||
| β 1 | 25.5 | CH3 | 1.58, s | CO, α, β2 | βMe1 | 25.4 | CH3 | 1.57, s |
| β 2 | 24.2 | CH3 | 1.50, s | CO, α, β1 | βMe2 | 24.2 | CH3 | 1.50, s |
| Iva10 | Iva10 | |||||||
| CO | 174.3 | C | CO | 174.4 | C | |||
| NH | 7.74, s | CO (U9), CO, α, β2 | NH | 7.74, s | ||||
| α | 59.5 | C | α | 59.5 | C | |||
| β 1 | 21.6 | CH3 | 1.54, s | α, CO, β2 | β 1 | 21.6 | CH3 | 1.48, s |
| β 2 | 28.7 | CH2 | 2.25, m; 1.66, m | α, γ, β1 | β 2 | 28.7 | CH2 | 2.25, m; 1.66, m |
| γ | 6.5 | CH3 | 0.87, m | β2, α | γ | 6.4 | CH3 | 0.87, m |
| Iva11 | Aib11 | |||||||
| CO | 174.4 | C | CO | 175.0 | C | |||
| NH | 7.73, s | CO (J10), CO, α, β2 | NH | 7.83, s | ||||
| α | 59.7 | C | α | 55.9 | C | |||
| β 1 | 21.6 | CH3 | 1.49, s | α, CO, β2 | βMe1 | 24.8 | CH3 | 1.52, s |
| β 2 | 28.3 | CH2 | 2.31, m; 1.66, m | α, γ, β1 | βMe2 | 24.2 | CH3 | 1.43, s |
| γ | 6.9 | CH3 | 0.87, m | β2, α | ||||
| Hyp12 * | Hyp12 | |||||||
| CO | 171.9 | C | CO | 172.0 | C | |||
| α | 61.7 | CH | 4.61, dd (7.6, 10.5) | CO, β, γ | α | 61.7 | CH | 4.61, dd (7.6, 10.5) |
| β | 36.2 | CH2 | 1.88, m;1.82, m | CO, γ | β | 36.2 | CH2 | 1.88, m;1.82, m |
| γ | 69.4 | CH | 4.45, brs | δ, β | γ | 69.4 | CH | 4.45, brs |
| δ | 55.7 | CH2 | 3.88, d (13.3); 3.86, d (13.3) | γ | δ | 55.7 | CH2 | 3.88, d (13.3); 3.86, d (13.3) |
| Iva13 | Iva13 | |||||||
| CO | 175.1 | C | CO | 175.2 | C | |||
| NH | 7.37, s | CO (O12), CO, α, β2 | NH | 7.34, s | ||||
| α | 59.5 | C | α | 59.6 | C | |||
| β 1 | 21.9 | CH3 | 1.45, s | α, CO, β2 | β 1 | 21.9 | CH3 | 1.47, s |
| β 2 | 28.8 | CH2 | 2.21, m; 1.66, m | α, γ, β1 | β 2 | 28.8 | CH2 | 2.21, m; 1.66, m |
| γ | 6.7 | CH3 | 0.86, m | β2, α | γ | 5.7 | CH3 | 0.86, m |
| Ile14 | Ile14 | |||||||
| CO | 172.0 | C | CO | 172.1 | C | |||
| NH | 7.74, d (6.1) | CO (J13), CO, α, β | NH | 7.74, d (6.1) | ||||
| α | 59.5 | CH | 4.39, m | CO, β, γ, γ' | α | 59.5 | CH | 4.39, m |
| β | 36.4 | CH | 1.98, m | β, γ, γ', CO | β | 36.4 | CH | 1.98, m |
| γ | 28.5 | CH2 | 1.72, m; 1.32, m | β, δ, α, γ' | γ | 28.4 | CH2 | 1.72, m; 1.32, m |
| γ' | 8.6 | CH3 | 0.93, s | β, δ, α, γ | γ' | 8.8 | CH3 | 0.94, m |
| δ | 13.9 | CH3 | 0.80, m | γ, β | δ | 14.0 | CH3 | 0.79, m |
| Mannitol | Mannitol | |||||||
| 1 | 60.9 | CH2 | 3.42, dd (2.9, 11.4); 3.66, dd (3.8, 11.4) | C-2, C-3 | 1 | 60.9 | CH2 | 3.42, dd (2.9, 11.4); 3.66, dd (3.8, 11.7) |
| 2 | 69.4 | CH | 3.89, dd (3.8, 8.5) | C-1, C-3 | 2 | 69.9 | CH | 3.89, dd (3.8, 8.5) |
| 3 | 72.3 | CH | 5.24, d (8.5) | CO (I14), C-1, C-2, C-4 | 3 | 72.4 | CH | 5.24, d (8.5) |
| 4 | 70.5 | CH | 3.84, dd (8.5, 12.3) | C-3, C-5 | 4 | 69.7 | CH | 3.84, dd (8.5, 12.3) |
| 5 | 70.7 | CH | 3.55, dd (5.7, 12.3) | C-4, C-6 | 5 | 70.5 | CH | 3.55, dd (5.7, 12.3) |
| 6 | 60.9 | CH2 | 3.64, dd (5.7, 11.4); 3.81, dd (3.8, 11.4) | C-5, C-4 | 6 | 60.9 | CH2 | 3.64, dd (5.7, 11.4); 3.81, dd (3.8, 11.4) |
Assignments may be interchanged.
U=Aib, O=Hyp, L=Leu, J=Iva, I=Ile.
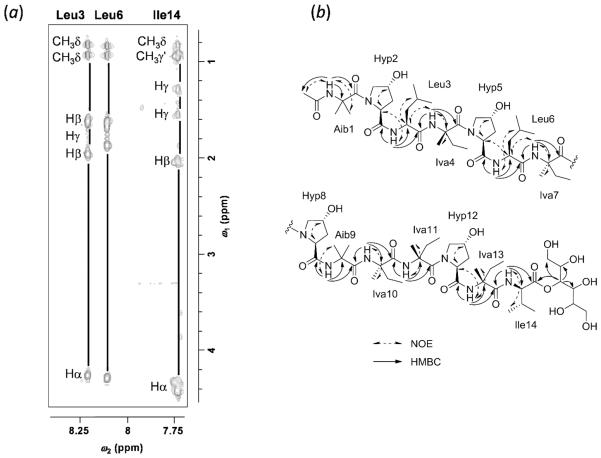
The amino acid sequences of compound 3 and related analogue 1 were examined by positive HRESIMS and HRESIMS/MS using Higher-Energy Collisional Dissociation (HCD).20,21 The full-scan HRESIMS spectra of 3 and 1 (Figure 1) exhibited several common in-source ions; in particular, b-type ions22,23 at m/z 1160.6924 and 1174.7075 suggested the presence of Aib-Hyp and Iva/Val-Hyp labile bonds, corresponding to the formation of the undecapeptides Ac-Aib1-Hyp2-Leu3-Iva4-Hyp5-Leu6-Iva7-Hyp8-Aib9-Iva10-Aib11 and Ac-Aib1-Hyp2-Leu3-Iva4-Hyp5-Leu6-Iva7-Hyp8-Aib9-Iva10-Iva/Val11 for 3 and 1, respectively. From HRESIMS/MS spectra using HCD, a series of fragments (Figure 2) was observed via sodium ion adducts [M + Na]+ at m/z 1689.9517 and 1703.9677 of 3 and 1, respectively, confirming the sequence of each peptaibol. Based on the exact mass of the ion fragments a11*, the presence of Iva/Val11 in 1, instead of Aib11 as seen in 3, was established. The replacement of Aib by Iva or Val has been reported, and some examples include chrysospermins B and D,24 trichorzianines,25 and asperelines,26 among others.
Figure 1.
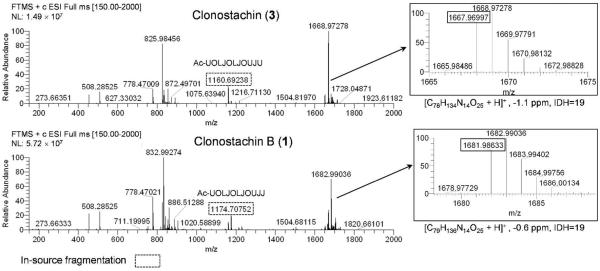
Full-scan HRESIMS spectra of peptaibols 3 and 1 (Source voltage: 4.5 kV; Tube lens voltage: 115 V; Capillary voltage: 46 V).
Figure 2.
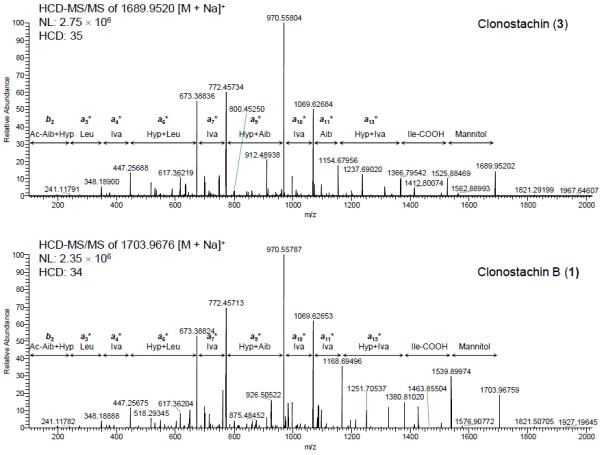
Positive HRESIMS/MS mass fragmentation patterns (HCD) of peptaibols 3 and 1, exhibiting bn or an* [an + Na] ions.
The composition and absolute configuration of the residues was derived from the total acid hydrolysis of 3 and 1 followed by examination using a 10 min Marfey's-UPLC method (Figure S15).21,27,28 Thus, the residues in 3 were confirmed to be Aib (3), l-Hyp (4), l-Leu (2), d-Iva (2), l-Iva (2), and l-Ile (1), as reported previously.17 As expected, compound 1 showed an additional d-Iva11 (3) and one less Aib11 residue. Finally, the complete amino acid sequence of peptaibols 1 and 3 was established by reference to those confirmed for 3 by Edman degradation,17 along with interpretation of a suite of high field (950 MHz) NMR experiments. For peptaibol 1, the assignments of the 1H and 13C chemical shifts of the amino acid residues (Table 1) were complemented by 2D NOESY, TOCSY, and HMBC experiments (Figure 3). For instance, the complete spin systems of Leu3, Leu6, and Ile14 (NH and α-, β-, γ-, and δ-H resonances of the side chains) were clearly observed in the TOCSY spectrum (Figure 3). Ten NH singlets were attributed to the Aib, Leu, Iva, and Ile residues, which were confirmed by the HMBC correlations through 2JCH and 3JCH couplings between NH resonances and the carbonyl and α-C quaternary carbons around δC 172–174 and 51–59, respectively (Figure 3). Thus, HMBC correlations of the acetyl methyl carbon at δC 22.0 and the NH at δH 8.69 to the carbonyl carbon at δC 175.1 indicated the acetylated N-terminus of the Aib1 residue. In addition, the interactions of NH (δH 8.24, Leu3) with δC 172.9 (C=O, Hyp2) and 173.0 (C=O, Leu3), NH (δH 7.76, Iva4) with C=O (Leu3) and δC 174.4 (C=O, Iva4), NH (δH 8.14, Leu6) with δC 173.2 (C=O, Hyp5) and 173.3 (C=O, Leu6), NH (δH 7.75, Iva7) with C=O (Leu6) and δC 174.3 (C=O, Iva7), NH (δH 7.95, Aib9) with δC 170.1 (C=O, Hyp8) and 173.2 (C=O, Aib9), NH (δH 7.74, Iva10) with C=O (Aib9) and δC 174.3 (C=O, Iva10), NH (δH 7.73, Iva11) with C=O (Iva10) and δC 174.4 (C=O, Iva11), NH (δH 7.37, Iva13) with δC 171.9 (C=O, Hyp12) and 175.1 (C=O, Iva13), and NH (δH 7.74, Ile14) with C=O (Iva13) and δC 172.0 (C=O, Ile14) established the sequence of Ac-Aib1-Hyp2-Leu3-Iva4-Hyp5-Leu6-Iva7-Hyp8-Aib9-Iva10-Iva11-Hyp12-Iva13-Ile14. Finally, the mannitol residue was shown to form an ester bond at the carboxy terminus (C=O, Ile14) with the methine proton at δH 5.24 on the basis of HMBC correlations. In summary, the structure of 3 was verified, while the structure of the related analogue 1 was elucidated. So as to be consistent with the literature, the trivial name clonostachin B was ascribed to the latter.
Figure 3.
(a) Expansion of the TOCSY spectrum (in methanol-d3: ω2 = 7.7–8.3 ppm, ω1 = 0.8–4.4 ppm), and (b) Key NOESY and HMBC correlations observed in 1
Compounds 2, 4, and 5 were isolated as colorless powders. The main metabolite 4 was identified as the unusual tetramic acid glycoside named virgineone, recently isolated by Ondeyka and co-workers from a saprotrophic fungus Lachnum virgineum using the Candida albicans fitness test.29 Compound 5, the aglycone of 4, was also reported by the same research group, but as a hydrolysis product of 4. In this project 5 was isolated as a natural product or perhaps by hydrolysis during isolation, and the NMR and HRMS data were in agreement with those reported previously.29 The configuration at positions C-2 and C-26 for compounds 4 and 5 was established as 2S and 26S by comparison of the NMR spectra and optical rotation values (see Supporting Information) with those reported for the synthetic isomers.30
Compound 2 was assigned the molecular formula of C40H61NO12 by HRESIMS, revealing an index of hydrogen deficiency of 11, which was one more than that of compound 4. The NMR spectra of 2 (Table 2) revealed nearly identical structural features to those found in virgineone (4), except for the methine signal for C-2 (δC 61.8) in 4, which was replaced with a quaternary carbon at δC 169.4 in 2. The side chain linked at C-4 was identical to that of 4, including the presence of a β-mannose moiety (elucidated by means of analyses of proton and COSY spectra and coupling constants as reported by Wangun et al.31) at the end of the aliphatic chain, one methyl carbon (δC 14.9), one aliphatic ketone (δC 213.1), and 17 methylene carbons in the aliphatic region (attributable to C-8–C-15 and C-17–C-25 through the analyses of the 2D NMR spectroscopic data), an anomeric methine (δH 4.51, δC 100.7), five oxymethines, and two oxymethylenes. The HMBC correlation data for the anomeric proton H-1" and the methyl group H3-28 of 2 were essentially identical to those observed in 4, confirming the glycosidation at C-27 and the connectivity of the pyrrolone system with the aliphatic chain. The 1H NMR spectrum of 2 (Table 2) also showed a typical AA'BB' spin system attributable to a para-hydroxy-benzene moiety at δH 6.71 (H-4'/H-6') and 7.05 (H-3'/H-7'), which was further supported by COSY and HMBC experiments. The phenolic carbon C-5' exhibited HMBC correlations to H-3'/7' and H-4'/6', and the correlations from H-3'/7' to C-1' (δC 42.2) confirmed the position of the para-hydroxy-benzene residue at C-2. Finally, the HRESIMS analysis of 2 showed a major fragment ion at m/z 586.3724, ascribed to loss of the mannose moiety, and key fragment ions at m/z 370.2176 and 398.1975 due to cleavage of the carbon bonds before (C-15/C-16) and after (C-16/C-17) the C-16 keto group, respectively, as observed for 4.29 Compound 2, therefore, was ascribed the trivial name 1,2-dehydrovirgineone (2).
Table 2.
1H (500 MHz) and 13C (125 MHz) NMR Assignments in Methanol-d4 of Compound 2
| position | δC type | δH mult (J in Hz) | HMBC (H→C) | |
|---|---|---|---|---|
| 1 | N | |||
| 2 | 169.4 | C | ||
| 3 | 203.9 | C | ||
| 4 | 107.4 | C | ||
| 5 | 172.4 | C | ||
| 6 | 208.5 | C | ||
| 7 | 42.3 | CH | 3.63, m | 4, 6, 9, 28 |
| 8 | 33.1 | CH2 | 1.44, m | 9 |
| 9 | 25.3 | CH2 | 1.20–1.30, m | 8, 10–13 |
| 10–13 | 29.3 | CH2 | 1.20–1.30, m | 9, 10–13, 14, 15 |
| 14 | 26.7 | CH2 | 1.27, m | 12, 13, 15 |
| 15 | 42.1 | CH2 | 2.41, dd (7.4) | 13, 14, 16 |
| 16 | 213.1 | C | ||
| 17 | 42.1 | CH2 | 2.41, dd (7.4) | 16, 18, 19 |
| 18 | 23.5 | CH2 | 1.55, m | 16, 17, 19 |
| 19–24 | 29.3 | CH2 | 1.20–1.30, m | |
| 25 | 32.3 | CH2 | 1.30, m; 1.65, m | 23–24, 26, 27 |
| 26 | 70.3 | CH | 3.73, m | |
| 27 | 73.6 | CH2 | 3.42, dd (6.0, 9.8); 3.84, dd (2.3, 10.1) | 1”, 26 |
| 28 | 14.9 | CH3 | 1.06, d (6.8) | 6, 7, 8 |
| 1' | 42.2 | CH2 | 3.59, s | 2, 2', 5, 3'/7' |
| 2' | 124.6 | C | ||
| 3'/7' | 130.1 | CH | 7.05, d (8.6) | 1', 3'/7', 5' |
| 4'/6' | 115.0 | CH | 6.71, d (8.6) | 3'/7, 4;/6, 5' |
| 5' | 156.4 | C | ||
| 1” | 100.7 | CH | 4.51, brs | 2”, 3”, 27 |
| 2” | 71.1 | CH | 3.87, dd (1.0, 3.4) | 3”, 4” |
| 3” | 73.9 | CH | 3.43, dd (3.4, 9.7) | 1”, 2”, 4” |
| 4” | 67.0 | CH | 3.54, t (9.5) | 3”, 5”, 6” |
| 5” | 76.9 | CH | 3.18, ddd (2.3, 6.3, 9.5) | 4”, 6” |
| 6” | 61.5 | CH2 | 3.69, dd (6.3, 12.0); 3.87, dd (2.8, 11.3) | 3”, 4”, 5” |
Compound 7 was isolated as a colorless oil and its molecular formula C15H28O3 was assigned by HRESIMS (m/z 257.2110 [M + H]+). The NMR spectra and [α]D − 48.7 value of 7 were consistent to those observed for the bisabolane sesquiterpene, 5,6-dihydroxybisabolol.32 This compound was isolated as one of the major products of the bioconversion of (−)-α-bisabolol by Aspergillus niger by Miyazawa and co-workers.32 However, this represents the first report of compound 7 as a natural product.
The antibacterial (including several MRSA isolates) and antifungal properties of all compounds but 6 were examined as described previously.21,33,34 The tetramic acid derivatives, 1,2-dehydrovirgineone (2), virgineone (4), and virgineone aglycone (5), showed promising antimicrobial activity against S. aureus and several MRSA isolates, and compound 4 displayed moderate antifungal activity against Candida albicans, Cryptococcus neoformans, and A. niger (Table 3); these results were in agreement with those reported previously.29 None of the compounds tested were active against E. coli and several strains of Mycobacterium (data not shown).
Table 3.
Antimicrobial Assay Data for Compounds 2, 4, and 5
| Target microorganism | Antimicrobial activity (MIC in μg/mL) |
|||
|---|---|---|---|---|
| 2 | 4 | 5 | Positive control | |
| S. aureus ATCC 6538a | 14.0 | 7.2 | 2.8 | 0.25 |
| MRSA DA 1a | 14.0 | 7.2 | 2.8 | 0.25 |
| MRSA ATCC 43300a | 14.0 | 7.2 | 2.8 | 0.25 |
| MRSA GU 34380a | 14.0 | 7.2 | 2.8 | 0.50 |
| MRSA GU 34864a | 14.0 | 7.2 | 2.8 | 0.38 |
| MRSA GU 3636la | 28.0 | 7.2 | 5.5 | 0.75 |
| MRSA GU 53016a | 28.0 | 7.2 | 2.8 | 0.38 |
| MRSA GU 52300a | 28.0 | 7.2 | 5.5 | 0.50 |
| C. albicans b | >55 | 14.4 | >88 | 25 |
| C. neoformans b | >55 | 14.4 | >88 | 25 |
| A. niger b | >55 | 14.4 | >88 | 100 |
The positive controls were either
vancomycin
amphotericin B
In summary, a series of biogenetically diverse and bioactive compounds were isolated from a Bionectria sp. (MSX 47401). The structure of the new peptaibol clonostachin B (1) and the known clonostachin (3) were fully elucidated by HRESIMS/MS using HCD, Marfey's analysis and high resolution NMR (950 and 700 MHz) studies. Compound 1 represents the second peptaibol containing an ester-linked sugar alcohol (mannitol) instead of an amide-linked amino alcohol. Additionally, a new tetramic acid, 1,2-dehydrovirgineone (2) was identified along with the new natural product virgineone aglycone (5), and the known virgineone (4), the isocoumarin AGI-7 (6), and the sesquiterpene 5,6-dihydroxybisabolol (7). This represents the first report of compounds 5 and 7 as natural products. All compounds but 6 were tested for antibacterial (including several MRSA isolates) and antifungal activities. Compounds 2, 4 and 5, showed promising antimicrobial activity against S. aureus and several MRSA isolates, whereas 1, 3 and 7 were inactive (MIC >118 μg/mL). Interestingly, both peptaibols 1 and 3, as well as the tetramic acid derivatives 2 and 4, incorporated a sugar residue (mannitol and β-mannose, respectively).
Experimental Section
General Experimental Procedures
Optical rotations, UV and CD spectra were recorded on a Rudolph Research Autopol III polarimeter (Rudolph Research), a Varian Cary 100 Bio UV–vis spectrophotometer (Varian Inc.), and an Olis DSM 17 CD spectrophotometer (Olis), respectively. NMR experiments were conducted in CDCl3, methanol-d4 or methanol-d3 with presaturation of the OH peak at δH 4.9 ppm, using a JEOL ECA-500 (operating at 500 MHz for 1H, 125 MHz for 13C; JEOL Ltd.), a Bruker Ultrashield Plus 950 MHz with Avance III console and equipped with a QNP style Cryoprobe (operating at 950.3 MHz for 1H; Bruker BioSpin Corp.), a Bruker Ultrashield Plus 600 MHz with Avance III and equipped with a QNP style Cryoprobe (operating at 600 MHz for 1H and 150 MHz for 13C; Bruker BioSpin Corp.), or an Agilent 700 MHz DD2 NMR system equipped with a 5 mm Enhanced Cold Probe (operating at 700 MHz for 1H and 175 MHz for 13C; Agilent, Inc.). HRESIMS data were measured using an electrospray ionization source coupled to a LTQ Orbitrap XL system (Thermo Fisher Scientific) and equipped with an HCD cell. Data were collected in both positive and negative ionization modes via a liquid chromatographic/autosampler system that consisted of an Acquity UPLC system [Waters BEH C18 column (1.7 μm; 50 × 2.1 mm); Waters Corp.). Flash chromatography was conducted with a CombiFlash Rf system using a RediSep Rf Si-gel Gold column (both from Teledyne-Isco). Analytical and preparative HPLC were carried out on a Varian Prostar HPLC system equipped with Prostar 210 pumps and a Prostar 335 photodiode array detector, with data collected and analyzed using Galaxie Chromatography Workstation software (version 1.9.3.2, Varian Inc.). For preparative (250 × 21 mm), semipreparative (250 × 10 mm), and analytical (250 × 4.6 mm) HPLC, Gemini-NX 5 μm or Synergi Max-RP 80 4 μm columns (all from Phenomenex) were utilized. For UPLC analysis, a BEH C18 (1.7 Qm; 50 × 2.1 mm; Waters Corp.) column was used. Reference standards of amino acids and Marfey's reagent were obtained from Sigma-Aldrich. All other reagents and solvents were obtained from Fisher Scientific and were used without further purification.
Producing Organism and Fermentations
Mycosynthetix fungal strain 47401 was isolated by Dr Barry Katz in May of 1990 from leaf litter and leaves collected in a humid mountain forest. The growth conditions for the solid phase fermentations were as described previously12,35 and outlined in the Supporting Information. For molecular identification of strain MSX 47401, the Internal Transcribed Spacer (ITS) regions 1 and 2 and 5.8S nrDNA were sequenced.36 Detailed methodology for DNA extraction, PCR amplification, and sequencing are described in the Supporting Information. The combined ITS and LSU sequence was deposited in the GenBank (accession no. KC222025). The complete ITS sequence including both spacers and the 5.8S region of MSX 47401 (~470 base pairs) was compared with GenBank's database37 using the BLAST search.38 BLAST search and Maximum Likelihood (ML) analyses revealed phylogenetic affinities of MSX 47401 with members of the order Hypocreales. Further multiple sequence alignment and additional phylogenetic analysis indicated that MSX 47401 shared only affinities with two strains of Bionectria rossmaniae (AF358227 and AF210665) with significant bootstrap support values (Figure S1).
Extraction and Isolation
To the large-scale solid fermentation culture of MSX 47401 (2.8 L Fernbach flask containing 150 g rice and 300 ml H2O, which was inoculated using seed cultures grown in the YESD media and incubated at 22 °C for 14 days; Supporting Information), 500 mL of 1:1 MeOH/CHCl3 were added and the mixtures were shaken for 16 h on a reciprocating shaker. The solution was filtered and equal volumes of H2O and CHCl3 were added to the filtrate to bring the total volume to 2 L; the mixture was stirred for 2 h and then transferred into a separatory funnel. The bottom layer was drawn off and evaporated to dryness. The extract was defatted by stirring vigorously for 30 min in a mixture of 100 mL 1:1 MeOH-CH3CN and 100 mL hexane, and then partitioned in a separatory funnel. The bottom layer was collected and evaporated to dryness. The defatted large scale extract (1.3 g) was adsorbed onto a minimal amount of Celite 545 (Acros Organics) and dried with mixing via a mortar and pestle. This material was fractionated via flash chromatography on a 40 g RediSep Rf Gold Si-gel column, using a gradient solvent system of hexane-CHCl3-MeOH at a 40 mL/min flow rate and 53.3 column volumes over 63.9 min. Fractions were collected every 25 mL and pooled according to UV and ELSD profiles. From fractions 56–71 (F-4), which were combined and evaporated (96 mg), a precipitate (compound 6; 2.8 mg) was isolated (>75% inhibition of the three cell lines at 20 μg/mL). Preparative HPLC purification (Synergi column) of fraction 6 (F-6, 450 mg) using a linear gradient (40 to 85% of CH3CN-0.1% aqueous formic acid over 30 min at a flow rate of 21.2 mL/min) yielded eight subfractions. Purification of sub-fraction 2 (24 mg) by semipreparative HPLC (Gemini column, various ratios of CH3CN in 0.1% aqueous formic acid) afforded compounds 1 (1.5 mg) and 3 (2.1 mg). Preparative HPLC (Gemini column, various ratios of CH3CN in 0.1% aqueous formic acid) of sub-fraction 3 (99.0 mg) and further purification of the subsequent fractions by semipreparative HPLC (Gemini column, various ratios of CH3CN in 0.1% aqueous formic acid) afforded compounds 2 (0.8 mg) and 4 (62.1 mg). A second large-scale solid fermentation culture of MSX 47401 was extracted (515 mg), and fractionated in the same fashion as shown above. Preparative HPLC (Gemini column with a linear gradient of 20 to 100% of CH3CN-0.1% aqueous formic acid over 45 min at a flow rate of 21.2 mL/min) of sub-fractions 3 (70 mg) and 4 (66 mg) yielded compound 4 (9.3 mg) from the former and compounds 5 (3.6) mg and 7 (3.1 mg) from the latter.
Clonostachin B (1): white powder; +3.7 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 279.0 (2.90), 221 (3.33) nm; CD (MeOH) λmax (Δε) 225 (+1.0), 237 (−2.2), and 292 (−0.9); 1H NMR (methanol-d3, 950.30 MHz) and 13C NMR (methanol-d3, 238.95 MHz), see Table 1; HRESIMS m/z 1681.9863 [M + H]+ (calcd for C79H137N14O25 1681.9874).
1,2-Dehydrovirgineone (2): colorless powder; +3.6 (c 0.08, MeOH) UV (MeOH) λmax (log ε) 349 (3.03), 284 (3.23), 226 (3.19) nm; CD (MeOH) λmax (Δε) 219 (+0.8), 251 (−0.4), and 306 (+0.2); 1H NMR (methanol-d4, 500 MHz) and 13C NMR (methanol-d4, 125 MHz), see Table 2; HRESIMS m/z 748.4246 [M + H]+ (calcd for C40H62NO12 748.4267).
Marfey's Analysis of 1 and 3
The method was based on Ayers et al.21 (see Supporting Information) with the following modifications. UPLC conditions were 15–80% CH3CN in H2O over 10 min using a BEH column with the eluent monitored at 340 nm.
Cytotoxicity Assay
The cytotoxicity measurements against the MCF-739 human breast carcinoma (Barbara A Karmanos Cancer Center), NCI-H46040 human large cell lung carcinoma [HTB-177, American Type Culture Collection (ATCC)], and SF-26841 human astrocytoma (NCI Developmental Therapeutics Program) cell lines were performed as described in detail previously.12
Antimicrobial Assay
MIC measurements were performed as described,21,33,34 with the single modification of dilution of cultures in growth medium (10- to 100,000-fold). Undiluted cultures and dilutions were used as inocula for MIC measurements. All measurements were made in duplicate.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by P01 CA125066 from the National Cancer Institute/National Institutes of Health, Bethesda, MD, USA. Mycology technical support was provided by B. Darveaux and M. Lawrence (Mycosynthetix). The authors acknowledge the technical assistance of M. D. Williams (Virginia Tech) in measuring MICs and thank Dr. K. Knagge of the David H. Murdock Research Institute, Kannapolis, NC, for the 950 MHz NMR data. The high resolution mass spectrometry data were acquired in the Triad Mass Spectrometry Laboratory at the University of North Carolina at Greensboro.
Footnotes
Supporting Information. Information about the producing organism and its fermentation, the HRMS data and [α]D values for compounds 4–6, a phylogram of the most likely tree, NMR spectra for compounds 1–7, and Marfey's analysis of 1 and 3. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Blackwell M. Am. J. Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- (2).Hawksworth DL. Mycol. Res. 2001;105:1422–1432. [Google Scholar]
- (3).Dictionary of Natural Products. Chapman & Hall/CRC Chemical Database. http://www.chemnetbase.com.
- (4).Berdy J. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- (5).Misiek M, Hoffmeister D. Planta Med. 2007;73:103–115. doi: 10.1055/s-2007-967104. [DOI] [PubMed] [Google Scholar]
- (6).van den Berg MA, Albang R, Albermann K, Badger JH, Daran JM, Driessen AJ, Garcia-Estrada C, Fedorova ND, Harris DM, Heijne WH, Joardar V, Kiel JA, Kovalchuk A, Martin JF, Nierman WC, Nijland JG, Pronk JT, Roubos JA, van der Klei IJ, van Peij NN, Veenhuis M, von Dohren H, Wagner C, Wortman J, Bovenberg RA. Nat. Biotechnol. 2008;26:1161–1168. doi: 10.1038/nbt.1498. [DOI] [PubMed] [Google Scholar]
- (7).Ito T, Odake T, Katoh H, Yamaguchi Y, Aoki M. J. Nat. Prod. 2011;74:983–988. doi: 10.1021/np100859a. [DOI] [PubMed] [Google Scholar]
- (8).Pearce C. J. Adv. Pharm. Technol. Res. 2011;2:136–137. doi: 10.4103/2231-4040.85523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Orjala J, Oberlies NH, Pearce CJ, Swanson SM, Kinghorn AD. In: Bioactive Compounds from Natural Sources. Natural Products as Lead Compounds in Drug Discovery. 2nd ed. Tringali C, editor. Taylor & Francis; London, UK: 2012. pp. 37–63. [Google Scholar]
- (10).These results were retrieved from the Mycosynthetix database, which covers previous research on this organism going back over 30 years.
- (11).Kinghorn AD, Carcache de Blanco EJ, Chai HB, Orjala J, Farnsworth NR, Soejarto DD, Oberlies NH, Wani MC, Kroll DJ, Pearce CJ, Swanson SM, Kramer RA, Rose WC, Fairchild CR, Vite GD, Emanuel S, Jarjoura D, Cope FO. Pure Appl. Chem. 2009;81:1051–1063. doi: 10.1351/PAC-CON-08-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Ayers S, Graf TN, Adcock AF, Kroll DJ, Matthew S, Carcache de Blanco EJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. J. Nat. Prod. 2011;74:1126–1131. doi: 10.1021/np200062x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Figueroa M, Graf TN, Ayers S, Adcock AF, Kroll DJ, Yang J, Swanson SM, Munoz-Acuna U, Carcache de Blanco EJ, Agrawal R, Wani MC, Darveaux BA, Pearce CJ, Oberlies NH. J. Antibiot. 2012;65:559–564. doi: 10.1038/ja.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Lee JH, Park YJ, Kim HS, Hong YS, Kim KW, Lee JJ. J. Antibiot. 2001;54:463–466. doi: 10.7164/antibiotics.54.463. [DOI] [PubMed] [Google Scholar]
- (15).Kumagai H, Masuda T, Ishizuka M, Takeuchi T. J. Antibiot. 1995;48:175–178. doi: 10.7164/antibiotics.48.175. [DOI] [PubMed] [Google Scholar]
- (16).Salloum RM, Jaskowiak NT, Mauceri HJ, Seetharam S, Beckett MA, Koons AM, Hari DM, Gupta VK, Reimer C, Kalluri R, Posner MC, Hellman S, Kufe DW, Weichselbaum RR. Cancer Res. 2000;60:6958–6963. [PubMed] [Google Scholar]
- (17).Chikanishi T, Hasumi K, Harada T, Kawasaki N, Endo A. J. Antibiot. 1997;50:105–110. doi: 10.7164/antibiotics.50.105. [DOI] [PubMed] [Google Scholar]
- (18).Whitmore L, Wallace BA. Nucleic Acids Res. 2004;32:D593–D594. doi: 10.1093/nar/gkh077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Whitmore L, Wallace BA. Eur. Biophys. J. 2004;33:233–237. doi: 10.1007/s00249-003-0348-1. [DOI] [PubMed] [Google Scholar]
- (20).Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Nat. Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- (21).Ayers S, Ehrmann BM, Adcock AF, Kroll DJ, Carcache de Blanco EJ, Shen Q, Swanson SM, Falkinham JO, 3rd, Wani MC, Mitchell SM, Pearce CJ, Oberlies NH. J. Pept. Sci. 2012;18:500–510. doi: 10.1002/psc.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Daniel JF, Filho ER. Nat. Prod. Rep. 2007;24:1128–1141. doi: 10.1039/b618086h. [DOI] [PubMed] [Google Scholar]
- (23).Mohamed-Benkada M, Montagu M, Biard JF, Mondeguer F, Verite P, Dalgalarrondo M, Bissett J, Pouchus YF. Rapid Commun. Mass. Spectrom. 2006;20:1176–1180. doi: 10.1002/rcm.2430. [DOI] [PubMed] [Google Scholar]
- (24).Woon-Hyung Y, Bong-Sik Y, Young-Sook K, Sang-Jun L, Ik-Dong Y, Kab-Sig K, Eun-Kyung P, Jong-Chull L, K. YH. Plant. Pathol. J. 2002;18:18–22. [Google Scholar]
- (25).El Hajji M, Rebuffat S, Lecommandeur D, Bodo B. Int. J. Pept. Protein. Res. 1987;29:207–215. doi: 10.1111/j.1399-3011.1987.tb02247.x. [DOI] [PubMed] [Google Scholar]
- (26).Ren J, Xue C, Tian L, Xu M, Chen J, Deng Z, Proksch P, Lin W. J. Nat. Prod. 2009;72:1036–1044. doi: 10.1021/np900190w. [DOI] [PubMed] [Google Scholar]
- (27).Marfey P. Carlsberg Res. Commun. 1984;49:591–596. [Google Scholar]
- (28).Motohashi K, Inaba K, Fuse S, Doi T, Izumikawa M, Khan ST, Takagi M, Takahashi T, Shin-ya K. J. Nat. Prod. 2011;74:1630–1635. doi: 10.1021/np200386c. [DOI] [PubMed] [Google Scholar]
- (29).Ondeyka J, Harris G, Zink D, Basilio A, Vicente F, Bills G, Platas G, Collado J, Gonzaez A, de la Cruz M, Martin J, Kahn JN, Galuska S, Giacobbe R, Abruzzo G, Hickey E, Liberator P, Jiang B, Xu D, Roemer T, Singh SB. J. Nat. Prod. 2009;72:136–141. doi: 10.1021/np800511r. [DOI] [PubMed] [Google Scholar]
- (30).Yajima A, Ida C, Taniguchi K, Murata S, Katsuta R, Nukada T. Tetrahedron Lett. 2013;54:2497–2501. [Google Scholar]
- (31).Wangun HV, Dahse HM, Hertweck C. J. Nat. Prod. 2007;70:1800–1803. doi: 10.1021/np070245q. [DOI] [PubMed] [Google Scholar]
- (32).Miyazawa M, Funatsu Y, Kameoka H. Chem. Express. 1992;7:217–220. [Google Scholar]
- (33).Falkinham JO, 3rd, Macri RV, Maisuria BB, Actis ML, Sugandhi EW, Williams AA, Snyder AV, Jackson FR, Poppe MA, Chen L, Ganesh K, Gandour RD. Tuberculosis. 2012;92:173–181. doi: 10.1016/j.tube.2011.12.002. [DOI] [PubMed] [Google Scholar]
- (34).Williams AA, Sugandhi EW, Macri RV, Falkinham JO, 3rd, Gandour RD. J. Antimicrob. Chemother. 2007;59:451–458. doi: 10.1093/jac/dkl503. [DOI] [PubMed] [Google Scholar]
- (35).Sy-Cordero AA, Graf TN, Adcock AF, Kroll DJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. J. Nat. Prod. 2011;74:2137–2142. doi: 10.1021/np2004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. Proc. Natl. Acad. Sci. USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. Nucleic Acids Res. 2012;40:D48–D53. doi: 10.1093/nar/gkr1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- (39).Soule HD, Vazguez J, Long A, Albert S, Brennan M. J. Natl. Cancer Inst. 1973;51:1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- (40).Carney DN, Gazdar AF, Bunn PA, Jr., Guccion JG. Stem Cells. 1982;1:149–164. [PubMed] [Google Scholar]
- (41).Rosenblum ML, Gerosa MA, Wilson CB, Barger GR, Pertuiset BF, de Tribolet N, Dougherty DV. J. Neurosurg. 1983;58:170–176. doi: 10.3171/jns.1983.58.2.0170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.