Abstract
The MDM2-p53 feedback loop is crucially important for restricting p53 level and activity during normal cell growth and proliferation, and is thus subjected to dynamic regulation in order for cells to activate p53 upon various stress signals. Several ribosomal proteins, such as RPL11, RPL5, RPL23, RPL26, or RPS7, have been shown to play a role in regulation of this feedback loop in response to ribosomal stress. Here, we identify another ribosomal protein S14, which is highly associated with 5q-syndrome, as a novel activator of p53 by inhibiting MDM2 activity. We found that RPS14, but not RPS19, binds to the central acidic domain of MDM2, like RPL5 and RPL23, and inhibits its E3 ubiquitin ligase activity toward p53. This RPS14-MDM2 binding was induced upon ribosomal stress caused by actinomycin D or mycophenolic acid. Overexpression of RPS14, but not RPS19, elevated p53 level and activity, leading to G1 or G2 arrest. Conversely, knockdown of RPS14 alleviated p53 induction by these two reagents. Interestingly, knockdown of either RPS14 or RPS19 caused a ribosomal stress that led to p53 activation, which was impaired by further knocking down the level of RPL11 or RPL5. Together, our results demonstrate that RPS14 and RPS19 play distinct roles in regulating the MDM2-p53 feedback loop in response to ribosomal stress.
Keywords: RPS14, p53, MDM2, RPL11, RPL5, RPS19, ribosomal stress
Introduction
The tumor suppressor p53 plays an essential role in guarding the genome and preventing tumorigenesis by stopping cell proliferation, inducing apoptosis, and blocking metastasis mostly via its transcriptional activity (1, 2). Due to these cytotoxic activities, the oncoprotein MDM2, which is encoded by a p53’s transcriptional target gene (3, 4), is utilized by cells to monitor p53 functions. MDM2 via its N-terminal domain directly binds to both of the N- and C-termini of p53 and deactivates this protein by either abrogating its transcriptional activity (5, 6) or ubiquitinating it and mediating its proteasomal turnover (7–9). This inhibition of p53 by MDM2 has been gracefully verified by TP53 and mdm2 double knockout studies (10, 11).
Thus, under stress conditions, multiple cellular mechanisms can be awakened to reactivate p53 by blocking this feedback loop (12). Recently, an emerging new signaling pathway has been gradually appreciated to untie the MDM2-p53 loop in response to a type of stress, called ribosomal or nucleolar stress (13, 14). This stress can be caused by any chemical reagents or genetic, molecular and cellular events that disrupt each step of ribosomal biogenesis. These reagents and events include treatment of cells with low dose of actinomycin D (Act D) (15), 5-fluorouracil (16, 17), or mycophenolic acid (MPA) (18), serum starvation or contact inhibition (19), impairment of 40S or 60S ribosomal biogenesis by knockdown of either ribosomal protein (RP) S6 (20) or RPL29 and RPL30 (21), defects in 18S or 28S RNA processing (22), malfunction of nucleolar proteins, such as Bop1 (23), involved in ribosome biogenesis, inhibition of B23 (also known as nucleophosmin) activity by ARF (24), or ablation of nucleostemin by siRNAs (25), and knockdown of PAK1IP1, a nucleolar protein important for rRNA processing (26). Interestingly, it has been recently shown that ribosomal stress can also be induced by genotoxic insults, such as cisplatin or UV irradiation, probably through degradation of RPL37 (27). Upon ribosomal stress, several RPs, such as RPL11 (19, 28–30), RPL5 (31), RPL23 (32, 33), RPL26 (34), RPS7 (35, 36), RPS27 (37) or RPS27a (38), associate with MDM2 and inhibit MDM2-mediated ubiquitination and degradation of p53. Furthermore, the MDM2C305F, which is defective in RPL11- and RPL5- binding in vitro (39) and in vivo (40), has been shown to impair the p53 response to ribosomal stress in animals (40), strongly demonstrating the essential role of RP-MDM2 interactions in activation of p53 upon ribosomal stress.
The ribosomal stress-p53 pathway is also possibly linked with the pathogenesis of some human genetic diseases, such as myelodisplastic syndrome (MDS) (41). For instance, mutations of RPS19 are associated with Diamond-Blackfan anemia (DBA) (42), a congenital erythroblastopenia with strikingly absent or decreased erythroid precursors. Also, deletion of one allele of another RP-encoding gene, RPS14, results in 5q-syndrome with a characteristic defect in erythroid differentiation (43). In line with these human genetic studies, RPS14 haploinsufficiency in mice also led to a 5q-syndrome-like phenotype with bone marrow disorders, which can be completely rescued by further deleting TP53 in the animals (44), suggesting that p53 plays a role in the pathogenesis of this subtype of MDS (41, 44). Recently, mutations of several MDM2-binding RPs, such as RPL5, RPL11 (45, 46) or RPS7 (45), have also been found in DBA patients. These studies prompted us to explore whether DBA-associated RPS19 or 5q-syndrome-associated RPS14 may play a role in regulating the MDM2-p53 feedback loop as well.
In this study, we reveal differential regulation of this feedback loop by RPS14 and RPS19. RPS14, but not RPS19, interacted with MDM2, inhibited MDM2-mediated ubiquitination and degradation of p53, and induced p53-dependent cell cycle arrest in response to ribosomal stress.
Results
RPS14 interacts with MDM2 in cells and in vitro
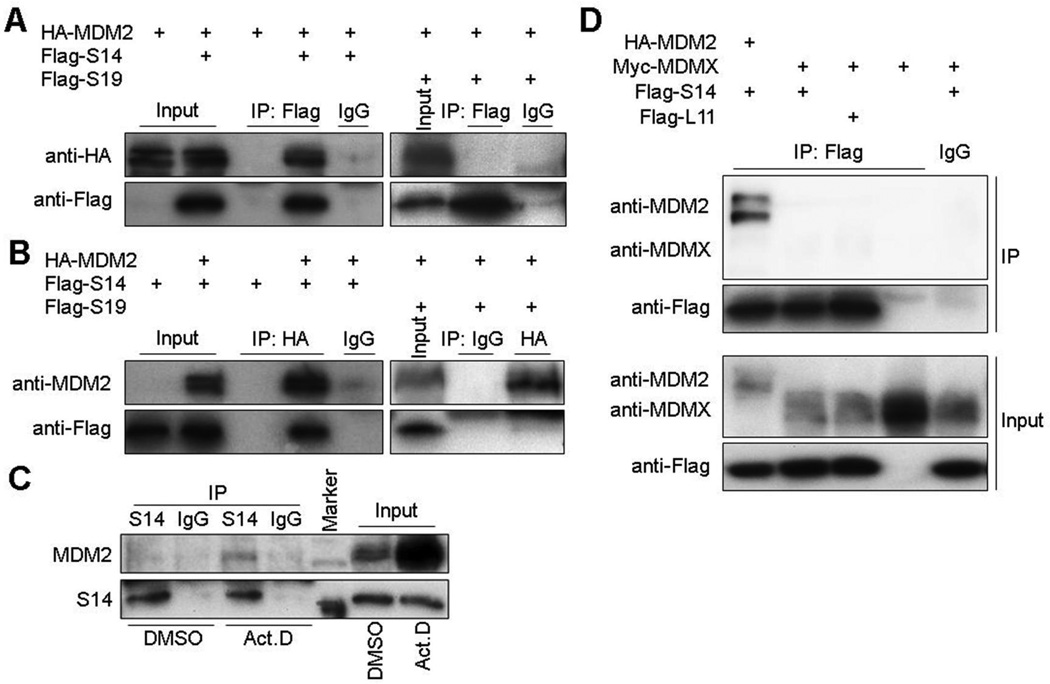
The fact that mutations of the MDM2-interacting RPL11, RPL5 and RPS7 have recently been shown to be associated with DBA (45, 46) hints that DBA-linked RPS19 and 5q-syndrome-associated RPS14 might also interact with MDM2. To test this idea, we first introduced ectopic Flag-RPS14 or Flag-RPS19 alone or together with HA-MDM2 into 293 cells and then performed a set of reciprocal co-IP-IB assays. Interestingly, we found only Flag-RPS14, but not Flag-RPS19, could be co-immunoprecipitated with HA-MDM2 in these co-IP-IB assays (Fig. 1A and B). Consistent with these results, endogenous RPS14 also bound to endogenous MDM2 in response to ribosomal stress caused by actinomycin D treatment of HCT116 cells (Fig. 1C). These results indicate that RPS14, but not RPS19, can associate with MDM2 in cells as well upon ribosomal stress. In addition, nucleolar RPS14 molecules were released to the nucleoplasm upon ribosomal stress (Fig. S1), which probably favored the interaction of RPS14 with MDM2, as MDM2 mainly localized in the nucleoplasm (Fig. S1). Although it has been reported that ribosomal proteins, L5, L11 and L23, selectively bind to MDM2, but not MDMX (16), we wondered if RPS14 associates with MDMX or not. To test this idea, ectopic Myc-MDMX was introduced into H1299 cells alone or together with Flag-S14 or Flag-L11. As shown in Fig. 1D, like RPL11, RPS14 did not bind to MDMX.
Figure 1. RPS14 interacts with MDM2 in cells.
(A) (B) Exogenous RPS14 interacts with exogenous MDM2 in 293 cells. Cells were transfected with HA-MDM2, Flag-RPS14, or Flag-RPS19 plasmids and harvested for IP (0.5 mg total proteins) using anti-Flag, anti-HA, or mouse IgG, followed by IB with the antibodies as indicated. (C) Endogenous RPS14 interacts with endogenous MDM2 in HCT116 cells. Cells lysates (1 mg) were prepared from HCT116 cells after treatment with Act D for 8 h. IP was conducted with anti-RPS14 or rabbit IgG followed by IB with anti-MDM2 or anti-RPS14. (D) Exogenous RPS14 does not interact with exogenous MDMX in H1299 cells. Cells were transfected with combinations of HA-MDM2, Myc-MDMX, Flag-S14 or Flag-L11 plasmids and harvested for co-IP-IB assays using the antibodies as indicated. All the experiments were repeated more than 2 times and true to the rest of data.
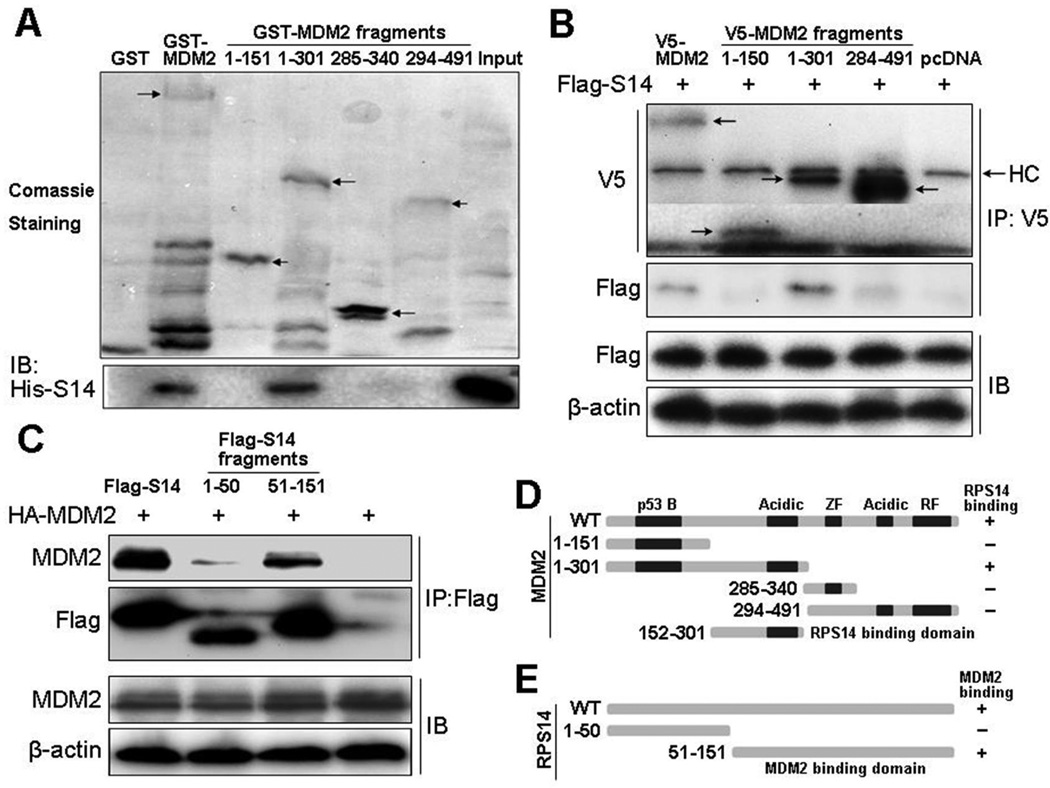
Since other 40S subunits, such as RPS7 (35, 36), RPS3 (47), RPS27 (37) and RPS27a (38), have been reported to bind to MDM2, the association of RPS14 with MDM2 in cells might be indirect via these known MDM2-binding RPs. To determine whether RPS14 directly binds to MDM2 and if so, to which domain of MDM2 it binds, we conducted a set of GST fusion protein association assays by using His-S14 and GST-MDM2 deletion fusion proteins purified from bacteria (Fig. 2A). GST-MDM2, but not GST alone, interacted with His-S14 directly. We also found that His-S14 bound to residues 1–301, but not 1–151, 285–340, or 294–491, of MDM2. This result indicates that like RPL5 (31), RPS14 also directly contacts with the central acidic domain of MDM2 (Fig. 2D) which was confirmed by co-IP-IB assays following introduction of Flag-RPS14 and V5-MDM2 fragments into 293 cells (Fig. 2B).
Figure 2. Mapping the MDM2-RPS14 binding domains.
(A) RPS14 directly binds to the central acidic domain of MDM2 in vitro. Purified GST, GST-MDM2, or GST-MDM2-fragments immobilized on glutathione beads were incubated with 500 ng of bacterially purified His-RPS14. Bound proteins were blotted with anti-His. Arrows indicate the GST-MDM2 or GST-MDM2-fragments. (B) RPS14 binds to the central acidic domain of MDM2 in cells. Cells were transfected with Flag-RPS14, V5-MDM2, or deletion mutants of V5-MDM2 plasmids and harvested for IP (0.5 mg total proteins) with anti-V5, followed by IB with the antibodies as indicated. Arrows indicate the V5-MDM2, V5-MDM2-fragments or heavy chains (HC). (C) MDM2 binds to the C-terminus of RPS14 in cells. Cells were transfected with HA-MDM2, Flag-RPS14, or deletion mutants of Flag-RPS14 plasmids and harvested for IP (0.5 mg total proteins) with anti-Flag, followed by IB with the antibodies as indicated. (D) Schematic presentation of the MDM2 domain(s) that binds to RPS14. (E) Schematic presentation of the RPS14 domain(s) that binds to MDM2.
To test whether RPS14 competes with RPL5 or RPL23 for association with MDM2, we introduced combinations of ectopic HA-MDM2, Flag-L5, GFP-L23 or Flag-S14 into H1299 cells. As shown in Fig. S2, RPS14 slightly reduced the level of the RPL5-MDM2 interaction, because both of the RPs bound to the same central acidic domain of MDM2. However, the RPL23-MDM2 interaction was not affected by overexpressing RPS14, as RPL23 preferentially bound to the C-terminal acidic domain of MDM2 (31).
Furthermore, to map the MDM2 binding domain of RPS14, we introduced HA-MDM2 along with Flag-RPS14, Flag-RPS14/1–50 or Flag-RPS14/51–151 into 293 cells through transient transfection and conducted co-IP-IB assays. As shown in Figs. 2C and 2E, MDM2 preferred interacting with the C-terminus (from amino acid 51–151), but not N-terminus (from aa 1–50), of RPS14. Taken together, these results demonstrate that RPS14 binds to MDM2 directly in vitro and in cells.
Ectopic expression of RPS14 stabilizes p53 by inhibiting MDM2-mediated p53 ubiquitination
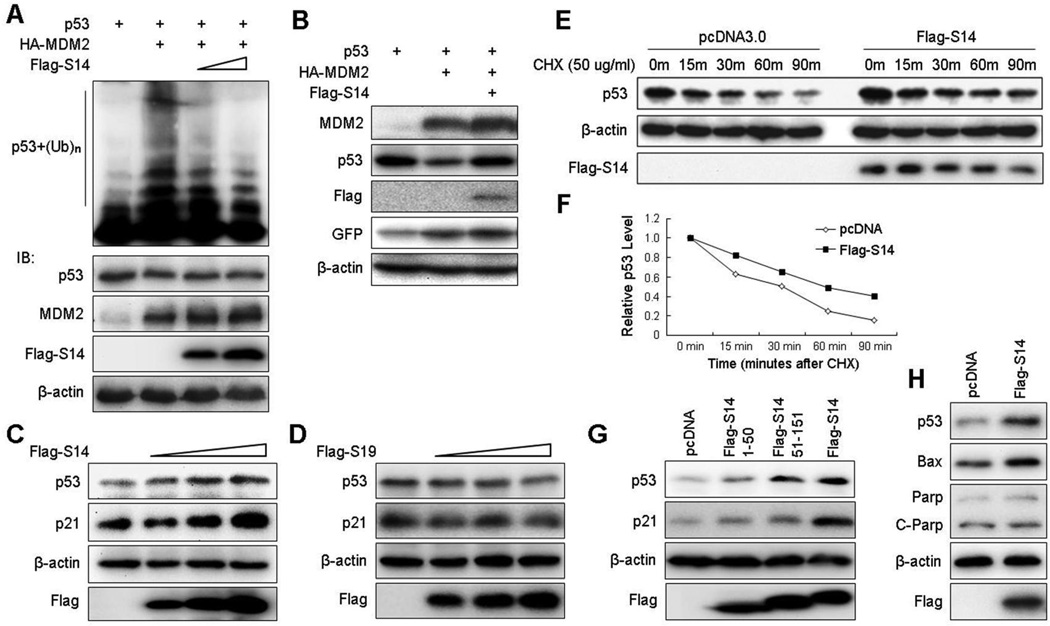
It has been previously shown that all of the RPs that interact with MDM2 stabilize p53 by inhibiting MDM2 E3 ligase activity to p53 (28, 31–36, 47). To test if RPS14 would also do so, we introduced combinations of His-Ub, p53, HA-MDM2, and Flag-RPS14 plasmids into H1299 cells to test p53 ubiquitination in the cells. As shown in Fig. 3A, MDM2 ubiquitinated p53, whereas ectopic Flag-RPS14 inhibited this ubiquitination in a dose-dependent manner. This inhibition was not due to variation of sample loading, as the p53 level in each lane was approximately equivalent (middle panel of Fig. 3A). Consistent with this result, Flag-RPS14 overcame the MDM2-mediated degradation of exogenous p53 in H1299 cells (Fig. 3B) as well as of endogenous p53 and induced p53 activity in a dose-dependent fashion in A549 cells (Fig. 3C). Also Flag-RPS14 markedly extended the half-life of endogenous p53 (Figs. 3E–3F). By contrast, overexpression of Flag-RPS19 did not show any significant effect on p53 level and activity in A549 cells (Fig. 3D), which is consistent with the results of Figs. 1A–1B. These results indicate that RPS14, but not RPS19, alleviates MDM2-mediated p53 ubiquitination and degradation, leading to p53 activation. This effect was perhaps through direct binding of RPS14 to MDM2, as overexpression of wild type RPS14 induced p53 and p21 levels more markedly than did its N-terminal or C-terminal fragment (aa 1–50) (Fig. 3G), which was with a reduced ability to bind to MDM2 (Fig. 2C). In addition to p21, another p53 target, Bax, which is involved in the apoptotic pathway, was also slightly induced by overexpressing RPS14 (Fig. 3H). However, the level of an apoptosis indicator, cleaved-PARP, was not elevated, probably because the free forms of ribosomal proteins, such as extra RPS14 molecules, would only cause modest stress (ribosomal stress), which might not need to induce apoptosis as a terminating outcome. Although it has been well documented that RPs activate p53 by inhibiting MDM2’s E3 ligase activity, we wondered whether RPS14 could interrupt the MDM2-p53 interaction or not. We tested this idea by performing a set of protein-protein binding competition assays using both cell-based and in vitro systems with purified proteins. As shown in Fig. S3, RPS14 had no effect on the MDM2-p53 interaction in cells by co-IP-IB assays (Fig. S3A) or in vitro by GST pull-down assays (Fig. S3B). These results suggest that RPS14 can lead to p53 stabilization by inhibiting MDM2-mediated p53 ubiquitination without interfering with the MDM2-p53 interaction.
Figure 3. RPS14 stabilizes p53 by inhibiting MDM2-mediated p53 ubiquitination.
(A) Ectopic expression of RPS14 inhibits MDM2-mediated p53 ubiquitination in cells. H1299 cells were transfected with combinations of plasmids encoding p53, HA-MDM2 or Flag-RPS14 in the presence of the His-Ub plasmid. Cells were treated with MG132 for 8 h before harvesting. Cell lysates were subjected to an ubiquitination assay followed by IB with anti-p53. The expression levels of p53, HA-MDM2 and Flag-RPS14 are shown in the lower panels. (B) Ectopic expression of RPS14 stabilizes exogenous p53 in H1299 cells. Cells were transfected with combinations of p53, HA-MDM2 or Flag-RPS14 in the presence of pEGFP plasmids as a control and harvested 36 h post-transfection for IB with antibodies as indicated. (C) RPS14 stabilizes endogenous p53 in A549 cell. Cells were transfected with an increasing amount of the Flag-RPS14 plasmid and harvested 36 h after transfection for IB with antibodies as indicated. (D) RPS19 does not stabilize endogenous p53 in A549 cell. The assay as that in panel C was conducted except the RPS19 plasmid was used here. (E) RPS14 extends the half-life of endogenous p53. A549 cells were transfected with pcDNA3 or Flag-RPS14 and treated with cycloheximide (CHX) at 36 h post-transfection, and harvested at different time points for IB with antibodies as indicated. (F) A graphic presentation of the result from panel E. (G) wild-type RPS14 and its C-terminus (amino acids 51–151), but not its N-terminus (aa 1–51), drastically induced p53 in A549 cells. Cells were transfected with pcDNA3, Flag-RPS14/1–50, Flag-RPS14/51–151, or Flag-RPS14 plasmids and harvested 36 h after transfection for IB with antibodies as indicated. (H) RPS14 slightly induces a p53 apoptotic target, Bax, but not the cleaved-Parp. A549 cells were transfected with pcDNA3 or Flag-S14 plasmids and harvested for IB with antibodies as indicated.
Ectopic expression of RPS14 induces p53-dependent cell cycle arrest and growth inhibition
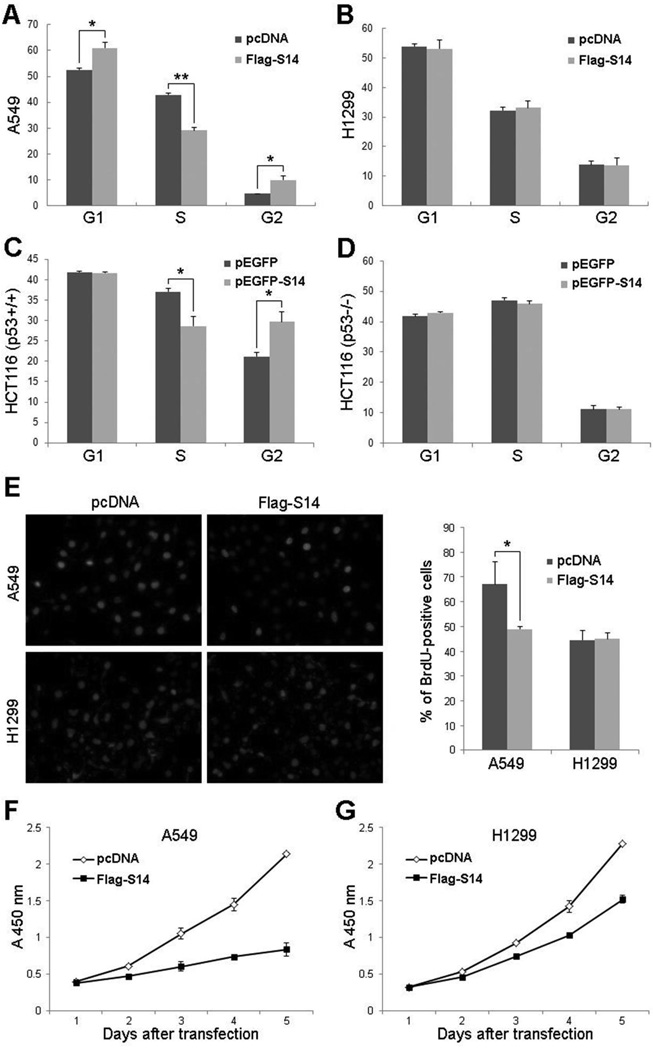
Previous studies by us (31, 32) and others (28, 33) have shown that overexpression of RPL11, RPL5, or RPL23 inhibits p53-dependent cell proliferation through induction of p21. Since RPS14 also induced p21 via p53 (Fig. 3C), we tested whether RPS14 has a similar effect on cell growth. To do so, we introduced pcDNA or Flag-RPS14 plasmids into A549 or H1299 cells and performed FACS analysis. As shown in Fig. 4, ectopic expression of RPS14 significantly inhibited the cell cycle progression at both of the G1 and G2 phases in A549 cells (Fig. 4A), whereas overexpression of RPS14 in p53-deficient H1299 cells had no effect on the cell cycle progression (Fig. 4B). The same experiments were performed employing HCT116 cells harboring wild-type or null p53 except for using pEGFP-RPS14 and pEGFP empty vector as a control (Figs. 4C and 4D). Interestingly, overexpression of RPS14 induced G2 arrest in this colon cancer cell line in a p53-dependent fashion (Figs. 4C and 4D). Consistent with the above results, RPS14 also inhibited cell proliferation as measured by BrdU incorporation assays (Fig. 4E). To test the effect of ectopically expressed RPS14 on cell growth, we conducted cell viability assays using Cell Counting Kit-8 (CCK-8). Cells were seeded into 96-well plates at 24 h post-transfection and the absorbance at 450 nm was measured every 24 hours. As shown in Fig. 4F, RPS14 dramatically suppressed cell growth in A549 cells during 5-days long culture. Interestingly, RPS14 also moderately suppressed cell growth in p53-deficient H1299 cells (Fig. 4G), which might be due to the inhibition of c-Myc by RPS14 (data not shown). These results indicate overexpression of RPS14 can indeed induce p53-dependent cell cycle arrest and consequently inhibit cell proliferation.
Figure 4. RPS14 induces p53-dependent cell cycle arrest.
(A) A549 cells were transfected with pcDNA3 or Flag-RPS14 and harvested 30 h after transfection for FACS analysis. The mean percentage of cells arrested in the G1 and G2 phases obtained from three separate experiments is presented. (B) H1299 cells were transfected with pcDNA3 or Flag-RPS14 and harvested 30 h after transfection for FACS analysis. The mean percentage of cells in each phases obtained from three separate experiments is presented. The same experiments as shown in (A) and (B) were performed in HCT116p53+/+ cells (C) and HCT116p53−/− cells (D) except the pEGFP and pEGFP-RPS14 plasmids were used here. The mean percentages of cells in each phases obtained from three separate experiments are presented. (E) RPS14 induced p53-dependent growth arrest was determined by BrdU incorporation assays (left panel). Cells were transfected with pcDNA3 or Flag-RPS14 and treated with 10 µM BrdU for 5 h, followed by BrdU and DAPI staining. The percentage of BrdU-positive cells is shown in the right panel. (F) (G) RPS14 inhibits cell growth as measured in cell viability assays. A549 cells (F) or H1299 (G) cells were transfected with pcDNA3 or Flag-S14 plasmids and seeded into 96-well plates at 24 h post-transfection. WST-8 was added to each well and cells were incubated at 37 °C for additional 2–3 h before absorbance at 450 nm was measured using a microplate reader every 24 h during 5-days long culture. Bars indicate standard deviations. “*” indicates P<0.05, “**” indicates P<0.005.
Ablation of endogenous RPS14 by siRNAs impairs ribosomal stress-triggered p53 activation, though induces p53, too
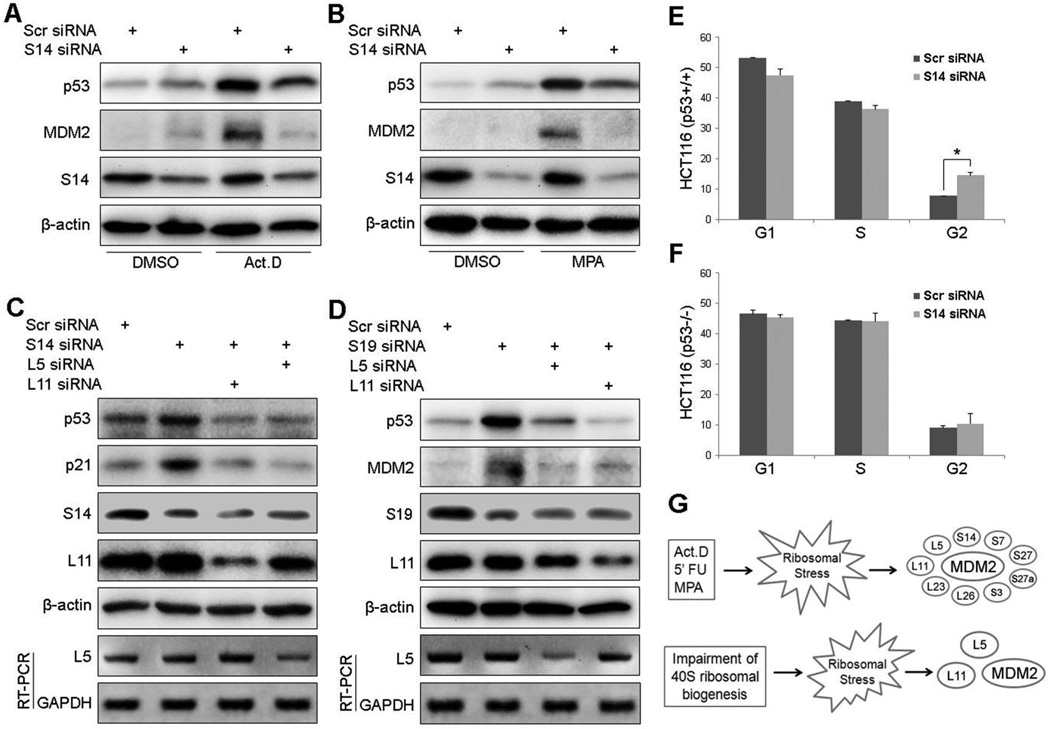
To demonstrate the physiological impact of endogenous RPS14 on the ribosomal stress-p53 pathway, we employed siRNAs to knock down endogenous RPS14. As shown in Figs. 5A and B, knockdown of RPS14 in HCT116 or A549 cells induced p53 compared to the scrambled siRNA control, as expected (41, 44). Consistently, depletion of RPS14 caused p53-dependent G2-arrest in HCT116 cells (Figs. 5E and 5F). Nevertheless, while cells were under ribosomal stress, such as treatment with 5 nM actinomycin D (Fig. 5A) or 10 µM mycophenolic acid (MPA) (Fig. 5B), the reduction of RPS14 by siRNAs partially lessened the induction of p53 by these reagents as reflected in the decrease of the p53 level and activity (as measured by MDM2 level). These results clearly demonstrate that the ribosomal stress-caused p53 activation in part requires RPS14 in cells.
Figure 5. Regulation of p53 activity by knocking down endogenous RPS14 or RPS19.
(A) and (B) RPS14 knockdown activates p53 but also inhibits ribosomal stress-induced p53 activation. HCT116p53+/+ cells (A) and A549 cells (B) were transfected with scrambled siRNAs or RPS14 siRNAs. Cells were treated with 5 nM actinomycin D (Act D) or 10 µM mycophenolic acid (MPA) before harvesting for IB with antibodies as indicated. (C) RPS14 knockdown results in RPL5- and RPL11-dependent p53 activation. A549 cells were transfected with combinations of scrambled siRNAs, RPS14 siRNAs, RPL5 siRNAs or RPL11 siRNAs. Expression of p53, p21, RPS14, RPL11 and β-actin were detected by IB using antibodies as indicated. Messenger RNA levels of RPL5 and GAPDH were detected by RT-PCR. (D) Knockdown of RPS19 results in RPL5- and RPL11-dependent p53 activation. The same experiment as shown in panel C was conducted except the RPS19 siRNAs were used here. Expression of p53, MDM2, RPS19, RPL11 and β-actin were detected by IB using antibodies as indicated. Messenger RNA levels of RPL5 and GAPDH were detected by RT-PCR. (E) (F) Knockdown of RPS14 causes p53-dependent cell cycle arrest. HCT116p53+/+ cells (E) and HCT116p53−/− cells (F) were transfected with scrambled siRNAs or RPS14 siRNAs for 48 h, followed by FACS analysis. The mean percentages of cells in each phase obtained from three separate experiments are presented. Bars indicate standard deviations. “*” indicates P<0.05. (G) A schematic model for p53 activation by general ribosomal stress (upper panel) or 40S subunit perturbation (lower panel).
To test whether this requirement of RPS14 for p53 activation is only responsive to ribosomal stress or not, we conducted the same experiments as shown in Figs. 5A and B except for using Doxorubicin as a DNA damage inducer. Interestingly, ablation of RPS14 impaired the induction of MDM2, although failed to affect p53, in response to DNA damage stress (Fig. S4). Furthermore, we found that the half-life of MDM2 upon both ribosomal stress and DNA damage decreased in response to RPS14 ablation regardless of whether p53 half-life reduced or not (Fig. S5). At last, overexpression of RPS14 in p53 null H1299 cells led to the increase of MDM2 levels in a dose-dependent manner, but independently of p53 (Fig. S6A), and this increase was not at RNA levels (data not shown). The induction of MDM2 levels was also reported when RPS7 (36) or RPL11 (48) was overexpressed. However, overexpression of RPS14, like RPL11 (48), resulted in the accumulation of ubiquitinated forms of MDM2 (Fig. S6B), suggesting that RPS14 may regulate MDM2 stability via a post-ubiquitination mechanism similar to that for RPL11 (48). Together with the above results (Figs. 1–5 and S1–S6), these data suggest that RPS14 regulates the stability of p53 and that of MDM2 through distinct mechanisms.
Ablation of endogenous RPS14 or RPS19 results in RPL5- and RPL11- dependent p53 activation
The RPS19 gene is one of the mostly mutated RP genes in DBA patients with the defect in erythroid differentiation (42) analogous to 5q-syndrome patients with RPS14 haploinsufficiency (41). The p53 activation has been associated with these ribosomopathies (41). Also, the results of Figs. 5A and 5B showed that knockdown of RPS14 indeed induced p53 in cells. It is likely that haploinsufficiency of each of the RPs might give rise to ribosomal stress and activate p53 through an RPL5- and/or RPL11 dependent mechanism. To test this idea, we performed siRNA-mediated ablation of RPS14 or RPS19. Indeed, reduction of either RPS14 or RPS19 by siRNAs activated p53 and increased the level of the p53 targets, p21 and MDM2 (Figs. 5C and 5D, lane 1 and 2). Further knockdown of RPL5 or RPL11 by siRNAs markedly inhibited the p53 level that was induced by RPS14 or RPS19 knockdown (Figs. 5C and 5D, lane 3 and 4). These results show that p53 activation induced by knockdown of RPS14 or RPS19 requires RPL5 and RPL11, which is in accordance with recent reports (20, 21).
Discussion
Several RPs have been recently identified as critical regulators of this MDM2-p53 loop in response to ribosomal stress. Here we reveal RPS14, which is highly associated with 5q-syndrome (43, 44), as another novel player of this ribosomal stress-p53 pathway. Supporting this statement are several lines of evidence. First, RPS14 directly interacted with MDM2 in cells and in vitro via the MDM2’s central acidic domain (Figs. 1 and 2). Also, like RPL5 (31), it inhibited MDM2-mediated p53 ubiquitination and degradation, extending p53’s half-life (Fig. 3). Additionally, RPS14 induced p53-dependent G1 and G2 arrest (Fig. 4). Conversely, ablation of RPS14 by siRNAs hampered the p53 activation induced by ribosomal stress (Figs. 5A and 5B) but not DNA damage (Fig. S4). These results clearly demonstrate that RPS14 is a new player of the ribosomal stress-p53 pathway.
However, interestingly, knockdown of RPS14 also led to p53 activation (Fig. 5A and 5B), similar to the case of RPL23 (32, 33). This p53 activation was RPL5- and RPL11-dependent (Fig. 5C), which is consistent with a recent study in human blood cells (49). Although overexpression of RPS19 did not affect p53 (Figs. 1 and 3), its knockdown, similar to RPS14 knockdown, also activated p53 dependently of RPL11 and RPL5 (Fig. 5D). These results together with studies by others (20, 49) suggest a testable model in the future, i.e., reducing the level of each of these 40S subunits may cause p53-activating ribosomal stress by interfering with 40S assembly.
Identification of RPS14 as another MDM2 suppressor, in addition to more than half a dozen of previously and recently reported MDM2 regulators as aforementioned, in response to ribosomal stress raises one simple question. Why would mammalian cells need so many RPs to overcome the negation of p53 by MDM2? One possibility would be that MDM2 might work as a multiple subunit complex, such as a homohexamer (50, 51), to inactivate p53 in cells. Thus, individual RPs would need to work together as a sub-ribosomal complex or to independently bind to different MDM2 molecules in the homohexameric complex in order to efficiently inactivate MDM2 and consequently activate p53 (Fig. 5G). Alternatively, different MDM2-binding RPs may act at different time points in a sequential manner to inactivate MDM2 upon ribosomal stress dependent on which RP is released early post-stress. Also, different ribosomal stress signals may utilize different MDM2-binding RPs to inhibit MDM2. For example, as aforementioned (Figs. 5C, 5D and 5G), RPS14 or RPS19 knockdown induced p53 by employing RPL11 and RPL5 to inactivate MDM2, as RPS14 would not be available in this case (Fig. 5G), similar to the case of RPS6 knockdown (20). Interestingly, impairing the 60S biogenesis by knocking down RPL30 and RPL29 also required RPL11 and RPL5 for p53 activation (21). Since the cancer-derived mutation C305F in the zinc finger domain of MDM2, which disrupted RPL11- and RPL5-MDM2, but not RPL23-MDM2, interaction, attenuated the p53 activation induced by Act.D, 5-FU, and MPA in vivo (40), it is likely that RPL11 and RPL5 may play a central role in the choreographic formation of the MDM2-interating sub-ribosomal complex in response to ribosomal stress.
However, most of the known MDM2-binding RPs bind to the central acidic domain of MDM2 except RPL11 (31–35, 52). This highlights the importance of this domain, in addition to Zinc finger domain, of MDM2 in sensing ribosomal stress. It remains to be investigated whether any modifications or mutations in this acidic domain might affect the binding of specific RPs to MDM2. Multiple sites of the central acidic domain of MDM2 have been shown to be phosphorylated by casein kinase I in response to genotoxic stresses (53), leading to enhanced binding of SCFβ-TRCP E3 ligase (53) to MDM2. As previously reported, multiple genotoxic insults could also cause ribosomal stress (14), so, may the modifications influence the interaction between RPs and MDM2? Inversely, would the binding of RPs to the central domain of MDM2 interfere with the binding of SCFβ-TRCP E3 ligase to MDM2, leading to stabilization of MDM2 (53)? In addition, it has been shown that ribosomal stress elevated p53 acetylation levels (54), as thus, it remains to be tested if MDM2 could be acetylated under ribosomal stress and, if so, acetylated MDM2 would affect the RP-MDM2 interactions or not. Recently, neddylation of RPL11 was shown to regulate RPL11 stability and its ability to activate p53 in response to ribosomal stress (55). By contrast, RPL26 or RPS7 was reported to be ubiquitinated by MDM2 for degradation (36, 52). Finally, it would be interesting to see if mutations in the central acidic domain of MDM2, which might disrupt the RP-MDM2 interactions, could be identified in human cancers. Of note, RPS14, like RPL11 (48) and RPS7 (36), could also induce the protein level of MDM2 via a post-ubiquitination mechanism independently of p53 (Figs. S4–S6). This finding implies that these RPs might block the recruitment of MDM2 to the 26S proteosomal machinery by interfering with the interaction of MDM2 with the lid subunits, such as S2 (56), of the 26S proteasome. These are enticing questions for future studies.
Materials and Methods
Cell lines, plasmids, and antibodies
Human 293, H1299, A549 and HCT116 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 50 U/ml penicillin and 0.1 mg/ml streptomycin at 37°C in a 5% CO2 humidified atmosphere. Flag-tagged RPS14 and RPS19 expression plasmids were generated by inserting the full-length cDNAs amplified by RT–PCR from HCT116 cells into the pcDNA3-2Flag vectors. The primers used were: P1: 5’-CGCGGATCCATGGCACCTCGAAAGGGGA-3’ and P2: 5’-CCGGAATTCTCACAGACGGCGACCACGGCGA-3’ for RPS14, P3: 5’ CGCGGATCCATGCCTGGAGTTACTGTAA 3’ and P4: 5’ CCGGAATTCCTAATGCTTCTTGTTGGCAGCT 3’ for RPS19. The His-tagged RPS14 plasmid for bacterial expression was generated by engineering the RPS14 full-length cDNAs from pcDNA3-2Flag-RPS14 to the pET-30a vector using BamHI and EcoRI restriction sites. A set of Flag-tagged deletion mutant RPS14 expression plasmids were generated by inserting the nucleotides 1–150 or 151–453 of full-length cDNAs into the pcDNA3-2Flag vectors. The primers used were: P1 and P5: 5' CCGGAATTCTCACTTGCCAGAAAGATCAGTGA 3' for RPS14/1–50; P6: 5' CGCGGATCCGAAACCATCTGCCGTGTGAC 3' and P2 for RPS14/51–151. EGFP-tagged RPS14 was generated using primers: P7: 5' CCGGAATTCTATGGCACCTCGAAAGGGGA 3' and P8: 5' CGCGGATCCTCACAGACGGCGACCACGGCGA 3'. Flag-L5, Flag-L11 GFP-L23, HA-MDM2, GST-MDM2, deletion mutants of GST- MDM2 and V5-MDM2, Myc-MDMX, His-Ub, p53 pEGFP-p53 and GST-p53 coding plasmids have been described previously (31, 32). Anti-L5 (31), anti-L11 (57), anti-HA (12CA5),anti-MDM2 (2A10 and 4B11) (31, 32) and anti-MDMX (8C6) (56) antibodies have been previously described. Anti-Flag (Sigma), anti-HA (F-7, Santa Cruz Biotechnology), anti-p21 (M-19, Santa Cruz Biotechnology), anti-Bax (N-20, Santa Cruz Biotechnology), anti-Parp (Cell Signaling), anti-p53 (DO-1, Santa Cruz Biotechnology), anti-RPS14 (H-130, Santa Cruz Biotechnology), anti-RPS19 (WW-4, Santa Cruz Biotechnology) and anti-BrdU (IIB5, Santa Cruz Biotechnology) were commercially purchased.
GST fusion protein association assay
His-tagged RPS14 proteins were expressed in E. coli, purified through a Ni-NTA (QIAGEN) column, and eluted with 0.5 M imidazole. Protein-protein interaction assays were conducted as described previously by using fusion protein-containing glutathione beads (58). Purified His-tagged RPS14 proteins were incubated with the glutathione-Sepharose 4B beads (Sigma) containing 500 ng of GST-MDM2/1–491, GST-MDM2/1–151, GST-MDM2/1–301, GST-MDM2/285–340, GST-MDM2/294–491, or GST. Thirty minutes after incubation at room temperature, the mixtures were washed twice in lysis buffer containing 10% glycerol and once in SNNTE buffer. Bound proteins were analyzed on a 15% SDS gel and detected by IB with the anti-RPS14 polyclonal antibody.
Transient transfection, immunoblot and co-immunoprecipitation analyses
Cells were transfected with plasmids as indicated in figure legends using TransFectin lipid reagent following the manufacturer’s protocol (Bio-Rad). The cells were harvested at 30 to 48 h post-transfection and lysed in lysis buffer consisting of 50 mM Tris/HCl (pH 8.0), 0.5% Nonidet P-40 (NP-40), 1 mM EDTA, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), 0.25 µg/ml pepstatin A, and 1 mM leupeptin. Equal amounts of clear cell lysate (20 to 50 µg) were used for immunoblot (IB) analysis as described previously (59). Immunoprecipitation (IP) was conducted by using antibodies as indicated in the figure legends and described previously (60). Beads were washed twice with lysis buffer, once with SNNTE buffer (50 mM Tris-HCl (pH 7.4), 5 mM EDTA, 1% Nonidet P-40, 500 mM NaCl, and 5% sucrose). Bound proteins were detected by IB with antibodies as indicated in the figure legends.
In vivo ubiquitination assay
H1299 cells were transfected with His-Ub (1.5 µg), p53 (0.5 µg), HA-MDM2 (1 µg), or Flag-RPS14 expression plasmids as indicated in Fig. 3. At 48 h after transfection, cells were harvested and split into two aliquots, one for IB and the other for ubiquitination assays. In vivo ubiquitination assays were conducted as previously described (32). Eluted proteins were analyzed by IB with monoclonal p53 antibodies.
Cell cycle analysis
Cells transfected with pcDNA, Flag-RPS14, pEGFP, pEGFP-RPS14, scrambled siRNAs or RPS14 siRNAs as indicated in the figure were fixed and stained in 500 µl of propidium iodide (PI, Sigma) stain buffer (50 µg/ml PI, 200 µg/ml RNase A, 0.1% Triton X-100 in phosphate-buffered saline) at 37 °C for 30 min. The cells were then analyzed for DNA content using a BD Biosciences FACScan flow cytometer. Data were analyzed using the CellQuest and Modfit software programs.
BrdU incorporation assays
BrdU incorporation assays were conducted as described previously (57). Cells were incubated in the presence of 10 µM of BrdU for 5 h. Cells were then fixed with 95% of ethanol and 5% of acetic acid, treated with 2M HCl containing 1% Triton X-100, and stained with the monoclonal anti-BrdU antibody (Santa Cruz Biotechnology), followed by staining with Alexa Fluor 546 (red) goat anti-mouse antibodies and DAPI. Stained cells were analyzed under a Zeiss Axiovert 25 fluorescent microscope.
Cell viability assays
To assess the long term cell growth, the Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies Inc., Gaithersburg, Maryland) was used according to the manufacturer’s instructions. Cell suspensions were seeded at 3,000 cells per well in 96-well culture plates at 24 h post-transfection. Cell growth inhibition was determined by adding WST-8 at a final concentration of 10% to each well, and the absorbance of the samples was measured at 450 nm using a Microplate Reader (Molecular Device, SpecrtraMax M5e) after culture for 5 days.
Supplementary Material
Acknowledgements
We want to thank Steven Ellis for reagents and discussion. This work was supported in part by NIH-NCI grants CA095441, CA 079721 and CA129828 to H.L.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Sharpless NE, DePinho RA. p53: good cop/bad cop. Cell. 2002;110(1):9–12. doi: 10.1016/s0092-8674(02)00818-8. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 3.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes & development. 1993;7(7A):1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 4.Juven T, Barak Y, Zauberman A, George DL, Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8(12):3411–3416. [PubMed] [Google Scholar]
- 5.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362(6423):857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 6.Poyurovsky MV, Katz C, Laptenko O, Beckerman R, Lokshin M, Ahn J, et al. The C terminus of p53 binds the N-terminal domain of MDM2. Nature structural & molecular biology. 2010;17(8):982–989. doi: 10.1038/nsmb.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 8.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs SY, Adler V, Buschmann T, Wu X, Ronai Z. Mdm2 association with p53 targets its ubiquitination. Oncogene. 1998;17(19):2543–2547. doi: 10.1038/sj.onc.1202200. [DOI] [PubMed] [Google Scholar]
- 10.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378(6553):203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 11.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378(6553):206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 12.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer cell. 2009;16(5):369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Molecular cell. 2010;40(2):216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashcroft M, Taya Y, Vousden KH. Stress signals utilize multiple pathways to stabilize p53. Molecular and cellular biology. 2000;20(9):3224–3233. doi: 10.1128/mcb.20.9.3224-3233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. The EMBO journal. 2006;25(23):5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. The Journal of biological chemistry. 2007;282(11):8052–8059. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- 18.Sun XX, Dai MS, Lu H. Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. The Journal of biological chemistry. 2008;283(18):12387–12392. doi: 10.1074/jbc.M801387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. The EMBO journal. 2004;23(12):2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, et al. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nature cell biology. 2009;11(4):501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun XX, Wang YG, Xirodimas DP, Dai MS. Perturbation of 60 S ribosomal biogenesis results in ribosomal protein L5- and L11-dependent p53 activation. The Journal of biological chemistry. 2010;285(33):25812–25821. doi: 10.1074/jbc.M109.098442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holzel M, Orban M, Hochstatter J, Rohrmoser M, Harasim T, Malamoussi A, et al. Defects in 18 S or 28 S rRNA processing activate the p53 pathway. The Journal of biological chemistry. 2010;285(9):6364–6370. doi: 10.1074/jbc.M109.054734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Molecular and cellular biology. 2001;21(13):4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, et al. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Molecular cell. 2003;12(5):1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 25.Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Molecular and cellular biology. 2008;28(13):4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu W, Qiu Z, Gao N, Wang L, Cui H, Qian Y, et al. PAK1IP1, a ribosomal stress-induced nucleolar protein, regulates cell proliferation via the p53-MDM2 loop. Nucleic acids research. 2011;39(6):2234–2248. doi: 10.1093/nar/gkq1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Llanos S, Serrano M. Depletion of ribosomal protein L37 occurs in response to DNA damage and activates p53 through the L11/MDM2 pathway. Cell cycle. 2010;9(19):4005–4012. doi: 10.4161/cc.9.19.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer cell. 2003;3(6):577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki M, Kawahara K, Nishio M, Mimori K, Kogo R, Hamada K, et al. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nature medicine. 2011;17(8):944–951. doi: 10.1038/nm.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, et al. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Molecular and cellular biology. 2003;23(23):8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. The Journal of biological chemistry. 2004;279(43):44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 32.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Molecular and cellular biology. 2004;24(17):7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin A, Itahana K, O'Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Molecular and cellular biology. 2004;24(17):7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Wang J, Yuan Y, Zhang W, Guan W, Wu Z, et al. Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic acids research. 2010;38(19):6544–6554. doi: 10.1093/nar/gkq536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, et al. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26(35):5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, et al. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Molecular cell. 2009;35(3):316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong X, Zhao Y, He H, Sun Y. Ribosomal protein S27-like and S27 interplay with p53-MDM2 axis as a target, a substrate and a regulator. Oncogene. 2011;30(15):1798–1811. doi: 10.1038/onc.2010.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun XX, DeVine T, Challagundla KB, Dai MS. Interplay between ribosomal protein S27a and MDM2 protein in p53 activation in response to ribosomal stress. The Journal of biological chemistry. 2011;286(26):22730–22741. doi: 10.1074/jbc.M111.223651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Molecular and cellular biology. 2007;27(3):1056–1068. doi: 10.1128/MCB.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS, et al. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer cell. 2010;18(3):231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115(16):3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nature genetics. 1999;21(2):169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 43.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451(7176):335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, Lane AL, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nature medicine. 2010;16(1):59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gazda HT, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, Schneider H, et al. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. American journal of human genetics. 2008;83(6):769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Petrtylova K, Mihal V, et al. Identification of mutations in the ribosomal protein L5 (RPL5) and ribosomal protein L11 (RPL11) genes in Czech patients with Diamond-Blackfan anemia. Human mutation. 2009;30(3):321–327. doi: 10.1002/humu.20874. [DOI] [PubMed] [Google Scholar]
- 47.Yadavilli S, Mayo LD, Higgins M, Lain S, Hegde V, Deutsch WA. Ribosomal protein S3: A multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA repair. 2009;8(10):1215–1224. doi: 10.1016/j.dnarep.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai MS, Shi D, Jin Y, Sun XX, Zhang Y, Grossman SR, et al. Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. The Journal of biological chemistry. 2006;281(34):24304–24313. doi: 10.1074/jbc.M602596200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, et al. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117(9):2567–2576. doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poyurovsky MV, Priest C, Kentsis A, Borden KL, Pan ZQ, Pavletich N, et al. The Mdm2 RING domain C-terminus is required for supramolecular assembly and ubiquitin ligase activity. The EMBO journal. 2007;26(1):90–101. doi: 10.1038/sj.emboj.7601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. The EMBO journal. 2007;26(1):102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Molecular cell. 2008;32(2):180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, et al. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer cell. 2010;18(2):147–159. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133(4):612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO reports. 2009;10(10):1132–1139. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jin Y, Zeng SX, Sun XX, Lee H, Blattner C, Xiao Z, et al. MDMX promotes proteasomal turnover of p21 at G1 and early S phases independently of, but in cooperation with, MDM2. Molecular and cellular biology. 2008;28(4):1218–1229. doi: 10.1128/MCB.01198-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dai MS, Arnold H, Sun XX, Sears R, Lu H. Inhibition of c-Myc activity by ribosomal protein L11. The EMBO journal. 2007;26(14):3332–3345. doi: 10.1038/sj.emboj.7601776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin Y, Zeng SX, Dai MS, Yang XJ, Lu H. MDM2 inhibits PCAF (p300/CREB-binding protein-associated factor)-mediated p53 acetylation. The Journal of biological chemistry. 2002;277(34):30838–30843. doi: 10.1074/jbc.M204078200. [DOI] [PubMed] [Google Scholar]
- 59.Zeng X, Li X, Miller A, Yuan Z, Yuan W, Kwok RP, et al. The N-terminal domain of p73 interacts with the CH1 domain of p300/CREB binding protein and mediates transcriptional activation and apoptosis. Molecular and cellular biology. 2000;20(4):1299–1310. doi: 10.1128/mcb.20.4.1299-1310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zeng SX, Dai MS, Keller DM, Lu H. SSRP1 functions as a co-activator of the transcriptional activator p63. The EMBO journal. 2002;21(20):5487–5497. doi: 10.1093/emboj/cdf540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.