Abstract
Expression of p16INK4A (p16 positive) is highly correlated with human papilloma virus (HPV) infection in head and neck squamous cell carcinoma (HNSCC), however, p16-positivity is not limited to HPV positive tumors and therefore, not a perfect surrogate for HPV. p16 survival outcomes are best documented for the oropharyngeal site (OP); non-OP sites such as the oral cavity (OC), larynx, and hypopharynx (HP) are understudied. The goal of this study was to evaluate p16 in the context of HPV16 and examine p16 survival outcomes in HPV16 positive and HPV16 negative site-specific HNSCC. p16 and HPV16 status were determined by immunohistochemistry and qPCR respectively, on 80 primary HNSCC from four sites: OC, OP, larynx and HP. p16 expression was different across sites (p<0.001), was more frequent in OP than non-OP sites (p<0.0001), and was different between Caucasian Americans (CA) and African Americans (AA) (p=0.031), similar to HPV (p=0.013). p16 was associated with marital status (p=0.008) and smoking (p=0.014). p16 positive patients had improved survival (similar to HPV16 positive cases). Patients with p16 negative/HPV16 negative status had the worst survival for all sites combined as well as for OP. p16 status is an important prognostic indicator in both OPSCC and non-OPSCC and the p16 positive/HPV16 negative group is likely a distinct subgroup lacking any HPV genotype. Cohorts with larger representations of non-OP sites examining multiple molecular markers will be key to deciphering and dissecting out p16’s role as a useful prognostic indicator when assessed in combination with HPV status.
Keywords: p16 expression, HPV, head and neck cancer, non-oropharyngeal sites
1. Introduction
In the United States, approximately 52,610 new cases of head and neck squamous cell carcinoma (HNSCC) are expected in 2012 with an estimated 11,500 deaths from cancers of the oral cavity, pharynx, and larynx (American Cancer Society [ACS], 2012). Despite considerable efforts, the 5-year survival rate for HNSCC has not changed significantly. In addition to tobacco and alcohol (Hashibe et al., 2007), epidemiological and laboratory evidence now warrant the conclusion that the human papilloma virus (HPV) is a causative agent for some HNSCC (Gillison & Lowy, 2004) and an independent risk factor for oropharyngeal HNSCC (OPSCC) (Ang et al., 2010). A systematic review of 5046 patients with HNSCC reported an overall prevalence of HPV infection of 25.9%. The prevalence of HPV infection was significantly higher among patients with OPSCC (35.6%) than among those with oral (23.5%) or laryngeal (24.0%) SCC (Kreimer et al., 2005). The biologic significance of HPV, underscored by the improved prognosis for patients with HPV positive HNSCC relative to HPV negative HNSCC (Ang et al., 2010), is due in part to a better therapeutic response to chemo radiotherapy (Fakhry et al., 2008).
In addition to the above mentioned risk factors, an important gene product that is involved in HNSCC pathogenesis is the p16INK4a (p16) protein, made by the p16INK4a (CDKN2A) gene located at 9p21. p16 is a cyclin-dependent kinase inhibitor that inhibits pRb phosphorylation and blocks cell cycle progression at the G1 to S check point (Zhang et al., 1999). Loss of p16 expression by deletion, mutation, or hypermethylation is common in HNSCC (Worsham et al., 2006) and is associated with worse prognosis in HNSCC (Namazie et al., 2002). Also, tobacco/alcohol-associated HNSCC appears to be associated with p16 downregulation (as an early event) and TP53 gene mutations resulting in p53 overexpression (Psyrri & DiMaio, 2008). Loss of p16 is a potential druggable target. Recombinant adenovirus capable of directing a high level of p16 protein expression (Ad5-p16) demonstrated a significant antitumor effect of Ad5-p16 against human HNSCC in vivo (Rocco et al., 1998).
Similar to HPV positive status, p16 expression (p16 positive) has been correlated with improved outcome in OPSCC (Weinberger et al., 2004). Expression of p16 occurs as a result of functional inactivation of the retinoblastoma protein (pRb) by the HPV E7 protein. HPV viral oncogene products E6 and E7 play a key role in HPV-associated carcinogenesis, abrogating p53 and retinoblastoma (Rb) tumor suppressor functions, respectively (Munger et al., 2004). HPV positive HNSCC are generally associated with wild-type TP53, as opposed to tobacco-induced tumors, which are characterized by genetic alterations of the TP53 pathway (Braakhuis et al., 2004). The functional inactivation of Rb by E7 leads in turn to upregulation of p16. Thus HPV positive tumors are characterized by high expression of p16 (Fakhry et al., 2008; Smeets et al., 2007). Moreover, because transcription of the E7 oncogene is required for p16 upregulation, it has been suggested that carcinomas overexpressing p16 likely represent those tumors in which HPV has been involved in the carcinogenic process (von Knebel Doeberitz, 2002). Thus, there is good evidence that p16 positivity may be regarded as a biomarker for tumors harboring oncogenic HPV. Though p16 expression is highly correlated with HPV infection especially in OPSCC (Lassen et al., 2009), making it a useful surrogate marker for HPV infection, in non-oropharyngeal sites this has yet to be demonstrated. The goal of this study was to evaluate p16 status in the context of HPV16 and examine p16 survival outcomes in HPV16 positive and HPV16 negative site-specific HNSCC.
2. Materials and Methods
2.1 Cohort
The site-specific, retrospective pilot cohort of 80 primary HNSCC consisted of 10 HPV16 positive and 10 HPV16 negative cases each for four sites: oral cavity (OC), oropharynx (OP), larynx, and hypopharynx (HP). The 10 HPV16 positive and 10 HPV16 negative cases for each of the four investigated tumor sites were selected from a larger HNSCC cohort with already available HPV status by qPCR. Patients were diagnosed within the Henry Ford Health System between 1986 and 2003, and followed from 6-23 years (through 2009). Risk factor information of race (as self reported), age, gender, marital status, and smoking history was obtained through medical record abstraction. This study was approved by the Henry Ford Health System Institutional Review Board committee.
2.2 DNA Extraction
DNA was extracted from formalin-fixed tissue blocks. Upon hematoxylin and eosin staining, the pathologist determined tumor content in the tumor block. For tumor tissue blocks with 90% or more tumor, 5 micron tissue sections were cut directly into tubes for DNA extraction. If the tissue block indicated admixture of tumor and normal or other neoplastic lesions, tumor tissue was micro-dissected for DNA extraction and thus separated from non-tumor tissue. DNA was extracted as previously described (Raju et al., 2006).
2.3 p16 Immunohistochemistry (IHC) assay
IHC for p16 status was performed using the DAKO EnVision™ FLEX+ detection system together with the Autostainer Link instrument (DAKO Corp, Carpentaria, California) on paraffin embedded tissue. Antigen was retrieved using EnVision™ FLEX Target Retrieval Solution, High pH and p16 was detected using p16 mouse monoclonal antibody (clone 1E12E10, sc-81156, 1:1000 dilution; Santa Cruz Biotechnology, Inc, Santa Cruz, California). The EnVision™ FLEX+, Mouse, High pH, (LINK) Kit was used to perform the assay. It contains the substrate chromogen 3-3′-diaminobenzidine (DAB), which, on staining results in a brown-colored precipitate at the antigen site. For statistical analysis, samples are classified in a binary manner as either positive (any cells with nuclear and cytoplasmic staining) or negative (Figure 1).
Figure 1. p16 by IHC for two patient samples.
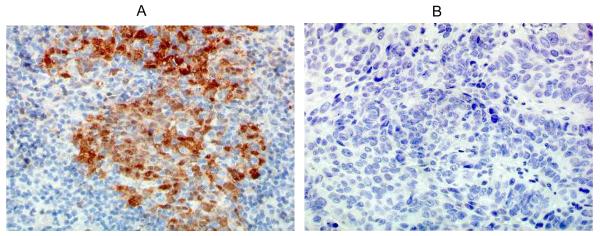
(A) Case 1 shows brown nuclear and cytoplasmic staining indicating p16 positive status; (B) Case 2 shows no staining indicating p16 negative status. 40X magnification. (IHC - immunohistochemistry)
2.4 HPV16 Detection by Quantitative PCR (qPCR)
Tumor HPV16 DNA concentrations were measured using a real-time quantitative PCR system (qPCR). An advantage of the qPCR system is that detection of HPV copy number reliably excludes contaminated samples and those with minimal HPV DNA content that are unlikely to be significant in tumorigenesis and it corresponds to FISH and Southern Blotting methods (Ha et al., 2002). Real-time PCR reactions were set up in a reaction volume of 20ul using the TaqMan Universal Master Mix II with UNG (Applied Biosystems, Foster City, CA). Specific primers and probes were designed to amplify a housekeeping gene, ß-globin (to standardize the input DNA), and the E6 region of HPV16. The primers and probes used were as follows: ß-globin: forward 5′-GTGCACCTGACTCCTGAGGAGA-3′, reverse 5′-CCTTGATACCAACCTGCCCAG-3′, 6FAM-5′-AAGGTGAACGTGGATGAAGTTGGTGG-3′-TAMRA; HPV16 E6 : forward 5′-GAGAACTGCAATGTTTCAGGACC-3′, reverse 5′-TGTATAGTTGTTTGCAGCTCTGTGC-3′, 6FAM-5′-CAGGAGCGACCCAGAAAGTTACCACAGTT-3′-TAMRA. DNA amplifications were carried out in a 96-well reaction plate format in a PE Applied Biosystems 7900HT Sequence Detector (Applied Biosystems, Foster City, CA). HPV16 viral copy number was determined using the CaSki (American Type Culture Collection, Manassas, VA) cell line genomic DNA reported to contain an integrated HPV type 16 genome (about 600 copies per cell, 6.6 pg of DNA/genome) in serial dilutions to develop a standard curve. Multiple water blanks and HPV16 positive controls (HPV16 positive HNSCC specimens) were included in every run. Both the HPV16 and ß-globin PCR reactions were carried out in duplicate. The qPCR cut-off value for HPV16 positive infection status of ≥0.03 (>3 HPV16 genome copy/100 cells) was determined as previously described (Stephen et al., 2012). The HPV16 status of this cohort was treated as a binary variable (positive vs negative) based on this cut-off value.
2.5 Statistical Analysis
Chi-square tests were used to test associations of p16 and HPV16 with the other risk factors (race, stage, gender, smoking, and tumor site). T-tests were used to test associations with age. Variables with p<0.05 were considered significant. Kaplan-Meier survival estimates were computed and plotted. Cox regression was used to estimate hazard ratios and test associations between p16 positive and HPV16 positive status and overall survival for all sites combined and for the OP subgroup.
3. Results
Race and stage distributions for this cohort were as follows: 40 Caucasian American (CA), 38 African American (AA), 2 other race; 19 early stage, 53 late stage and 8 unknown stage (Table 1).
Table 1.
Cohort Characteristics
| Characteristics | Total | p16+ve | p16−ve | p-value | HPV16+ve | HPV16−ve | p-value |
|---|---|---|---|---|---|---|---|
|
| |||||||
| N=80 | N=21 (%) | N=59 (%) | N=40 (%) | N=40 (%) | |||
| Gender | |||||||
| Male | 66 | 16 (24) | 50 (76) | 0.376 | 34 | 32 | 0.556 |
| Female | 14 | 5 (36) | 9 (64) | 6 | 8 | ||
| Age Group | |||||||
| ≤50 years | 12 | 5 (42) | 7 (58) | 0.302 | 8 (67) | 4 (33) | 0.293 |
| 51-65 years | 32 | 9 (28) | 23 (72) | 17 (53) | 15 (47) | ||
| >65 years | 36 | 7(19) | 29 (81) | 15 (42) | 21 (58) | ||
| Marital Status ∞ | |||||||
| Unmarried | 28 | 12 (43) | 16 (57) | 0.008† | 12 (43) | 16 (57) | 0.254 |
| Married | 46 | 7 (15) | 39 (85) | 26 (57) | 20 (43) | ||
| Smoking | |||||||
| Never smoker | 10 | 6 (60) | 4 (40) | 0.014† | 7 (70) | 3 (30) | 0.455 |
| Past smoker | 27 | 9 (33) | 18 (67) | 12 (44) | 15 (56) | ||
| Current smoker | 39 | 6 (15) | 33 (85) | 20 (51) | 19 (49) | ||
| Race * | |||||||
| Caucasian American (CA) | 40 | 15 (38) | 25 (62) | 0.031† | 26 (65) | 14 (35) | 0.013† |
| African American (AA) | 38 | 6 (16) | 32 (84) | 14 (37) | 24 (63) | ||
| p16 Status | |||||||
| Positive | 21 | 12 (57) | 9 (43) | 0.446 | |||
| Negative | 59 | 28 (47) | 31(53) | ||||
| HPV16 Status | |||||||
| Positive | 40 | 12 (30) | 28 (70) | 0.446 | |||
| Negative | 40 | 9 (23) | 31 (77) | ||||
| Sites | |||||||
| Oropharynx | 20 | 13 (65) | 7 (35) | <0.001† | 10 (50) | 10 (50) | 1.000 |
| Non-Oropharynx | 60 | 8 (13) | 52 (87) | 30 (50) | 30 (50) | ||
| Oral Cavity | 20 | 4 (20) | 16 (80) | 0.004† | 10 (50) | 10 (50) | |
| Larynx | 20 | 3 (15) | 17 (85) | 0.001† | 10 (50) | 10 (50) | |
| Hypopharynx | 20 | 1 (5) | 19 (95) | <0.001† | 10 (50) | 10 (50) | |
| Stage | |||||||
| Early | 19 | 2 (11) | 17 (89) | 0.073 | 8 (42) | 11 (58) | 0.792 |
| Late | 53 | 18 (34) | 35 (66) | 25 (47) | 28 (53) | ||
Marital status missing for 6 cases;
Other race (N=2) are p16 negative, HPV16 negative and are located in Hypopharynx;
significant p-values; OP - oropharynx; p16+ve: p16 positive, p16−ve: p16 negative, HPV16+ve: HPV16 positive, HPV16−ve: HPV16 negative
p16 expression was detected in 21/80 (26%) samples and was significantly different between OP and non-OP sites (p<0.0001) as well as among pairwise site to site comparisons with OP (Table 1). Among individual sites, p16 was positive in 1/20 HP, 3/20 larynx, 4/20 OC and 13/20 OP (Table 1) with a frequency of 65% in OP cases as compared to 13% in non-OP sites. p16 expression was significantly associated with marital status (p=0.008) and smoking (p=0.014). There was no association with gender and age. With respect to race, the frequency of p16 expression was different between AA and CA (p=0.031) across all sites. Race as CA was significantly associated with p16 expression in the OP site (p=0.004). For non-OP sites there was no difference for p16 in AA vs CA (13.3% s 14.3%). There is no association between race and site as OP vs non-OP (p=0.366). A breakdown of race with respect to site, p16 and HPV16 status is provided in Table 2. A trend towards an association was noted for p16 and stage (p=0.073).
Table 2.
Race, Site, p16 and HPV16 status
| Caucasian American | African American | Other | |
|---|---|---|---|
| Sites | |||
| Oropharynx | 12 | 8 | – |
| Hypopharynx | 8 | 10 | 2 |
| Larynx | 9 | 11 | – |
| Oral Cavity | 11 | 9 | – |
| p16 | |||
| Positive | 15 (OP=11, non-OP=4) | 6 (OP=2, non-OP=4) | – |
| Negative | 25 (OP=1, non-OP=24) | 32 (OP=6, non-OP=26) | 2 |
| HPV16 | |||
| Positive | 26 (OP=8, non-OP=18) | 14 (OP=2, non-OP=12) | – |
| Negative | 14 (OP=4, non-OP=10) | 24 (OP=6, non-OP=18) | 2 |
OP - oropharynx
There was no association of HPV16 with gender, age, marital status or smoking. For all sites combined, HPV16 was associated with race as CA (p=0.013). Race differences for HPV16 in OP (25% vs 67% for AA and CA, respectively) as well as in non-OP sites (40% vs 64% for AA and CA, respectively) are substantial; however, the p-values are not significant (p=0.170 and p=0.074 for OP and non-OP respectively). In a logistic regression model for stage, there was no interaction between HPV16 and p16 (p=0.924). In the OP site, HPV16 and p16 were marginally associated (p=0.057, 9/10 HPV16 positive OP tumors were also p16 positive).
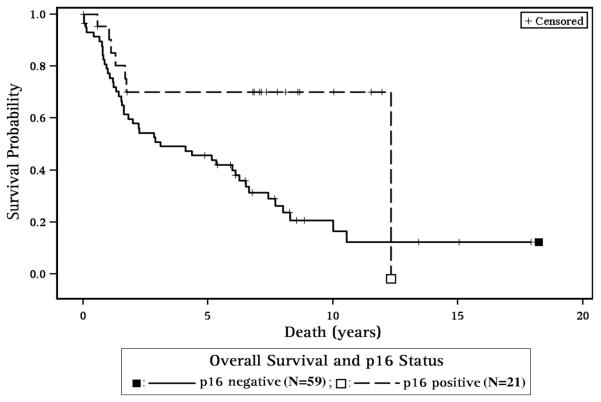
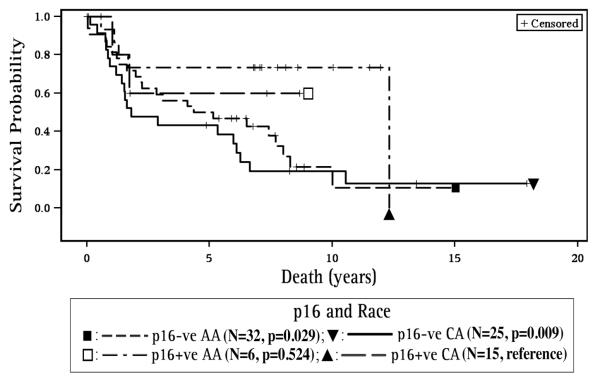
With regard to p16 and survival, p16 positive patients had improved overall survival for all sites combined (HR=0.33, 95% CI 0.15, 0.75, p=0.007, Figure 2, Table 3A) and for the OP site as a subgroup, (HR=0.22, 95% CI 0.06, 0.89, p=0.034) compared to p16 negative patients (Table 3B). For all sites combined (Table 3A), p16 negative CA and AA patients demonstrated poorer survival when compared to p16 positive CA (HR=3.78, 95% CI 1.40, 17, p=0.009 and HR=2.97, 95% CI 1.12, 7.85, p=0.029, respectively, Figure 3).
Figure 2. Survival and p16 status.
p16 positive patients had improved overall survival for all sites combined
Table 3.
p16, HPV16 and Survival
| A) All Sites Combined | N=80 | Hazard Ratio | Confidence Interval | p-value | |
|---|---|---|---|---|---|
|
| |||||
| p16 | p16+ve | 21/80 | 0.33 | 0.15 - 0.75 | 0.007* |
| HPV16 | HPV16+ve | 40/80 | 0.53 | 0.30 - 0.94 | 0.029* |
| p16/HPV | p16+ve/HPV16+ve | 12/80 | 1.00 (ref) | ||
| p16+ve/HPV16−ve | 9/80 | 6.97 | 1.33 - 36.41 | 0.021* | |
| p16−ve/HPV16+ve | 28/80 | 6.82 | 1.58 - 29.44 | 0.01* | |
| p16−ve/HPV16−ve | 31/80 | 8.67 | 2.04 - 36.85 | 0.003* | |
| p16 & Race | p16+ve CA | 15/78 | 1.00 (ref) | ||
| p16+ve AA | 6/78 | 1.71 | 0.33 - 8.86 | 0.524 | |
| p16−ve CA | 25/78 | 3.78 | 1.40 - 10.17 | 0.009* | |
| p16−ve AA | 32/78 | 2.97 | 1.12 - 7.85 | 0.029* | |
| HPV16 & Race | HPV16+ve CA | 26/78 | 1.00 (ref) | ||
| HPV16+ve AA | 14/78 | 1.56 | 0.63 - 3.86 | 0.333 | |
| HPV16−ve CA | 14/78 | 2.91 | 1.26 - 6.70 | 0.012* | |
| HPV16−ve AA | 24/78 | 1.79 | 0.86 - 3.73 | 0.121 | |
|
| |||||
| B) OP site only | N=20 | ||||
|
| |||||
| p16+ve | 13/20 | 0.22 | 0.06 - 0.89 | 0.034* | |
| HPV16+ve | 10/20 | 0.2 | 0.04 - 0.98 | 0.047* | |
| p16+ve/HPV16+ve | 9/20 | 1.00 (ref) | |||
| p16+ve/HPV16−ve | 4/20 | 6.98 | 0.61 - 79.47 | 0.117 | |
| p16−ve/HPV16+ve | 1/20 | 11.44 | 0.69 - 191.0 | 0.09 | |
| p16−ve/HPV16−ve | 6/20 | 10.3 | 1.20 - 88.33 | 0.033* | |
CA - Caucasian American, AA - African American, OP - Oropharynx,
significant p-value; p16+ve: p16 positive, p16−ve: p16 negative, HPV16+ve: HPV16 positive, HPV16−ve: HPV16 negative.
Figure 3. Survival with respect to p16 and Race.
p16 negative CA and AA patients demonstrated poorer survival when compared to p16 positive CA. (CA – Caucasian American, AA – African American, p16+ve – p16 positive, p16-ve – p16 negative,).
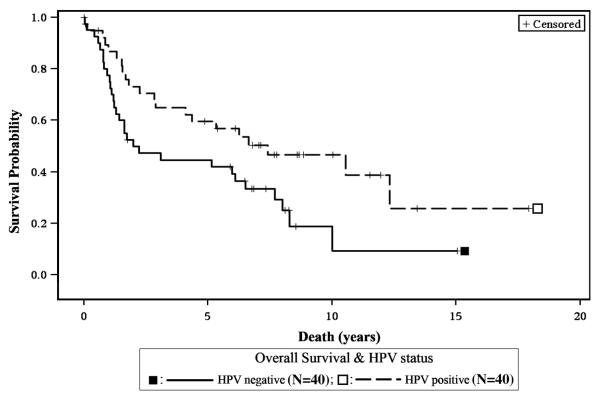
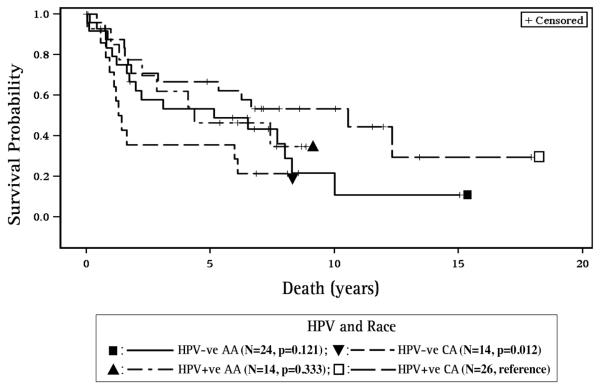
HPV16 positive patients also had better survival for the all sites combined group (HR=0.53, 95% CI 0.30, 0.94, p=0.029, Figure 4) and for the OP subgroup (HR=0.2, 95% CI 0.04, 0.98, p=0.047) when compared to HPV16 negative patients. Additionally, HPV16 positive CA had better survival when compared to HPV16 negative CA (p=0.012, Figure 5).
Figure 4. Survival and HPV16 status.
HPV16 positive patients had better overall survival for the all sites combined group.
Figure 5. Survival with respect to HPV16 and Race.
HPV16 positive CA had better survival when compared to HPV negative CA. (CA – Caucasian American, AA – African American, HPV16+ve – HPV16 positive, HPV16-ve – HPV16 negative).
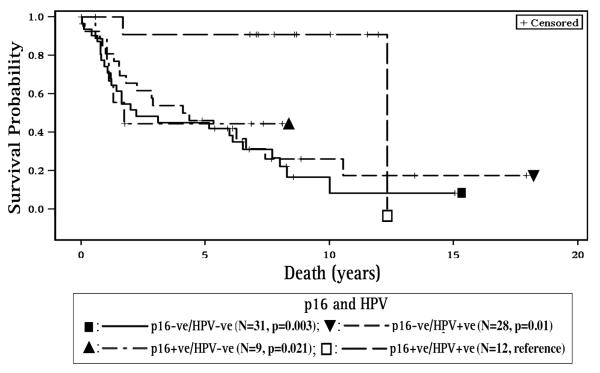
When compared to the p16 positive/HPV16 positive cases, patients with p16 positive/HPV16 negative and p16 negative/HPV16 positive had poorer survival (HR=6.97, 95% CI 1.33, 36.41, p=0.021 and HR=6.82, 95% CI 1.58, 29.44, p=0.010, respectively, Figure 6). p16 negative/HPV16 negative status had the worst survival for the all sites combined group (HR=8.67, 95% CI 2.04, 36.85, p= 0.003) and this was noted for the OP subgroup as well (HR=10.30, 95% CI 1.20, 88.33, p=0.033, Table 3B). The prognostic effects of HPV16 and p16 alone were also analyzed for individual non-OP sites (oral cavity, larynx, hypopharynx), but were not statistically significant.
Figure 6. Survival with respect to combined p16 and HPV16 status.
For all sites combined, when compared to the p16 positive/HPV16 positive cases, patients with p16 positive/HPV16 negative and p16 negative/HPV16 positive had poorer survival. p16 negative/HPV16 negative status had the worst survival. (p16+ve – p16 positive, p16-ve – p16 negative, HPV+ve – HPV16 positive, HPV-ve – HPV16 negative)
4. Discussion
Our exploratory study of a site-specific HNSCC cohort demonstrated a p16 prevalence of 26% with a significant difference between OP and non-OP sites as well as between each of the three non-OP and OP sites. p16 expression was significantly associated with site as OP, marital status and smoking. A trend towards an association between p16 and stage was noted. With regard to race, p16 expression was different between AA and CA across all sites. However, race as CA was significantly associated with p16 expression only in the OP site. In non-OP sites there was no difference in p16 expression with respect to race.
We found a lower prevalence of HPV16 in AA compared to CA, similar to a previous study by Settle et al. (2009a). Chernock et al. (2011) and Weinberger et al. (2010) detected significantly lower prevalence for HPV and p16 in AA vs CA concurring with our findings. Also, even though cases were pre-selected based on HPV16 status, p16 expression and HPV16 positive status appeared to be correlated (p=0.057) in the OP site, similar to other reports (Singhi & Westra, 2010; Shi et al., 2009; Lassen et al., 2009). There was no interaction between HPV16 and p16 with respect to stage, but in the OP site, HPV16 and p16 were marginally associated.
This current study indicates that p16 status is an important prognostic indicator in both OPSCC and non-OPSCC. Single marker analyses showed that patients with p16 positive status had improved survival similar to HPV16 positive cases for all sites combined and for the OP subgroup as well. Weinberger et al. (2004) also found that p16 was associated with improved prognosis as well as lower local recurrence rates in OPSCC. In our study, p16 negative status by itself was associated with poorer survival in both CA and AA for all sites combined, as well as for the OP subsite, similar to the study by Chernock et al. (2011), which demonstrated poorer disease free survival in AA and a trend toward poorer overall survival for AA.
Also, HPV16 negative status was associated with poorer survival for all sites combined and for the OP subgroup as well. With respect to HPV16 status and race, in the all sites combined group HPV16 negative CA had poorer survival than HPV16 positive and support studies that show better outcomes for HPV positive CA (Settle et al., 2009a; Chernock et al., 2011; Weinberger et al., 2010). A significant characteristic of HNSCC is its marked disparate unfavorable diagnosis and prognosis outcomes for AA (ACS, 2012; Hoffman et al., 1998; Shavers et al., 2003). There is no consensus on the causes of the differences in the higher incidence of and the mortality from HNSCC for AA when compared to CA, but they can include differences in access to care, stage at diagnosis, insurance status, attitudes of health providers, as well as HPV infection status (Settle et al., 2009b). Thus, in HNSCC, molecular subtyping for HPV is of clinical relevance, highlighting the importance of accounting for the influence of racial differences.
Prognostic outcomes for subgroups defined by both HPV16 and p16 status, indicate that patients with p16 negative/HPV16 negative status had the worst survival for all sites combined (HR=8.67, p= 0.003) as well as for the OP site (HR=10.30, p=0.033). Smith et al. (2010), while examining multiple molecular markers in a larger series of HNSCC, also found that p16 negative/HPV negative/p53 positive cases had poorer survival when compared to p16 positive/HPV positive/p53 negative.
We also found that discordant p16 and HPV16 status (p16 positive/HPV16 negative and p16 negative/HPV16 positive) conferred poorer survival but with the limited dataset it is difficult to determine whether p16 or HPV16 status is driving this effect. When testing solely for the HPV16 subtype, HPV16 negative status when accompanied by p16 positivity is often regarded as a false negative, and as likely harboring non-HPV16 genotypes. This is especially the case in OPSCC where p16 positivity is considered a surrogate for HPV, and in this context, p16 positive/HPV negative patients would demonstrate improved survival similar to p16 positive/HPV positive patients. However in our study, the hazard ratio for the p16 positive/HPV16 negative group (HR=6.97, p=0.021) indicate substantial deviation from the p16 positive/HPV16 positive referent group and suggests a distinct subgroup lacking any HPV genotype. The latter is further substantiated by Smith et al. (2010), where the p16 positive/HPV negative subgroup, with and without expression of p53, had hazard ratios of 4.6 (95% CI 1.4, 14.5) and 3.6 (95% CI 1.1, 11.1), respectively, when compared to the p16 positive/HPV positive/p53 negative referent group. The latter suggests that p16 expression can occur in the absence of HPV and is not necessarily a consequence of HPV infection.
Reports of HPV prevalence of 24% in laryngeal carcinomas and 23% in oral cavity carcinomas indicate that biologically relevant HPV infection also takes place outside of the oropharynx (Stephen et al., 2012; Kreimer et al., 2005). However, HNSCC studies that report on the significance of both HPV and p16 have focused on OPSCC, mainly due to small number of non-OPSCC cases. Thus, usefulness of p16 as a surrogate marker for HPV infection or as a prognostic marker is limited to OPSCC. However, it has been shown that even in OPSCC, 15% to 20% tend to be p16 positive/HPV16 negative (Smeets et al., 2007). We observed that p16 positive/HPV16 negative patients in the OP subgroup (4/20) did not show significant differences in survival when compared to the p16 positive/HPV16 positive reference group. However, in the all sites combined group (Table 3A), the p16 positive/HPV16 negative subgroup (9/80: 4 OP and 5 non-OP patients) had significantly poorer survival when compared to p16 positive/HPV16 positive referent. The latter suggests that in the absence of HPV16, p16 expression is likely due to non-HPV related genetic or epigenetic loss of pRb (Compton et al., 2011). Because HPV negative tumors have more mutations than HPV positive (Stransky et al., 2011), when assessing prognostic outcomes in HPV negative HNSCC, multiple markers will likely be key to providing more accurate outcomes, serving as potential therapeutic targets.
A limitation of this study is that in its exploratory design, the sample sizes are modest, especially for site subtypes. This is reflected in the very wide confidence intervals for the hazard ratio estimates shown in Table 3. However, despite the low precision, many of the associations are significant.
5. Conclusion
This study provides support for p16 status as an important prognostic factor in both OPSCC and non-OPSCC and supports the p16 positive/HPV16 negative group as likely a distinct subgroup lacking any HPV genotype. HNSCC cohorts with larger representations of non-OP sites examining multiple molecular markers will be key to deciphering and dissecting out p16’s role as a useful prognostic indicator when assessed in combination with HPV status.
Acknowledgements
Drs. Stephen and Worsham had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. This study was supported by R01 NIH DE 15990 (Dr. Worsham).
References
- American Cancer Society . Cancer Facts & Figures 2012. American Cancer Society; Atlanta: 2012. [Google Scholar]
- Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, Gillison ML. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. http://dx.doi.org/10.1056/NEJMoa0912217. PMid:20530316 PMCid:2943767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakhuis BJ, Snijders PJ, Keune WJ, Meijer CJ, Ruijter-Schippers HJ, Leemans CR, Brakenhoff RH. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst. 2004;96(13):998–1006. doi: 10.1093/jnci/djh183. http://dx.doi.org/10.1093/jnci/djh183. PMid:15240783. [DOI] [PubMed] [Google Scholar]
- Chernock RD, Zhang Q, El-Mofty SK, Thorstad WL, Lewis JS., Jr. Human papillomavirus-related squamous cell carcinoma of the oropharynx: a comparative study in whites and African Americans. Arch Otolaryngol Head Neck Surg. 2011;137(2):163–169. doi: 10.1001/archoto.2010.246. http://dx.doi.org/10.1001/archoto.2010.246. PMid:21339403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton AM, Moore-Medlin T, Herman-Ferdinandez L, Clark C, Caldito GC, Wang XI, Nathan CA. Human Papillomavirus in Metastatic Lymph Nodes from Unknown Primary Head and Neck Squamous Cell Carcinoma. Otolaryngol Head Neck Surg. 2011;145(1):51–57. doi: 10.1177/0194599811400385. http://dx.doi.org/10.1177/0194599811400385. PMid:21493313. [DOI] [PubMed] [Google Scholar]
- Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Gillison ML. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100(4):261–269. doi: 10.1093/jnci/djn011. http://dx.doi.org/10.1093/jnci/djn011. PMid:18270337. [DOI] [PubMed] [Google Scholar]
- Gillison ML, Lowy DR. A causal role for human papillomavirus in head and neck cancer. Lancet. 2004;363(9420):1488–1489. doi: 10.1016/S0140-6736(04)16194-1. http://dx.doi.org/10.1016/S0140-6736(04)16194-1. [DOI] [PubMed] [Google Scholar]
- Ha PK, Pai SI, Westra WH, Gillison ML, Tong BC, Sidransky D, Califano JA. Real-time quantitative PCR demonstrates low prevalence of human papillomavirus type 16 in premalignant and malignant lesions of the oral cavity. Clin Cancer Res. 2002;8(5):1203–1209. PMid:12006539. [PubMed] [Google Scholar]
- Hannon GJ, Beach D. p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature. 1994;371(6494):257–261. doi: 10.1038/371257a0. http://dx.doi.org/10.1038/371257a0. PMid:8078588. [DOI] [PubMed] [Google Scholar]
- Hashibe M, Brennan P, Benhamou S, et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99:777–789. doi: 10.1093/jnci/djk179. http://dx.doi.org/10.1093/jnci/djk179. PMid:17505073. [DOI] [PubMed] [Google Scholar]
- Hoffman HT, Karnell LH, Funk GF, Robinson RA, Menck HR. The National Cancer Data Base report on cancer of the head and neck. Arch Otolaryngol Head Neck Surg. 1998;124(9):951–962. doi: 10.1001/archotol.124.9.951. PMid:9738803. [DOI] [PubMed] [Google Scholar]
- Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer Epidemiol Biomarkers Prev. 2005;14(2):467–475. doi: 10.1158/1055-9965.EPI-04-0551. http://dx.doi.org/10.1158/1055-9965.EPI-04-0551. PMid:15734974. [DOI] [PubMed] [Google Scholar]
- Lassen P, Eriksen JG, Hamilton-Dutoit S, Tramm T, Alsner J, Overgaard J. Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol. 2009;27(12):1992–1998. doi: 10.1200/JCO.2008.20.2853. http://dx.doi.org/10.1200/JCO.2008.20.2853. PMid:19289615. [DOI] [PubMed] [Google Scholar]
- Munger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, Grace M, Huh K. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78(21):11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. http://dx.doi.org/10.1128/JVI.78.21.11451-11460.2004. PMid:15479788 PMCid:523272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namazie A, Alavi S, Olopade OI, Pauletti G, Aghamohammadi N, Aghamohammadi M, Srivatsan ES. Cyclin D1 amplification and p16(MTS1/CDK4I) deletion correlate with poor prognosis in head and neck tumors. Laryngoscope. 2002;112(3):472–481. doi: 10.1097/00005537-200203000-00013. http://dx.doi.org/10.1097/00005537-200203000-00013. PMid:12148857. [DOI] [PubMed] [Google Scholar]
- Psyrri A, DiMaio D. Human papillomavirus in cervical and head-and-neck cancer. Nat Clin Pract Oncol. 2008;5(1):24–31. doi: 10.1038/ncponc0984. http://dx.doi.org/10.1038/ncponc0984. PMid:18097454. [DOI] [PubMed] [Google Scholar]
- Raju U, Mei L, Seema S, Hina Q, Wolman SR, Worsham MJ. Molecular classification of breast carcinoma in situ. Curr Genomics. 2006;7:523–532. doi: 10.2174/138920206779315719. http://dx.doi.org/10.2174/138920206779315719. PMid:17375183 PMCid:1828915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco JW, Li D, Liggett WH, Jr, Duan L, Saunders JK, Jr, Sidransky D, O’Malley BW., Jr p16INK4A adenovirus-mediated gene therapy for human head and neck squamous cell cancer. Clin Cancer Res. 1998;4(7):1697–704. PMid:9676844. [PubMed] [Google Scholar]
- Settle K, Posner MR, Schumaker LM, Tan M, Suntharalingam M, Goloubeva O, Cullen KJ. Racial survival disparity in head and neck cancer results from low prevalence of human papillomavirus infection in black oropharyngeal cancer patients. Cancer Prev Res. 2009a;2(9):776–781. doi: 10.1158/1940-6207.CAPR-09-0149. http://dx.doi.org/10.1158/1940-6207.CAPR-09-0149. PMid:19641042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settle K, Taylor R, Wolf J, Kwok Y, Cullen K, Carter K, Suntharalingam M. Race impacts outcome in stage III/IV squamous cell carcinomas of the head and neck after concurrent chemoradiation therapy. Cancer. 2009b;115(8):1744–1752. doi: 10.1002/cncr.24168. http://dx.doi.org/10.1002/cncr.24168. PMid:19208429. [DOI] [PubMed] [Google Scholar]
- Shavers VL, Harlan LC, Winn D, Davis WW. Racial/ethnic patterns of care for cancers of the oral cavity, pharynx, larynx, sinuses, and salivary glands. Cancer Metastasis Rev. 2003;22(1):25–38. doi: 10.1023/a:1022255800411. http://dx.doi.org/10.1023/A:1022255800411. PMid:12716034. [DOI] [PubMed] [Google Scholar]
- Shi W, Kato H, Perez-Ordonez B, Pintilie M, Huang S, Hui A, Liu FF. Comparative prognostic value of HPV16 E6 mRNA compared with in situ hybridization for human oropharyngeal squamous carcinoma. J Clin Oncol. 2009;27(36):6213–6221. doi: 10.1200/JCO.2009.23.1670. http://dx.doi.org/10.1023/A:1022255800411. PMid:12716034. [DOI] [PubMed] [Google Scholar]
- Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer. 2010;116(9):2166–2173. doi: 10.1002/cncr.25033. PMid:20186832. [DOI] [PubMed] [Google Scholar]
- Smeets SJ, Hesselink AT, Speel EJ, Haesevoets A, Snijders PJ, Pawlita M, Brakenhoff RH. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer. 2007;121(11):2465–2472. doi: 10.1002/ijc.22980. http://dx.doi.org/10.1002/ijc.22980. PMid:17680565. [DOI] [PubMed] [Google Scholar]
- Smith EM, Rubenstein LM, Hoffman H, Haugen TH, Turek LP. Human papillomavirus, p16 and p53 expression associated with survival of head and neck cancer. Infect Agent Cancer. 2010;5(4) doi: 10.1186/1750-9378-5-4. PMid:20181227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen JK, Chen KM, Shah V, Havard S, Lu M, Schweitzer VP, Worsham MJ. Human Papillomavirus Outcomes in an Access-to-Care Laryngeal Cancer Cohort. Otolaryngol Head Neck Surg. 2012;146(5):730–738. doi: 10.1177/0194599811434482. http://dx.doi.org/10.1177/0194599811434482. PMid:22267491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Grandis JR. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333(6046):1157–1160. doi: 10.1126/science.1208130. http://dx.doi.org/10.1126/science.1208130. PMid:21798893 PMCid:3415217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Knebel Doeberitz M. New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur J Cancer. 2002;38(17):2229–2242. doi: 10.1016/s0959-8049(02)00462-8. http://dx.doi.org/10.1016/S0959-8049(02)00462-8. [DOI] [PubMed] [Google Scholar]
- Weinberger PM, Merkley MA, Khichi SS, Lee JR, Psyrri A, Jackson LL, Dynan WS. Human papillomavirus-active head and neck cancer and ethnic health disparities. Laryngoscope. 2010;120(8):1531–1537. doi: 10.1002/lary.20984. http://dx.doi.org/10.1002/lary.20984. PMid:20564751 PMCid:3051373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger PM, Yu Z, Haffty BG, Kowalski D, Harigopal M, Sasaki C, Psyrri A. Prognostic significance of p16 protein levels in oropharyngeal squamous cell cancer. Clin Cancer Res. 2004;10(17):5684–5691. doi: 10.1158/1078-0432.CCR-04-0448. http://dx.doi.org/10.1158/1078-0432.CCR-04-0448. PMid:15355894. [DOI] [PubMed] [Google Scholar]
- Worsham MJ, Chen KM, Tiwari N, Pals G, Schouten JP, Sethi S, Benninger MS. Fine-mapping loss of gene architecture at the CDKN2B (p15INK4b), CDKN2A (p14ARF, p16INK4a), and MTAP genes in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2006;132(4):409–415. doi: 10.1001/archotol.132.4.409. http://dx.doi.org/10.1001/archotol.132.4.409. PMid:16618910. [DOI] [PubMed] [Google Scholar]
- Zhang HS, Postigo AA, Dean DC. Active transcriptional repression by the Rb-E2F complex mediates G1 arrest triggered by p16INK4a, TGFbeta, and contact inhibition. Cell. 1999;97(1):53–61. doi: 10.1016/s0092-8674(00)80714-x. http://dx.doi.org/10.1016/S0092-8674(00)80714-X. [DOI] [PubMed] [Google Scholar]