Abstract
In Drosophila melanogaster, two chromosome-specific targeting and regulatory systems have been described. The male-specific lethal (MSL) complex supports dosage compensation by stimulating gene expression from the male X-chromosome, and the protein Painting of fourth (POF) specifically targets and stimulates expression from the heterochromatic 4th chromosome. The targeting sites of both systems are well characterized, but the principles underlying the targeting mechanisms have remained elusive. Here we present an original observation, namely that POF specifically targets two loci on the X-chromosome, PoX1 and PoX2 (POF-on-X). PoX1 and PoX2 are located close to the roX1 and roX2 genes, which encode noncoding RNAs important for the correct targeting and spreading of the MSL-complex. We also found that the targeting of POF to PoX1 and PoX2 is largely dependent on roX expression and identified a high-affinity target region that ectopically recruits POF. The results presented support a model linking the MSL-complex to POF and dosage compensation to regulation of heterochromatin.
Keywords: Painting of fourth, dosage compensation, heterochromatin, epigenetics, Drosophila melanogaster
The evolution of sex chromosomes, for example the X and Y chromosome pairs found in mammals and flies, makes it more difficult for balanced gene expression to be achieved. Although some genes located on the X-chromosome are expressed in a sex-specific manner, most genes require equal expression in males and females (Vicoso and Bachtrog 2009; Stenberg and Larsson 2011). The gradual degeneration of the proto-Y chromosome causes an increasing requirement to equalize gene expression between a single X (in males) and two X-chromosomes (in females) and to balance this level of expression with that of the two sets of autosomal chromosomes (Oliver 2007; Vicoso and Bachtrog 2009; Prestel et al. 2010a; Stenberg and Larsson 2011). This requirement has led to the evolution of dosage-compensation mechanisms. Such mechanisms must equalize the transcriptional activities of the two X-chromosomes in the homogametic sex with that of the single X-chromosome in the heterogametic sex, as well as balancing the relative expression levels between sex chromosomes and autosomes (Gupta et al. 2006; Nguyen and Disteche 2006; Deng et al. 2011). In fruit flies, both these requirements are solved by a 2-fold increase in gene expression that is restricted to the single male X-chromosome. This 2-fold increase is the result of a combination of a general buffering effect exerted on monosomic regions or chromosomes (Stenberg et al. 2009; Zhang et al. 2010; Lundberg et al. 2012) and an increase in expression from the male X-chromosome mediated by the male-specific lethal (MSL) complex (Hamada et al. 2005; Deng et al. 2009; Prestel et al. 2010a; Stenberg and Larsson 2011). The MSL-complex consists of two partly redundant noncoding RNAs, roX1 and roX2, together with at least five MSL proteins (MSL1, MSL2, MSL3, MLE, and MOF) that “paint” the dosage-compensated male X-chromosome. MOF mediates acetylation of H4 at lysine 16 (H4K16ac), and enrichment for H4K16ac on the male X-chromosome is believed to cause decondensation of the chromatin fiber, which at least partly explains the hypertranscription of this chromosome (Gelbart and Kuroda 2009; Prestel et al. 2010a). The MSL-complex not only recruits MOF to the male X-chromosome but also constrains the activation potential of MOF (Prestel et al. 2010b; Sun et al. 2013). The male specificity of the MSL-complex is accomplished by the MSL2 protein, which is only formed in males and is rate limiting for the formation of the complex (Kelley et al. 1995, 1997; Bashaw and Baker 1997).
Understanding how the MSL-complex correctly targets the ~1000 active genes on the male X-chromosome is a key challenge in this area of research; targeting is currently thought to be initiated by sequence-dependent binding of the complex to 100−200 nucleation sites on the X-chromosome, which are termed chromatin entry sites (CES) or high-affinity sites (HAS) (Alekseyenko et al. 2008; Straub et al. 2008, 2013). Two of the strongest nucleation sites are the roX1 and roX2 loci (Kelley et al. 1999). Targeting to nucleation sites is followed by spreading to neighboring genes; this is dependent on active transcription (Sass et al. 2003; Larschan et al. 2007), MSL-complex concentration (Dahlsveen et al. 2006), level of affinity (Straub et al. 2008; Lucchesi 2009), and probably also sequence composition (Philip et al. 2012).
However, the dosage compensation mediated by the MSL-complex is not the only chromosome-wide compensatory mechanism that has been described in Drosophila. There is also the protein Painting of fourth (POF), which in Drosophila melanogaster specifically targets the 4th chromosome (Larsson et al. 2001; Johansson et al. 2007a, 2012). POF binds to nascent RNA from actively transcribed genes on the 4th chromosome and increases levels of expression of these genes (Johansson et al. 2012). The compensation mediated by POF is sufficient to allow survival of haplo-4 flies (Johansson et al. 2007a). The specific targeting of POF to the 4th chromosome, the evolutionary connections between the 4th chromosome, the male X-chromosome and dosage compensation, and the stimulatory effect of POF on gene expression all support the hypothesis that POF originates from a dosage compensatory system (Larsson et al. 2001, 2004; Larsson and Meller 2006; Johansson et al. 2007a,b; Stenberg et al. 2009; Stenberg and Larsson 2011). Although dosage compensation has probably been required, and thus evolved, on a gene-by-gene or block-of-genes basis, the existence of systems like the MSL-complex that act across a whole chromosome argues that some of the functions were pre-existing and were recruited to compensate the X-chromosome as the proto-Y chromosome started to degenerate (Stenberg and Larsson 2011). The targeting mechanisms of these systems and the relationship between them are therefore important issues to study in order to understand chromosome evolution and how balanced genome expression is established.
Here we provide a further link between these two chromosome-wide targeting systems by exploring an original observation that POF targets two loci on the X-chromosome in a female-specific context. The targeting of these loci depends on roX expression, and by constructing transgenes and examining their effects we have identified a POF high-affinity recruitment element.
Material and Methods
Fly strains and genetic crosses
Flies were cultivated and crossed at 25º in vials containing potato mash-yeast-agar. roX1ex6, roX1SMC40, and roX1ex84A strains were obtained from Victoria Meller (Wayne State University, Detroit, MI) and are described in Deng and Meller (2009). roX1 roX2 double-mutant males were selected as non-green fluorescent protein males from a y w roX1ex6 Df(1)roX252 P[w+4Δ4.3]/ FM7i, P[w+mC ActGFP]JMR3 stock obtained from Yongkyu Park (New Jersey Medical School, Newark, NJ). roX1 roX2 double-mutant females larvae were selected from the cross: y w roX1ex6 Df(1)roX252 / Binsincy; P[w+4Δ4.3]/+ × y w roX1ex6 Df(1)roX252 / Dp(1;Y)Bs y+ v+; P[w+4Δ4.3]/+. Overexpression of roX1 and roX2 was performed using y w; [w+ hsp83:roX1] and y w; [w+ hsp83:roX2]34B, respectively. Female larvae overexpressing msl2 were selected from a w; P[w+ hsp83:msl2] msl3/TM6B stock obtained from Mitzi Kuroda (Harvard Medical School, Boston, MA). The P[w+ CkIIβ gDNA] transgenic line was obtained from Thomas Raabe (Universität Würzburg) and is described in Jauch et al. (2002). The duplications of the PoX1 and PoX2 loci used were w1118; Dp(1;3)DC112, PBac[y+mDint2 w+mC DC112]VK00033 (92 kb inserted at 3L:65B2 covering PoX1), w1118; Dp(1;3)DC244, PBac[y+mDint2 w+mC DC244]VK00033 (87 kb inserted at 3L:65B2 covering the three genes Ck2β, Hsc70-3, CG1578 in PoX2) and Dp(1;3)DC246, PBac[y+mDint2 w+mC DC246]VK00033 (102 kb inserted at 3L:65B2 covering the two genes SelG and CG1840 in PoX2) (Venken et al. 2009, 2010). To visualize Setdb1 on polytene chromosomes, we used the strain Setdb13HA (Seum et al. 2007), which contains a gene encoding hemagglutinin-tagged Setdb1, obtained from Carole Seum (University of Geneva). Oregon R was used as the wild-type strain.
Transgenic flies
To generate the P[w+ SelG CG1840] transgene, a genomic fragment was amplified with Long PCR Enzyme Mix (Thermo Scientific) using the primers 5′-GTGGATCCAAAAATGGTCTTGTTCCACA-3′, 5′-CGGCGGCCGCAATTTGGCGGAAGATTCAAA-3′ with Oregon R genomic DNA as a template. Thus, the fragment included both the SelG gene and the CG1840 gene as well as 1673-bp fragment upstream of SelG transcription start and 2733 bp downstream of the annotated transcript stop position of CG1840. The polymerase chain reaction (PCR) product was cut with BamHI/NotI and ligated into a P[w+ attB] vector. The latter, which is a pBluescript-based plasmid supplemented with an attB recombination site and a mini-white marker gene, was provided by M. Savitsky. Embryo microinjection into the Bl9750 strain was performed by BestGene (Inc).
RNA-fluorescent in situ hybridization (RNA-FISH) and immunostaining of polytene chromosomes
Immunostaining of polytene chromosomes was essentially as described previously (Johansson et al. 2012). We used primary antibodies against POF raised in rabbit, diluted 1:500 (Larsson et al. 2004), or raised in chicken, diluted 1:100 (Larsson et al. 2001); against HP1a (PRB291C, 1:400, Covance); and against HA (MMS 101R, 1:100, Covance, for detection of Setdb1.3HA). Goat or donkey antirabbit, antichicken, and antimouse conjugated with Alexa-Fluor555 or AlexaFluor488 (1:500; Molecular Probes) were used as secondary antibodies.
For RNA-FISH combined with immunostaining, salivary glands were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS), 0.3% Triton X-100 for 20 sec followed by 2−3 min in 50% acetic acid, 1% formaldehyde. Polytene chromosomes were squashed using high pressure treatment (Novikov et al. 2007), quick-frozen in liquid nitrogen, and stored in ethanol at −20° until required for use. The slides were rehydrated in an ethanol series (1 min each in 95%, 70%, and 30% ethanol), then incubated for 15 min in PBT (i.e., PBS, 0.1% Triton X-100), and the chromosomes were fixed in 3.7% formaldehyde in PBT for 15 min. The slides were then washed for 3× 3 min in PBT followed by prehybridization at 42° for 3−4 hr in hybridization mixture (5× saline sodium citrate (SSC), 5× Denhardts solution, 500 μg/mL cold DNA, 250 μg/mL transfer RNA, and 50% formamide).
The slides were then hybridized using a digoxigenin labeled antisense RNA probe against RE64691 mixed with hybridization mixture in a wet chamber at 42° overnight. After hybridization the slides were washed in 2× SSC at room temperature for 1 min followed by 2× 30 min in 50% formamide, 5× SSC, 10 mM dithiothreitol at 42°, 2× 30 min in 2× SSC at 42°, 1 hr in 0.1× SSC at room temperature, and finally 10 min in PBT at room temperature. The slides were transferred to blocking solution (PBS, 1% Roche blocking reagent) and incubated for 30 min at room temperature, then incubated for 1 hr at room temperature with primary antibody raised against POF (1:500 dilution) and sheep antidigoxigenin (0.4 μg/mL; Roche). The slides were then washed for 3× 2 minutes in PBS + 0.2% Tween-20 at room temperature. They were incubated in the secondary antibodies donkey antirabbit and donkey antisheep conjugated with AlexaFluor555 or AlexaFluor488 (Molecular Probes; diluted 1:500), together with DAPI (5 µg/mL), for 1 hr at room temperature. Finally, the slides were washed for 4× 5 min in PBS + 0.2% Tween-20 and mounted with Vectashield (Vector).
Chromatin immunoprecipitation (ChIP) and microarray analysis
For the POF ChIP-chip experiment, we dissected salivary glands from third instar larvae. The ChIP experiment was conducted as previously described (Johansson et al. 2007a,b) using 3 µL of anti-POF (Larsson et al. 2004) for precipitation. The purified ChIP and input DNA samples were amplified using a WGA2 GenomePlex Complete whole-genome amplification kit (Sigma-Aldrich) according to the recommendations of the supplier, and the amplified DNA was purified with a QIAquick PCR purification kit (QIAGEN). To verify that no amplification bias affected the enrichment profiles, we analyzed the ChIP DNA/input DNA ratio before and after amplification by using real-time PCR as previously described (Johansson et al. 2007b).
For tiling array analysis, the amplified ChIP DNA samples were fragmented, labeled, and hybridized to an Affymetrix Drosophila Genome 2.0 array. The signal intensity data generated were analyzed with Affymetrix Tiling Analysis Software (v. 1.1.02), using a 200-bp bandwidth as a smoothing parameter and setting a constraint of perfect match only. The enrichment profiles were produced as ChIP DNA/input DNA ratios expressed on a log2 scale. The complete data set is available at http://www.ncbi.nlm.nih.gov/geo/ (accession: GSE45402). The occupancy profiles obtained were visualized and analyzed using Integrated Genome Browser (7.0.1) (Nicol et al. 2009).
Gene identification
To verify the presence of a novel gene between the two genes Mnt and Rala, we sequenced the Expressed Sequence Tag clone RE64691 obtained from the Drosophila Genomics Resource Centre. To determine the 5′- and 3′-ends of the putative novel gene, we isolated poly(A)+ RNA from adult females using Dynabeads Oligo (dT)25 magnetic beads (Invitrogen) as previously described (Svensson et al. 2003) and verified the end sequences using the SMART-RACE cDNA-amplification kit (Clontech) according to a protocol provided by the supplier. The primers used were: RE64691 gene specific primer 1: 5′-CAGAAATCGAGTGACACACACAGG-3′, and primer 2: 5′-GCTAACATAAGCCCACATCCACAC-3′, the nested gene specific primer 1: 5′-ACTATAAGTCCCCCGTGATGACAG-3′ and the nested primer 2: 5′-GCAGATGGAGACGGAAAGAGTAGG-3′.
Results
POF binds to two loci, PoX1 and PoX2, on the X-chromosome in females
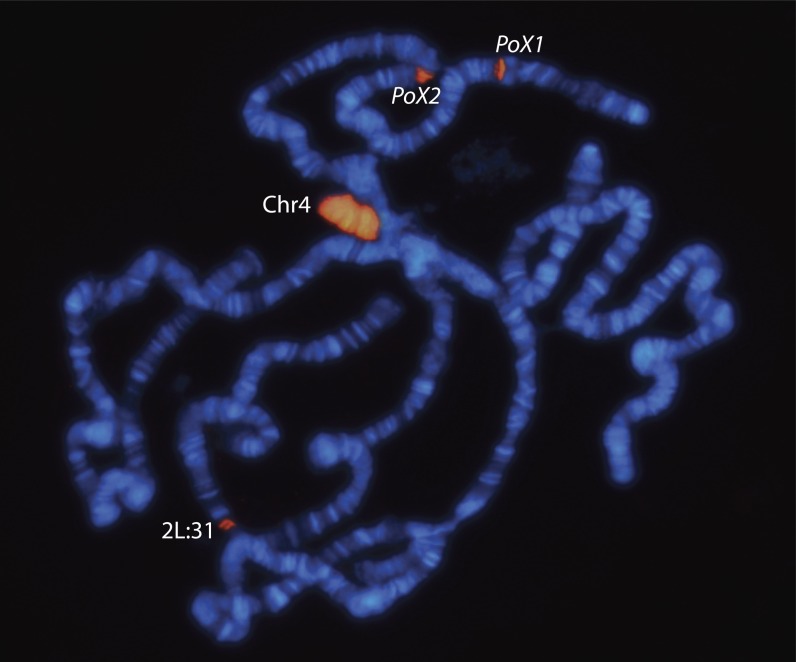
We have previously shown that POF binds with high specificity to the 4th chromosome in both males and females (Larsson et al. 2001; Johansson et al. 2007a,b; Figueiredo et al. 2012). With improved techniques for preparation and staining, we noticed that in good chromosome preparations from wild-type larvae two strong POF binding signals were visible on the X-chromosome. By sexing larvae we found that, out of the best 10−15 nuclei (fully polytenized and well spread) from each pair of salivary glands, 30−50% of all X-chromosomes in females showed one or two reproducible stained bands at cytological locations X:3E and X:10E−F (Figure 1). These bands were never detected in males. We named these loci PoX1 and PoX2 (POF-on-X) and found spreading of the POF binding in both loci (Figure 1 and Figure 2A). The presence and degree of spreading differed between nuclei, and we typically saw 1-3 bands decorated by POF in both PoX1 and PoX2. In addition to the two loci on the X-chromosome, we occasionally observed POF binding in parts of the 2L:31 region in both males and females (Figure 1) (Lundberg et al. 2013).
Figure 1.
POF binds two sites on the X-chromosome in females only. Immunostaining of polytene chromosomes from a wild-type female. In addition to binding on the 4th chromosome (Chr4), in this nucleus POF binds to two sites on the X-chromosome, PoX1 and PoX2 (seen in 30−50% of nuclei) and to region 2L:31 on the left arm of the second chromosome.
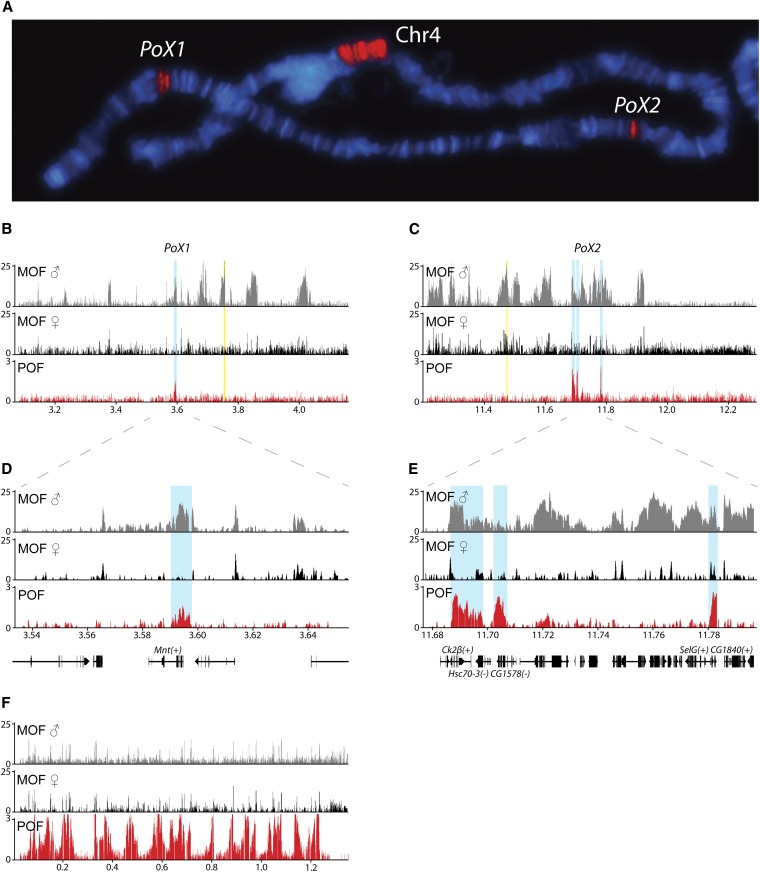
Figure 2.
POF binding profiles of the X-chromosome regions including PoX1 and PoX2. (A) Immunostaining of polytene chromosomes showing POF binding to the 4th chromosome (Chr4) and the two loci PoX1 and PoX2 as indicated. (B) POF binding profile from salivary gland tissue ChIP-chip. An ~1-Mb region including the PoX1 region (blue box) is shown. The binding profiles of MOF in males and females[from ChIP-seq (Conrad et al. 2012)] are shown for comparison. (C) POF binding profile in an ~1-Mb region including the PoX2 regions (blue boxes). The plots show the mean enrichment ratios obtained using a bandwidth of 200 bp for the POF ChIP-chip. Numbers on the x-axis denote chromosomal position along the chromosome in Mb. The y-axis shows ChIP enrichments as log2 ratios. PoX1 and PoX2 are located downstream of genes roX1 and roX2 (indicated by yellow boxes), respectively. The roX1 gene is transcribed from right to left and roX2 is expressed in the opposite direction. (D and E) High resolution profiles of enrichment at the PoX1 (D) and PoX2 (E) sites. The genes bound by POF are indicated; genes expressed from left to right are indicated by (+) and genes expressed in the opposite direction are indicated by (−). (F) Profile of POF binding on the 4th chromosome (~1.3 Mb) is shown for comparison.
PoX1 and PoX2 correspond to specific genes on the X-chromosome
We were intrigued by the fact that the two loci, PoX1 and PoX2, mapped cytologically close to roX1 (X:3F) and roX2 (X:10C) and decided to map these PoX loci more precisely by using ChIP-chip technology. We performed ChIP-chip analysis on chromatin extracts from salivary glands of 3rd instar larvae. The ChIP-chip profiles generated confirmed the cytological observations (Figure 2). The PoX1 locus corresponds to a strong binding peak/region ~200 kb downstream of roX1 (Figure 2B). In this region, POF is enriched within the 3′-end of the Mnt gene and in a distinct 2-kb region between the genes Mnt and Rala (Figure 2D). The PoX2 locus corresponds to three enriched regions covering the five genes Ck2β, Hsc70-3, CG1578, SelG, and CG1840, which are located ~200 kb downstream of roX2 (Figure 2, C and E). To test whether POF binding enrichment was correlated with enrichment for the MSL-complex, we plotted MOF data generated from salivary glands of males and females, which was available from (Conrad et al. 2012). Some, but not all, genes in the PoX1 and PoX2 loci that were enriched in POF, also were enriched in MOF (and presumably the MSL-complex) in males. In females, MOF bound to the promoters of Ck2β, SelG and CG1840 in the PoX2 locus (Figure 2, D and E). We conclude that, with respect to pattern of binding to the PoX1 and PoX2 loci, there is no obvious link between POF and the MSL-complex that distinguishes the targeted genes from other X-linked genes.
POF binds to a novel noncoding gene between Mnt and Rala
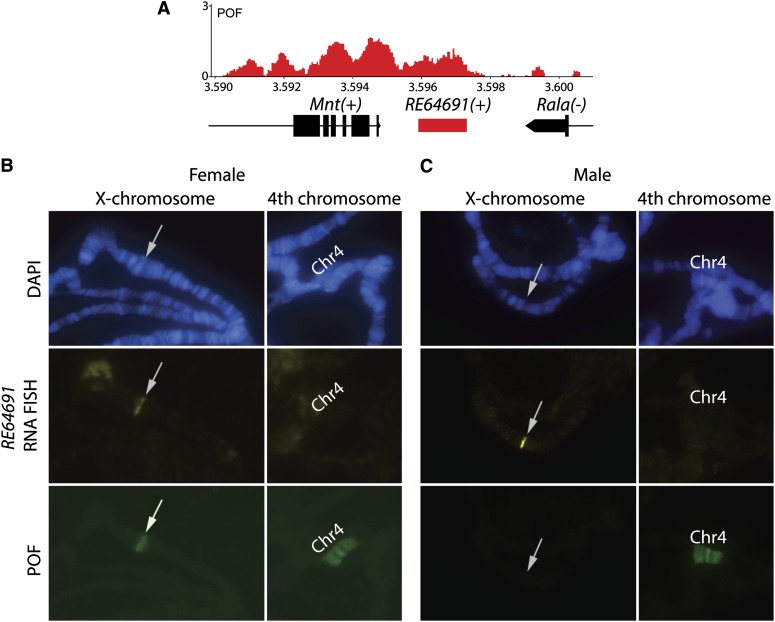
In the PoX1 locus, we found POF to be highly enriched within the 3′-end of Mnt and in a distinct region downstream of Mnt and Rala, which are transcribed in the opposite direction so that the region is downstream of both genes (Figure 3A). No gene had previously been annotated in this region, but a large number of expressed sequence tag (EST) cDNA clones are annotated according to the GBrowse genome browser at FlyBase (Tweedie et al. 2009). We sequenced one of these EST clones, RE64691, and confirmed using 5′- and 3′-RACE (i.e., rapid amplification of cDNA ends) that this EST cDNA clone probably corresponds to a novel gene as illustrated in Figure 3A. The longest putative ORF is 34 amino acids and it is therefore possible that RE64691 corresponds to a novel noncoding gene. Alternatively, RE64691 belongs to the class of genes with short ORFs encoding small bioactive peptides (Frith et al. 2006; Kastenmayer et al. 2006; Hanada et al. 2007) as exemplified by, e.g. the Drosophila gene tarsal-less (Galindo et al. 2007; Kondo et al. 2010).
Figure 3.
POF binds to a novel gene, RE64691, located between Mnt and Rala. (A) High-resolution enrichment profiling at the PoX1 site shows that POF binds to a novel gene (indicated by a red rectangle) represented by the EST clone RE64691. (B and C) Immunostaining combined with RNA-FISH shows that the gene RE64691 is expressed in both females (B) and males (C) but targeted by POF only in females (note that in contrast POF targets the 4th chromosome in both males and females). The RE64691 locus is indicated by an arrow. The tip of the X-chromosome and the 4th chromosome from one representative female nucleus are shown in (B) and from one male in (C). The 4th chromosome is indicated by “Chr4”.
Anyway, intrigued by the possibility that POF targets a gene encoding a noncoding RNA (ncRNA), and bearing in mind the fact that ncRNAs have been shown to be important for correct targeting of the MSL-complex, we conducted RNA-FISH experiments to probe whether RE64691 RNA was connected to POF targeting of chromosome 4. The RNA-FISH results showed that RE64691 is expressed in salivary glands in both males and females, but no enrichment of RE64691 RNA on the 4th chromosome was detected (Figure 3, B and C). The detection of RE64691 expression in both males and females indicated that expression per se could not explain the sex-specific targeting of PoX1. We therefore checked expression in salivary gland tissues of the five genes located in PoX2 by using RNA-seq data from (Graveley et al. 2011). The five genes located in PoX2 are expressed at similar levels in males compared to females (results not shown). We conclude that gene expression is not the determinant of the female specific targeting of POF to PoX1 and PoX2 and that RE64691 RNA is not enriched in POF-bound regions outside the RE64691 locus.
POF and HP1a colocalize at PoX loci
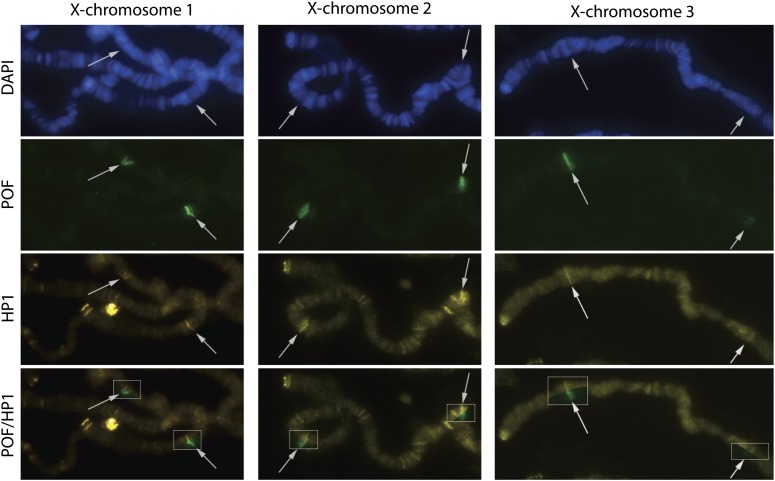
We have previously shown that POF and HP1a colocalize within the gene bodies of active genes on the 4th chromosome (Johansson et al. 2007a,b; Figueiredo et al. 2012). Indeed, no POF binding has been detected without HP1a enrichment. We therefore asked whether the PoX sites, which are bound only in females and not in all individual nuclei, are also targeted by HP1a. We observed that in the cases where POF binds to PoX1 and PoX2, weak but consistent enrichment for HP1a was present (Figure 4). We conclude that POF binding is accompanied by HP1a binding at all POF enriched sites including the female specific PoX1 and PoX2.
Figure 4.
POF and HP1a binding overlap on the PoX loci. POF (green) and HP1a (yellow) on the X-chromosome in three representative female nuclei. The proximal parts of the X chromosomes are shown, and the PoX1 and PoX2 loci are indicated with arrows. The boxes in the POF/HP1 row show the combined image. Note that levels of HP1a binding at PoX loci are above the background staining level.
POF binding is dependent on roX
Several lines of evidence have linked POF to dosage compensation and indicated that it has an evolutionary relationship with the MSL-complex (reviewed by (Stenberg and Larsson 2011)). We were therefore fascinated by the close proximity of PoX1 and PoX2 to roX1 and roX2, respectively. We asked whether the targeting by POF to PoX1 and PoX2 depended on roX1 and/or roX2. When we analyzed the best 10−15 nuclei in each chromosome preparation (fully polytenized chromosomes with a well spread X-chromosome from one pair of glands), 30-50% of these nuclei showed PoX1 and/or PoX2 targeting by POF (Table 1). Using the same criteria, we found, to our surprise, almost no POF targeting to PoX1 or PoX2 in roX1- or roX1 roX2-mutant female larvae (Table 1). Thus, the extent of POF targeting to PoX loci is dependent on roX; however, it does not require roX because in all roX mutant backgrounds tested we found individual nuclei (<5%) with clear targeting. Furthermore, overexpression of roX2 (but not roX1) in females using roX transgenes under a hsp83 promoter increased the targeting frequency. In contrast, expression of a partial MSL-complex in females decreased the targeting frequency (Table 1). We conclude that POF targeting to PoX loci in females is affected by but does not require roX.
Table 1. Frequency of PoX1 and PoX2 targeting by POF.
| Genotype |
PoX1 and PoX2 Targeting |
|
|---|---|---|
| Female | Male | |
| Wild type | + | − |
| roX1ex6 or roX1SMC40A or roX1ex84A | +/− | − |
| roX1ex6 roX2 | +/− | − |
| y w; P[w+ hsp83:roX2]34B | ++ | − |
| y w; P[w+ hsp83:roX1] | + | − |
| w; P[w+ hsp83:msl2] msl3 | +/− | ND |
| w1118; Dp(1;3)DC112 | +a | −a |
| w1118; Dp(1;3)DC244 | −a | −a |
| w1118; Dp(1;3)DC246 | +a | −a |
| w1118; P[w+ SelG CG1840] | ++a | ++a |
| roX1ex6 roX2; P[w+ SelG CG1840] | ++a | ++a |
More than 100 nuclei were scored for each genotype. +, 30−50% of all fully polytenized nuclei are stained at PoX1 and/or PoX2; −, no nuclei are stained at PoX1 and/or PoX2; +/−, <5% of all fully polytenized nuclei are stained at PoX1 and/or PoX2; ++, >50% of all fully polytenized nuclei are stained at PoX1 and/or PoX2; ND, not determined.
Refers to frequency of POF targeting to transgene or duplicated region.
Minimal POF recruitment element
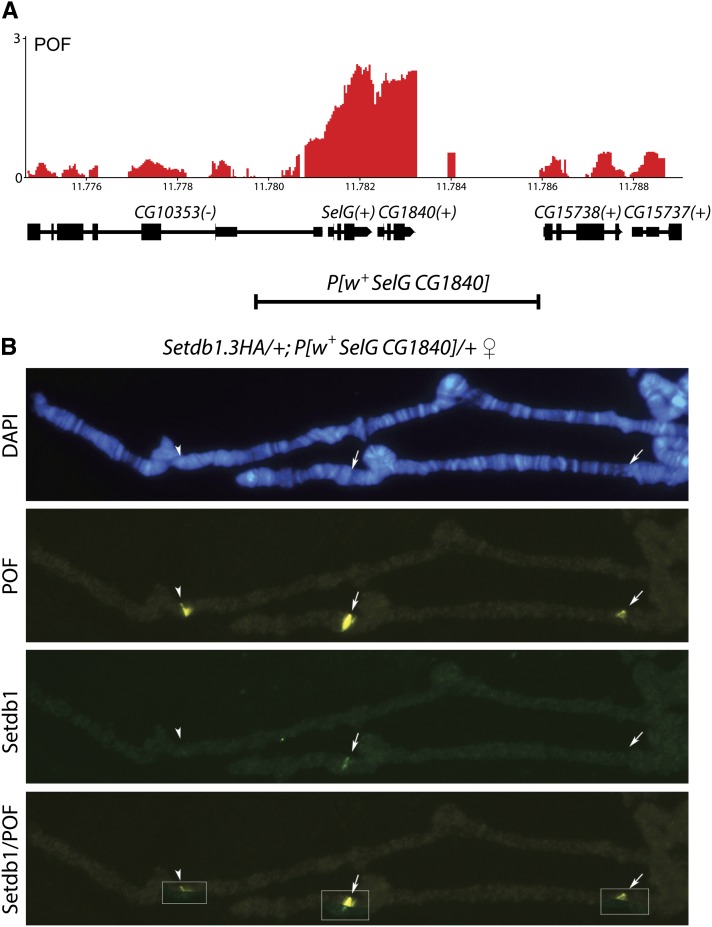
In contrast to the targeting of the MSL-complex, no high-affinity sites have previously been identified for POF binding. If the entire polytenized region of the 4th chromosome (~1.2 Mb) is translocated, it will not recruit POF if not under “heterochromatic pressure” (Johansson et al. 2007a). The PoX1 and PoX2 regions were therefore of interest as nonheterochromatic targets for POF binding, and we decided to dissect these regions further. First we tested the following duplications of the PoX1 and PoX2 loci: Dp(1;3)DC112 (92 kb covering PoX1), Dp(1;3)DC244 (87 kb covering the three genes Ck2β, Hsc70-3, CG1578 in PoX2), and Dp(1;3)DC246 (102 kb covering the two genes SelG and CG1840 in PoX2). These three duplications are all inserted in the same attP docking site in genomic region 3L:65B2 (Venken et al. 2010). Importantly, these duplications do not include roX1 or roX2 and will therefore reveal whether proximity to roX genes is required for targeting. In two of these duplications, DC112 and DC246, POF clearly targeted the duplicated regions at frequencies similar to those of the endogenous loci (30−50%). In contrast, in Dp(1;3)DC244, POF was not targeted to the duplicated region. In agreement with its behavior towards the endogenous loci, POF targeting to DC112 and DC246 was also to a large extent dependent on roX, i.e., POF was targeted to the duplicated region in <5% of nuclei in both roX1 roX2; Dp(1;3)DC112 and roX1 roX2; Dp(1;3)DC246 females (Table 1). To narrow down the region further, we tested flies transgenic for single genes or small regions. A 6-kb transgene including the genomic region covering the Ck2β gene, which has previously been described (Jauch et al. 2002), was tested for POF targeting. No targeting to the P[w+ CkIIβ gDNA] transgenic locus was detected, a result consistent with the lack of targeting in the duplication Dp(1;3)DC244, which covers Ck2β. We next cloned and generated transgenic flies with a 6-kb genomic region including the SelG and CG1840 genes targeted by POF in the PoX2 locus and in Dp(1;3)DC246 (Figure 5A). In P[w+ SelG CG1840] females we observed consistently strong targeting to the transgenic insert (Figure 5B). The targeting appears stronger than that at the endogenous site, since targeting is seen both in males and females and at a higher frequency (>50%) than that to the endogenous PoX loci (Table 1).
Figure 5.
POF binds to a transgenic minimal PoX site. (A) High-resolution enrichment profiling at the PoX1 site shows that POF binds to the two genes SelG and CG1840. The plots show the mean enrichment values obtained as log2 ChIP ratios. Numbers on the x-axis denote chromosomal position in megabases, and the y-axis shows ChIP enrichments. The genes are indicated; genes expressed from left to right are indicated by (+) and genes expressed in the opposite direction are indicated by (−). The extent of the 6.3-kb transgenic construct P[w+ SelG CG1840] is indicated. (B) POF (yellow) and Setdb1 (green) immunostaining of a Setdb1.3HA/+; P[w+ SelG CG1840] /+ female shows that POF and Setdb1 target both endogenous PoX1 and PoX2 (indicated by arrows) as well as the heterozygous transgene P[w+ SelG CG1840] inserted at position 3L:65B (arrowhead). The boxes in the Setdb1/HP1 row show the combined image. Setdb1 is visualized using the Setdb1.3HA hemagglutinin tagged Setdb1 and an anti-HA antibody.
Furthermore, targeting of POF to P[w+ SelG CG1840] in males was not abolished in the roX1 roX2 mutant background. In agreement with findings that we have previously reported for the 4th chromosome (Lundberg et al. 2013), POF and Setdb1 also colocalize on PoX loci and on the targeted P[w+ SelG CG1840] transgene (Figure 5B). We conclude that the 6-kb region inserted in P[w+ SelG CG1840] is a uniquely strong high-affinity recruitment site for POF targeting and is targeted independent of the genomic environment. The targeting of POF is also linked to the targeting by Setdb1 and HP1.
Discussion
Two chromosome specific targeting systems have been described in Drosophila melanogaster: the MSL-complex, which specifically targets the male X-chromosome, and POF, which targets the 4th chromosome in both males and females (Larsson and Meller 2006; Stenberg and Larsson 2011). Both the MSL-complex and POF target active genes and stimulate gene expression. The two systems stimulate gene expression levels to a comparable extent (Hamada et al. 2005; Johansson et al. 2007a; Deng et al. 2009; Stenberg et al. 2009; Zhang et al. 2010). High-affinity sites have been characterized for the MSL-complex and there are several published examples of short regions, including the roX1 and roX2 loci, that are capable of recruiting this complex when presented as transgenes (Kelley et al. 1999; Oh et al. 2004; Dahlsveen et al. 2006; Kind and Akhtar 2007; Alekseyenko et al. 2008). In contrast, until now no high-affinity sites for POF targeting have been identified. Translocated 4th chromosomes will not be targeted by POF, unless the proximal heterochromatic region is present and under conditions that favor heterochromatin formation (Johansson et al. 2007a). The characterization of POF targeting to PoX1 and PoX2 in females thus provides a unique opportunity to study the targeting of POF to a nonheterochromatic target and to further our understanding of the evolution of these two targeting systems.
POF binding to PoX1 and PoX2
Considering the evolutionary relationship between POF and the MSL-complex, it was intriguing to find POF targeting to two distinct regions on the X-chromosome, i.e., X:3E and X:10E-F. The apparent spreading of POF targeting in these two regions (resembling the spreading of the MSL-complex when it is targeted to roX transgenes) and the close location of these regions to roX1 and roX2 suggested a link with the MSL-complex and dosage compensation. It has been hypothesized that POF originated as a dosage compensation system, since POF targets the male X-chromosome in, for example, D. busckii and D. ananassae and in those species POF colocalizes with H4K16ac and the MSL-complex, respectively (Larsson et al. 2004; Stenberg and Larsson 2011). However, the targeting of POF to endogenous PoX1 and PoX2 in D. melanogaster is restricted to females. This sex-specific targeting is not caused by sex-specific expression of the targeted genes, since comparable expression levels of RE64691 as well as SelG and CG1840 are consistently found in male and female salivary glands.
PoX targeting depends on roX expression
Not only are the two targeted loci, PoX1 and PoX2, located in close proximity to roX1 and roX2, the targeting is also largely dependent on roX, because losses of roX1 alone or of roX1 and roX2 cause a clear decrease in the frequency of PoX targeting. Importantly, in all roX mutant conditions tested, we never found a complete loss of POF binding to the PoX sites. Therefore, roX is not absolutely required for PoX targeting but rather it enhances or stabilizes the interaction. The dependence of targeting on roX is not caused by the close proximity of the PoX loci to the corresponding roX loci, because in the duplications tested the PoX1 and PoX2 are located on another chromosome, i.e. chromosome arm 3L, and the roX genes are not included in the duplicated region. Despite this, two of the duplications show targeting by POF, comparable to that to the endogenous loci. Furthermore, targeting to these transgenic regions was found to be largely dependent on roX, which indicates that roX can act in trans to enhance or stabilize POF targeting. The most parsimonious model to explain these observations is that it is the roX ncRNA species that enhance or stabilize targeting of POF to these non-chromosome 4 targets. This model is supported by the fact that roX2 overexpression seems to further increase the frequency of targeting. However, it should be stressed that endogenous roX expression in females is reported to be at very low levels or absent. In females, roX1 RNA has been reported in early embryos but it appears to be lost midway through embryogenesis, whereas in males expression is maintained through adulthood (Meller et al. 1997; Meller 2003). roX2 RNA first appears a few hours after roX1, but only in male embryos (Meller 2003).
Identification of a high-affinity target for POF
No high-affinity sites for POF targeting have previously been identified (Johansson et al. 2007a). It therefore came as a surprise to us that a short (6-kb) region from PoX2 functions as a strong ectopic target for POF in both males and females. The nonsex-specific targeting of POF to the P[w+ SelG CG1840] transgene, in contrast to Dp(1;3)DC246 and endogenous PoX, may be explained by a competition of targeting between POF and the MSL-complex in males. This competition will be more pronounced at the endogenous PoX sites and the duplications as these are also targets for the MSL-complex in males. This finding is supported by the fact that on polytene chromosomes, Dp(1;3)DC246 is targeted by MSL-complex in males while the P[w+ SelG CG1840] transgene is not targeted (results not shown). A competition in targeting is also supported by the reduction frequency of PoX1 and PoX2 targeting by POF observed in females expressing a partial MSL-complex, i.e. w; P[w+ hsp83:msl2] msl3 females (Table 1). It is important to note that the targeting of POF to the P[w+ SelG CG1840] transgene was not caused by genomic location of this transgene since the same attP docking site (3L:65B2) was used as for the duplications of the PoX1 and PoX2 loci. The lack of targeting of POF to translocated parts of the 4th chromosome and the strong targeting to the PoX2 transgene suggest that the PoX regions may be POF targets that are functionally separable from the 4th chromosome genes. Since both Setdb1 and HP1a are detected on the transgene, it appears likely that POF recruitment leads to, or is connected with, the formation of GREEN (HP1a and H3K9me enriched) chromatin structure (Filion et al. 2010).
The targeting of POF to the 4th chromosome depends on its well-characterized heterochromatic nature and on the presence of HP1a and Setdb1. It is therefore important to note that links between the MSL-complex, roX1 and roX2 and heterochromatic regions have been reported previously, though they remain to be understood. It is known that in roX1 roX2 mutant males, the MSL-complex is still detected on the X-chromosome, albeit at a reduced number of sites, but binding is also found in the chromocenter and at a few reproducible sites on the 4th chromosome (Meller and Rattner 2002; Deng and Meller 2006; Johansson et al. 2011). In contrast to the X-chromosome, where the MSL-complex is believed to stimulate gene expression, loss of roX RNA reduces expression from genes located in the chromocenter and on the 4th chromosome (Deng et al. 2009). It has been suggested that roX RNAs participate in two distinct regulatory systems, the dosage compensation system and a system for the modulation of heterochromatin (Deng et al. 2009). Although the mechanism by which roX RNAs enhance binding of POF to PoX loci remains elusive, the observation supports a model linking dosage compensation to modulation of heterochromatin. Additional factors supporting a model linking heterochromatin to dosage compensation are the proposed binding of HP1a to the male X-chromosome (de Wit et al. 2005) and the fact that a reduction in the histone H3S10 kinase JIL-1 results in the spreading of heterochromatic markers (such as H3K9me2 and HP1a) along the chromosome arms, with the most marked increase taking place on the X-chromosomes (Zhang et al. 2006). The JIL1 kinase, which is believed to counteract heterochromatin formation, is highly enriched on the male X-chromosome (Jin et al. 1999, 2000; Regnard et al. 2011) and is reported to be loosely attached to the MSL-complex (Jin et al. 2000; Wang et al. 2013). It is noteworthy that POF, which targets genes in a heterochromatic environment, i.e., on the 4th chromosome, has an intrinsic ability to target the male X-chromosome, as seen in, e.g., D. ananassae, and the targeting to X-chromosome sites reported here is dependent on roX RNAs. At the same time the MSL-complex, which binds to and stimulates expression of genes on the male X-chromosome, has an intrinsic ability to target heterochromatin as seen in the roX1 roX2 mutant background. The link between these two systems is intriguing and promises to increase our understanding of balanced gene expression.
High-affinity targeting to the PoX1 and PoX2 loci therefore provides a novel system for further studies on targeting mechanisms involved in chromosome-wide gene regulation, the evolutionary relationship between POF and dosage compensation and the evolution of balanced gene expression, and the results favor a model involving not only the X-chromosome but also balance to heterochromatin.
Acknowledgments
We thank Victoria Meller, Yongkyu Park, Mitzi Kuroda, Thomas Raabe, and Carole Seum for fly lines; Karin Ekström for technical assistance; and Mikhail Savitsky for the P[w+ attB] vector. This work was supported by grants from the Swedish Research Council and Swedish Cancer Foundation (to J.L.).
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Alekseyenko A. A., Peng S., Larschan E., Gorchakov A. A., Lee O. K., et al. , 2008. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X-chromosome. Cell 134: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw G. J., Baker B. S., 1997. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell 89: 789–798 [DOI] [PubMed] [Google Scholar]
- Conrad T., Cavalli F. M., Vaquerizas J. M., Luscombe N. M., Akhtar A., 2012. Drosophila dosage compensation involves enhanced Pol II recruitment to male X-linked promoters. Science 337: 742–746 [DOI] [PubMed] [Google Scholar]
- Dahlsveen I. K., Gilfillan G. D., Shelest V. I., Lamm R., Becker P. B., 2006. Targeting determinants of dosage compensation in Drosophila. PLoS Genet. 2: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Meller V. H., 2006. roX RNAs are required for increased expression of X-linked genes in Drosophila melanogaster males. Genetics 174: 1859–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Meller V. H., 2009. Molecularly severe roX1 mutations contribute to dosage compensation in Drosophila. Genesis 47: 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Koya S. K., Kong Y., Meller V. H., 2009. Coordinated regulation of heterochromatic genes in Drosophila melanogaster males. Genetics 182: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Hiatt J. B., Nguyen D. K., Ercan S., Sturgill D., et al. , 2011. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 43: 1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Greil F., van Steensel B., 2005. Genome-wide HP1 binding in Drosophila: developmental plasticity and genomic targeting signals. Genome Res. 15: 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo M. L. A., Philip P., Stenberg P., Larsson J., 2012. HP1a recruitment to promoters is independent of H3K9 methylation in Drosophila melanogaster. PLoS Genet. 8: e1003061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion G. J., van Bemmel J. G., Braunschweig U., Talhout W., Kind J., et al. , 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M. C., Forrest A. R., Nourbakhsh E., Pang K. C., Kai C., et al. , 2006. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo M. I., Pueyo J. I., Fouix S., Bishop S. A., Couso J. P., 2007. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 5: e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelbart M. E., Kuroda M. I., 2009. Drosophila dosage compensation: a complex voyage to the X-chromosome. Development 136: 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M., et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Parisi M., Sturgill D., Nuttall R., Doctolero M., et al. , 2006. Global analysis of X-chromosome dosage compensation. J. Biol. 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada F. N., Park P. J., Gordadze P. R., Kuroda M. I., 2005. Global regulation of X chromosomal genes by the MSL-complex in Drosophila melanogaster. Genes Dev. 19: 2289–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Zhang X., Borevitz J. O., Li W. H., Shiu S. H., 2007. A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res. 17: 632–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauch E., Melzig J., Brkulj M., Raabe T., 2002. In vivo functional analysis of Drosophila protein kinase casein kinase 2 (CK2) β-subunit. Gene 298: 29–39 [DOI] [PubMed] [Google Scholar]
- Jin Y., Wang Y., Walker D. L., Dong H., Conley C., et al. , 1999. JIL-1: a novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Mol. Cell 4: 129–135 [DOI] [PubMed] [Google Scholar]
- Jin Y., Wang Y., Johansen J., Johansen K. M., 2000. JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associates with the male specific lethal (MSL) dosage compensation complex. J. Cell Biol. 149: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A. M., Stenberg P., Bernhardsson C., Larsson J., 2007a Painting of fourth and chromosome-wide regulation of the 4th chromosome in Drosophila melanogaster. EMBO J. 26: 2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A. M., Stenberg P., Pettersson F., Larsson J., 2007b POF and HP1 bind expressed exons, suggesting a balancing mechanism for gene regulation. PLoS Genet. 3: e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A. M., Allgardsson A., Stenberg P., Larsson J., 2011. msl2 mRNA is bound by free nuclear MSL-complex in Drosophila melanogaster. Nucleic Acids Res. 39: 6428–6439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A. M., Stenberg P., Allgardsson A., Larsson J., 2012. POF regulates the expression of genes on the fourth chromosome in Drosophila melanogaster by binding to nascent RNA. Mol. Cell. Biol. 32: 2121–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmayer J. P., Ni L., Chu A., Kitchen L. E., Au W. C., et al. , 2006. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 16: 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. L., Solovyeva I., Lyman L. M., Richman R., Solovyev V., et al. , 1995. Expression of msl-2 causes assembly of dosage compensation regulators on the X-chromosomes and female lethality in Drosophila. Cell 81: 867–877 [DOI] [PubMed] [Google Scholar]
- Kelley R. L., Wang J., Bell L., Kuroda M. I., 1997. Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature 387: 195–199 [DOI] [PubMed] [Google Scholar]
- Kelley R. L., Meller V. H., Gordadze P. R., Roman G., Davis R. L., et al. , 1999. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98: 513–522 [DOI] [PubMed] [Google Scholar]
- Kind J., Akhtar A., 2007. Cotranscriptional recruitment of the dosage compensation complex to X-linked target genes. Genes Dev. 21: 2030–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T., Plaza S., Zanet J., Benrabah E., Valenti P., et al. , 2010. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science 329: 336–339 [DOI] [PubMed] [Google Scholar]
- Larschan E., Alekseyenko A. A., Gortchakov A. A., Peng S., Li B., et al. , 2007. MSL-complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol. Cell 28: 121–133 [DOI] [PubMed] [Google Scholar]
- Larsson J., Meller V. H., 2006. Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 14: 417–431 [DOI] [PubMed] [Google Scholar]
- Larsson J., Chen J. D., Rasheva V., Rasmuson Lestander A., Pirrotta V., 2001. Painting of fourth, a chromosome-specific protein in Drosophila. Proc. Natl. Acad. Sci. USA 98: 6273–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J., Svensson M. J., Stenberg P., Mäkitalo M., 2004. Painting of fourth in genus Drosophila suggests autosome-specific gene regulation. Proc. Natl. Acad. Sci. USA 101: 9728–9733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi J. C., 2009. The structure-function link of compensated chromatin in Drosophila. Curr. Opin. Genet. Dev. 19: 550–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg L. E., Figueiredo M. L., Stenberg P., Larsson J., 2012. Buffering and proteolysis are induced by segmental monosomy in Drosophila melanogaster. Nucleic Acids Res. 40: 5926–5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg L. E., Stenberg P., Larsson J., 2013. HP1a, Su(var)3-9, SETDB1 and POF stimulate or repress gene expression depending on genomic position, gene length and expression pattern in Drosophila melanogaster. Nucleic Acids Res. 41: 4481–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller V. H., 2003. Initiation of dosage compensation in Drosophila embryos depends on expression of the roX RNAs. Mech. Dev. 120: 759–767 [DOI] [PubMed] [Google Scholar]
- Meller V. H., Rattner B. P., 2002. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL-complex. EMBO J. 21: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller V. H., Wu K. H., Roman G., Kuroda M. I., Davis R. L., 1997. roX1 RNA paints the X-chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88: 445–457 [DOI] [PubMed] [Google Scholar]
- Nguyen D. K., Disteche C. M., 2006. Dosage compensation of the active X-chromosome in mammals. Nat. Genet. 38: 47–53 [DOI] [PubMed] [Google Scholar]
- Nicol J. W., Helt G. A., Blanchard S. G., Raja A., Loraine A. E., 2009. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25: 2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikov D. V., Kireev I., Belmont A. S., 2007. High-pressure treatment of polytene chromosomes improves structural resolution. Nat. Methods 4: 483–485 [DOI] [PubMed] [Google Scholar]
- Oh H., Bone J. R., Kuroda M. I., 2004. Multiple classes of MSL binding sites target dosage compensation to the X-chromosome of Drosophila. Curr. Biol. 14: 481–487 [DOI] [PubMed] [Google Scholar]
- Oliver B., 2007. Sex, dose, and equality. PLoS Biol. 5: e340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip P., Pettersson F., Stenberg P., 2012. Sequence signatures involved in targeting the Male-Specific Lethal complex to X-chromosomal genes in Drosophila melanogaster. BMC Genomics 13: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestel M., Feller C., Becker P. B., 2010a Dosage compensation and the global re-balancing of aneuploid genomes. Genome Biol. 11: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestel M., Feller C., Straub T., Mitlöhner H., Becker P. B., 2010b The activation potential of MOF is constrained for dosage compensation. Mol. Cell 38: 815–826 [DOI] [PubMed] [Google Scholar]
- Regnard C., Straub T., Mitterweger A., Dahlsveen I. K., Fabian V., et al. , 2011. Global analysis of the relationship between JIL-1 kinase and transcription. PLoS Genet. 7: e1001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass G. L., Pannuti A., Lucchesi J. C., 2003. Male-specific lethal complex of Drosophila targets activated regions of the X-chromosome for chromatin remodeling. Proc. Natl. Acad. Sci. USA 100: 8287–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C., Reo E., Peng H., Rauscher F. J., Spierer P., et al. , 2007. Drosophila SETDB1 is required for chromosome 4 silencing. PLoS Genet. 3: e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg P., Larsson J., 2011. Buffering and the evolution of chromosome-wide gene regulation. Chromosoma 120: 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenberg P., Lundberg L. E., Johansson A. M., Rydén P., Svensson M. J., et al. , 2009. Buffering of segmental and chromosomal aneuploidies in Drosophila melanogaster. PLoS Genet. 5: e100302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T., Grimaud C., Gilfillan G. D., Mitterweger A., Becker P. B., 2008. The chromosomal high-affinity binding sites for the Drosophila dosage compensation complex. PLoS Genet. 4: e1000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T., Zabel A., Gilfillan G. D., Feller C., Becker P. B., 2013. Different chromatin interfaces of the Drosophila dosage compensation complex revealed by high-shear ChIP-seq. Genome Res. 23: 473–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Fernandez H. R., Donohue R. C., Li J., Cheng J., et al. , 2013. Male-specific lethal complex in Drosophila counteracts histone acetylation and does not mediate dosage compensation. Proc. Natl. Acad. Sci. USA 110: E808–E817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson M. J., Chen J. D., Pirrotta V., Larsson J., 2003. The ThioredoxinT and deadhead gene pair encode testis- and ovary-specific thioredoxins in Drosophila melanogaster. Chromosoma 112: 133–143 [DOI] [PubMed] [Google Scholar]
- Tweedie S., Ashburner M., Falls K., Leyland P., McQuilton P., et al. , 2009. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 37: D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J., Carlson J. W., Schulze K. L., Pan H., He Y., et al. , 2009. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat. Methods 6: 431–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J., Popodi E., Holtzman S. L., Schulze K. L., Park S., et al. , 2010. A molecularly defined duplication set for the X-chromosome of Drosophila melanogaster. Genetics 186: 1111–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B., Bachtrog D., 2009. Progress and prospects toward our understanding of the evolution of dosage compensation. Chromosome Res. 17: 585–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. I., Alekseyenko A. A., Leroy G., Elia A. E., Gorchakov A. A., et al. , 2013. Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat. Struct. Mol. Biol. 20: 202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Deng H., Bao X., Lerach S., Girton J., et al. , 2006. The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development 133: 229–235 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Malone J. H., Powell S. K., Periwal V., Spana E., et al. , 2010. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 8: e1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]