Abstract
Poor prognosis of hepatocellular carcinoma (HCC) associated with late diagnosis necessitates the development of early diagnostic biomarkers. We have previously delineated the landscape of DNA methylation in HCC patients unraveling the importance of promoter hypomethylation in activation of cancer- and metastasis-driving genes. The purpose of the present study was to test the feasibility that genes that are hypomethylated in HCC could serve as candidate diagnostic markers. We use high resolution melting analysis (HRM) as a simple translatable PCR-based method to define methylation states in clinical samples. We tested seven regions selected from the shortlist of genes hypomethylated in HCC and showed that HRM analysis of several of them distinguishes methylation states in liver cancer specimens from normal adjacent liver and chronic hepatitis in the Shanghai area. Such regions were identified within promoters of neuronal membrane glycoprotein M6-B (GPM6B) and melanoma antigen family A12 (MAGEA12) genes. Differences in HRM in the immunoglobulin superfamily Fc receptor (FCRL1) separated invasive tumors from less invasive HCC. The identified biomarkers differentiated HCC from chronic hepatitis in another set of samples from Dhaka. Although the main thrust in DNA methylation diagnostics in cancer is on hypermethylated genes, our study for the first time illustrates the potential use of hypomethylated genes as markers for solid tumors. After further validation in a larger cohort, the identified DNA hypomethylated regions can become important candidate biomarkers for liver cancer diagnosis and prognosis, especially in populations with high risk for HCC development.
Introduction
Aberrations in epigenetic modifications, particularly in DNA methylation patterns, have been linked to cancer development and progression in many studies in last decades [1], [2], [3]. Hypermethylation of tumor suppressor genes linked to transcriptional silencing, global DNA demethylation associated with genome rearrangements and instability, and recently reported promoter hypomethylation linked to activation of oncogenes and prometastatic genes are hallmarks of nearly all types of cancer [2], [3], [4], [5]. DNA hypermethylation of tumor suppressor genes has been shown to have diagnostic potential in several cancers [6], [7], [8], however our recent unraveling of the broad scope of hypomethylation in liver cancer suggested that potential biomarkers might be found in hypomethylated genes that play a critical role in driving cancer and cancer metastasis [4]. Identifying reliable biomarkers of HCC is of particular importance since the late onset of clinical symptoms accounts for a late diagnosis and high mortality rate. It is estimated that early detection of HCC increases cure rate from 5% to 80% [9].
We previously used a genome-wide approach to delineate DNA promoter methylation profiles in HCC tumors and revealed nearly 2,000 genes whose promoters were hypomethylated in tumors compared with matched adjacent normal tissue [4]. These genes were implicated in biological processes and pathways crucial for cancer development and invasion, which points to an important functional role of the observed alterations. Hypomethylation was observed in several gene families across chromosomes. The question arose whether it will specifically mark tumors and differentiate tumor samples from healthy tissue. In our previous work, we focused on comparison between differences in average DNA methylation across the entire promoter and anti-correlated changes in gene expression. We analyzed in detail 230 genes that were hypomethylated and induced in HCC tumors. The majority of these genes fell into a category of promoters with high CpG content. In the present study on DNA hypomethylation biomarkers for liver cancer, we first selected genes that were heavily hypomethylated in HCC samples as compared with matched adjacent normal tissue (≥2-fold change in promoter methylation based on the microarray analysis, P<10E-4). This group of genes was then screened for the most consistently hypomethylated probe (200–400 bp region, ≥1.5-fold change, P<10E-3) across the patients as determined by the microarray analysis. In order to increase the functional relevance of potential biomarkers, we subsequently limited this list to 47 genes that were induced in HCC tumors as measured by Affymetrix arrays (≥1.5-fold change, P<0.05, Table S1). Taking into account changes in promoter methylation, expression, and the extent of demethylation of the specific probes, we have chosen seven genes that demonstrated the highest promoter hypomethylation, the most hypomethylated probes across the patients and high induction of expression. In addition, we limited the list to genes with cancer-related functions based on publicly available datasets as detailed below. These seven hypomethylated genes as listed in Table 1 were selected to identify and optimize specific probes whose differential methylation would distinguish tumors from normal tissues using High Resolution Melting analysis (HRM). The HRM analysis is a simple PCR-based method easily translatable to clinical setting for detection of differences in DNA methylation patterns [10]. This method requires treatment of DNA with bisulfite that converts cytosine residues to uracil leaving 5-methylcytosine residues unaffected. Following amplification, it results in changes in DNA sequence where uracil is converted to thymidine and 5-methylcytosines remain as cytosines. Unmethylated DNA amplicon that contains thymidine instead of cytosine within CpG sequences exhibits different melting properties than methylated DNA. Following primer optimization, three probes corresponding to three genes NEURONAL MEMBRANE GLYCOPROTEIN M6-B (GPM6B), MELANOMA ANTIGEN FAMILY A 12 (MAGEA12), and IMMUNOGLOBULIN SUPERFAMILY FC RECEPTOR (FCRL1), were found to differentiate HCC from adjacent normal tissue and/or invasive tumors from non-invasive tumors in a Chinese set of samples as well as HCC from chronic hepatitis B infection (CHB) in samples from Bangladesh. GPM6B is involved in neural development and regulation of bone formation [11], [12]. Expression of this gene was shown in genome wide transcriptome profiling to serve as a predictor of brain tumors [13], [14] and lymphoid leukemias [15]. High expression of MAGEA12, a member of oncogenic MAGE family, was shown to be associated with bladder carcinoma [16], melanoma [17], breast cancer [18] and oral squamous cell carcinoma [19]. FCRL1 is a cell-surface membrane protein preferentially expressed on B cells regulating B cell activation and differentiation [20]. High expression of this gene was observed in metastatic melanomas [21] and in different types of leukemias [20]. None of these genes or their state of methylation was previously linked to HCC. MAGEA12 was shown before to be repressed in normal cells by promoter methylation and activated in cancer cells by demethylation [22], [23], whereas the role of GPM6B and FCRL1 methylation in promoter activity was not previously tested. Our present study validates for the first time overexpression of GPM6B, MAGEA12 and FCRL1 in HCC patients relative to normal tissue and investigates DNA methylation within their promoters as a potential diagnostic marker of liver cancer. Since early diagnosis of HCC increases the cure rate and survival, the identified epigenetic candidate biomarkers could have an impact on liver cancer therapy outcome after validation in a larger cohort.
Table 1. Functional analysis of top 7 genes whose promoters were significantly hypomethylated in liver cancer samples compared to matched adjacent normal tissue as assessed by genome-wide promoter microarray.
| Gene symbol | Gene name | Chromosome | Pathway/Function/Biological process |
| DLGAP5 | discs, large (Drosophila) homolog-associated protein 5 | 14 | Hepatoma up-regulated protein; regulation of cell cycle, cell adhesion, cell-cell signaling, proliferation and differentiation; positive regulation of mitotic metaphase/anaphase transition; phosphoprotein phosphatase activity |
| FCRL1 | immunoglobulin superfamily Fc receptor, gp42 | 1 | Regulation of cell proliferation, B-cell activation and differentiation; regulation of cancer cell growth; receptor activity |
| GPM6B | neuronal membrane glycoprotein M6-b | X | Involved in neural development, regulation of osteoblast function and bone formation, matrix vesicle release by osteoblasts; maintenance of the actin cytoskeleton; negative regulation of serotonin uptake |
| MAGEA12 | melanoma antigen family A, 12; cancer/testis antigen family 1, member 12 | X | Not very well known; May play a role in transformation and tumor progression; in vitro increases viability of cancer cells |
| MMP1 | matrix metallopeptidase 1 (interstitial collagenase) | 11 | Involved in the breakdown of extracellular matrix, blood coagulation, leukocyte migration, collagen catabolic process and cancer metastasis |
| SSX1 | sarcoma, synovial, X-chromosome-related 1; cancer/testis antigen family 5, member 1 | X | Regulation of transcription; transcription corepressor activity; involved in humoral and cellular immune responses in cancer patients; involved in the t(X;18) translocation found in all synovial sarcomas (fusion of the synovial sarcoma translocation gene on chromosome 18 to one of the SSX gene on chromosome X, the rsulting hybrid is responsible for transforming activity) |
| TPO | thyroid peroxidase | 2 | Involved in thyroid gland function, thyroid hormone generation, embryonic hemopoiesis |
Functional analyses were performed using GO database.
Materials and Methods
Ethics statement
All patients provided written informed consent, and the Ethics Committee from concerned institutions (Chinese National Human Genome Center at Shanghai, China and Bangabandhu Sheikh Mujib Medical University (BSMMU), Dhaka, Bangladesh) approved the study.
Patients and tissue samples
Cancerous and normal adjacent tissue samples were obtained from 12 patients with HCC and 1 patient with inflammatory pseudotumor (control patient) in Chinese National Human Genome Center at Shanghai, China (Dr. Ze-Guang Han) (Table 2). Bangladeshi cohort consisted of 4 HCC patients and 4 CHB patients in Bangabandhu Sheikh Mujib Medical University (BSMMU), Dhaka, Bangladesh (Table 2). The HCC samples were obtained at ultrasonogram guided fine needle aspiration, while the CHB samples were obtained at per-cutaneous liver biopsy using Tru-cut biopsy needle. In both cases local anaesthesia was used.
Table 2. Clinicopathological characteristics of patients from Chinese and Bangladeshi cohorts.
| Patient ID | Gender | Age | Cellular type | Differentiation stage | Portal vein infiltration | |
| Chinese | 1 | Male | 48 | HCC | Middle | Yes |
| 4 | Male | 31 | HCC | Low | Yes | |
| 5 | Female | 69 | HCC | Middle | Yes | |
| 6 | Male | 51 | HCC | Middle | Yes | |
| 7 | Male | 80 | HCC | Middle to low | Yes | |
| 8 | Male | 43 | HCC | High | No | |
| 9 | Male | 50 | HCC | Middle | No | |
| 10 | Male | 44 | HCC | High | No | |
| 11 | Male | 73 | HCC | Middle to low | No | |
| 12 | Female | 51 | HCC | Middle to high | No | |
| 13 | Male | 40 | Inflammatory pseudotumor | - | No | |
| 14 | Male | 52 | HCC | Middle | No | |
| 15 | Male | 31 | HCC | Middle | No | |
| Bangladeshi | 1 | Male | 22 | CHB | - | - |
| 2 | Male | 48 | HCC | - | - | |
| 3 | Male | 35 | CHB | - | - | |
| 4 | Male | 29 | CHB | - | - | |
| 5 | Male | 35 | HCC | - | - | |
| 6 | Male | 55 | HCC | - | - | |
| 8 | Male | 20 | CHB | - | - | |
| 9 | Male | 34 | HCC | - | - |
DNA isolation and bisulfite treatment of DNA
DNA from cancerous, normal adjacent, and CHB samples was isolated using standard phenol-chloroform extraction technique. Bisulfite conversion was performed as previously described [24]. Briefly, 2 µg of DNA was linearized with EcoRI and incubated for 3 h at 37°C followed by purification using the Quick Clean PCR Purification Kit (GenScript) according to the manufacturer's protocol. The purified DNA was then denatured with 3M NaOH and incubated for 15 minutes at 37°C. Freshly prepared 3.6M sodium bisulfite/1 mM hydroquinone mixture (pH 5.0) was added to denatured DNA and incubated for 2 minutes at 95°C and then 8 h at 55°C followed by 2 minutes at 95°C and 2 h at 55°C. DNA samples were then desalted and purified (Quick Clean PCR Purification Kit, GenScript). The purified DNA was again denatured with 3M NaOH and incubated for 15 minutes at 37°C. The solution was neutralized by addition of ammonium acetate to a final concentration of 10M and the DNA was precipitated with 95% ethanol and the pellet was resuspended in 50 µl of distilled water.
PCR amplification of bisulfite converted DNA
Amplification reactions contained 25 µg of bisulfite-treated genomic DNA, 0.4 µM forward and reverse primers listed in Table S2, 10 µl of 10× Light Cycler 480 SybrGreen I Master (Roche) in a final volume of 20 µl. Amplification was performed in a thermocycler using the following conditions: denaturation at 95°C for 10 min, amplification for 40 cycles at 95°C for 1 min, annealing temperature for 2 min 30 s, 72°C for 1 min, and final extension at 72°C for 5 min. As shown in Table S2, outside and nested primer sets were used for several probes in order to improve the efficiency of amplification. The amplified DNA was then subjected to high resolution melting (HRM).
High resolution melting analysis (HRM)
The amplified DNA was transferred to the Light Cycler 480 QPCR instrument (Roche) that enables HRM. Samples were gradually heated from 40°C for 1 min to 60°C for 15 s and finally DNA was melted at 95°C for 15 s. The melting of the PCR product induces a decrease in the fluorescence of SybrGreen since SybrGreen as a DNA intercalating dye is being released from double-stranded DNA following dissociation. The fluorescence signal is acquired during the melting phase and analyzed by the Light Cycler 480 software. The temperature of a melting peak of DNA amplicon is defined by the midpoint of the melt phase at which the rate of changes in fluorescence is the largest. This melting point depends on the sequence and the length of the amplicon and is specific to each product. As following bisulfite conversion methylated DNA after amplification contains CG base pairs, the melting temperature of an unmethylated version that contains TA base pairs is different. The methylated amplicon melts at higher temperature than when unmethylated and samples of different methylation state can be separated by comparing the melting curve peaks. Using 0% and 100% methylated control DNA, we showed that the tested probes reveal clear differences between unmethylated and fully methylated DNA. 0% methylated control was generated using whole genome amplification kit (Sigma), whereas 100% control was created by in vitro methylation reaction catalyzed by SssI methyltransferase in the presence of a methyl donor S-adenosyl-L methionine (SAM). Melting curve peaks in HCC tumors were compared with the average of melting peaks in normal adjacent tissues from 7 individuals with non-invasive HCC. This average curve was called here as normal adjacent reference (NorAdjRef).
Pyrosequencing
Specific bisulfite converted promoter sequences were amplified with HotStar Taq DNA polymerase (Qiagen) using biotinylated primers listed in Table S2. The biotinylated DNA strands were pyrosequenced in the PyroMarkTMQ24 instrument (Biotage, Qiagen) as previously described [25]. Data were analyzed using PyroMarkTMQ24 software.
RNA extraction and Quantitative real-time PCR (QPCR)
Total RNA was isolated using TRIzol (Invitrogen, Life Technologies, Carlsbad, CA, USA) according to the manufacturer's protocol. 1 µg of total RNA served as a template for cDNA synthesis using 20 U of AMV reverse transcriptase (Roche Diagnostics), as recommended by the manufacturer. The QPCR reaction was carried out in Light Cycler 480 machine (Roche) using 2 µl of cDNA, 400 nM forward and reverse primers listed in Table S2 and 10 µl of Light Cycler 480 SybrGreen I Master (Roche) in a final volume of 20 µl. Amplification was performed using the following conditions: denaturation at 95°C for 10 min, amplification for 60 cycles at 95°C for 10 s, annealing temperature for 10 s, 72°C for 10 s, and final extension at 72°C for 10 min. Quantification was performed using a standard curve and analyzed by the Roche LightCycler 480 software.
Statistical analysis
Statistical analysis was performed using unpaired two-sided t-test or Mann-Whitney U test. The Mann-Whitney U test was used for comparisons between two groups where sample sizes were small and it was therefore difficult to verify distributional assumptions. In each case, the test is accompanied by a scatter plot showing all values used in the test. Mean values are given ± S.D. Where necessary, P-values are adjusted for multiple tests using Bonferroni correction. For comparisons of four vs. four samples (Bangladeshi cohort), we also used a permutation test where the data were shuffled 10,000 times and the test statistic was recalculated for each shuffle. The R software environment for statistical computing was used for all statistical analyses except the permutation test where the Resampling Stats Add-in for Excel software was used (Stan Blank, Charles Seiter, Peter Bruce and Resampling Stats, Inc. 2000, the Institute for Statistics Education, USA).
Results
Testing and identification of candidate hypomethylated regions in HCC by HRM analysis
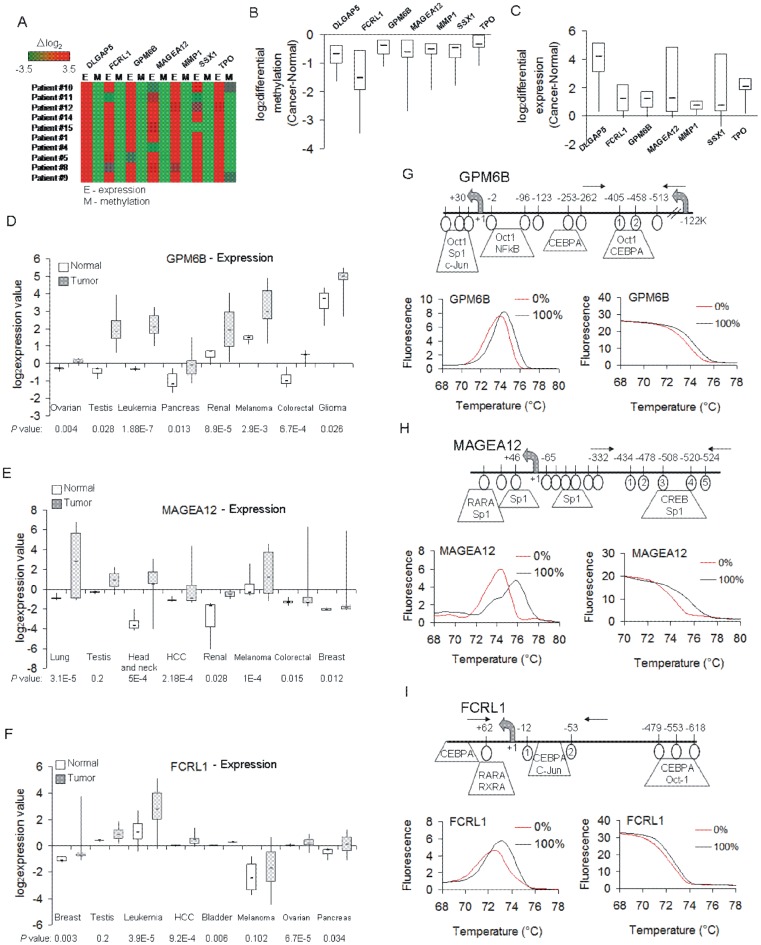
The goal of our study was to identify and optimize hypomethylated regions in HCC that could differentiate HCC from normal tissue using HRM. We applied several criteria that we predicted will increase the probability that the probes are ubiquitously differentially methylated in HCC. First, we selected seven genes (listed in Table 1) that were identified in our previous screen of hypomethylated genes in liver cancer using methylated DNA immunoprecipitation (MeDIP) and hybridization to genome-wide promoter oligonucleotide arrays. Second, the gene promoters contained probes with an average fold reduction in methylation between HCC and normal liver higher than or equal 2 and were highly statistically significant (P<10E-4). Third, hypomethylation of their promoters was also associated with overexpression in almost all the patients that were studied suggesting robustness and functional relevance of these DNA demethylation changes in HCC (Figure 1A–C). Fourth, the analysis of their functions based on publicly available datasets such as Gene Ontology (GO), KEGG, and NCBI reveals biological processes and pathways that are crucial for carcinogenesis such as regulation of cell cycle, cell adhesion, cell-cell signaling, proliferation and differentiation (Table 1). Fifth, the genes were previously associated with human cancers, in particular MAGEA12 [16], [17], [18], [19] however only DISCS LARGE (DROSOPHILA) HOMOLOG-ASSOCIATED PROTEIN 5 (DLGAP5) and MATRIX METALLOPEPTIDASE 1 (MMP1) have been previously reported to be up-regulated in HCC tumors [26], [27]. Sixth, using publicly available expression microarray data (Oncomine), we demonstrate that three of the genes that contained probes optimized for HRM analysis (see paragraph below) show increased expression in many different cancer types such as leukemia, ovarian and pancreatic cancer (GPM6B and FCRL1), renal and colorectal cancer (GPM6B and MAGEA12), HCC and breast cancer (MAGEA12 and FCRL1), testicular cancer and melanoma (significant overexpression of GPM6B in both cancers and MAGEA12 in melanoma) (Figure 1D–F). The data are consistent with a general role for these proteins in many different types of cancer and suggest their fundamental role in cancer progression and/or metastasis.
Figure 1. Hypomethylated and activated genes in HCC patients tested for biomarkers; characterization of optimized HRM probes corresponding to GPM6B, MAGEA12, and FCRL1.
(A) A heatmap showing log2 differential methylation (M) and expression (E) probe intensities for the indicated seven genes in every HCC patient samples as measured by microarrays. (B,C) Box plots of these methylation (B) and expression (C) differences in HCC patients (log2 [cancer-normal]). Negative methylation differences indicate hypomethylation in HCC and positive expression differences reflect higher expression in tumor samples as compared with matched adjacent normal tissue. (D,E,F) Cancer gene expression levels obtained from microarray data for GPM6B (D), MAGEA12 (E), and FCRL1 (F). The normal versus tumor gene expression data were obtained from Oncomine and are presented as log2-transformed median centered per array, and SD-normalized to 1 per array. All the presented changes are statistically significant (P<0.05) except MAGEA12 in testicular cancer and FCRL1 in testicular cancer and melanoma. (G,H,I) Melting curves and melting curve peaks for 0% and 100% methylated control DNA amplified for GPM6B (G), MAGEA12 (H) and FCRL1 (I) along with a map of promoters of the tested genes flanking the probes selected for HRM analysis. Horizontal arrows indicate the position of primers used for HRM. HRM for 0% and 100% methylated control DNA are shown. CpG sites that are located within the amplified probe are circled and numbered. The putative transcription factor binding sites are indicated as predicted by TransFac.
Three probes corresponding to GPM6B, MAGEA12, and FCRL1 optimized for HRM analysis of differential methylation in HCC
Several methylation-independent primer sets were subjected to optimization for HRM analysis (see primer sequences in Table S2). Control DNA that was methylated at 0% and 100% (standards) was amplified with the primers and melted in real-time. The melting pattern of the amplicons was established and analyzed. Melting curves for both standards were plotted in the same chart and exhibited different temperatures of melting peaks (Figure 1G–H). We then screened 12 HCC and 12 normal liver samples from the Shanghai cohort for methylation state within all 7 probes using designed primers. Four probes exhibited high heterogeneity of the methylation pattern in tumors, which was reflected in multiple melting peaks in HRM analysis for a single sample. Three other probes for GPM6B, MAGEA12, and FCRL1 showed a clear one-peak melting pattern and were used for further analyses as candidate DNA methylation markers. Overexpression of these genes was shown in other cancers, for instance higher expression of GPM6B was associated with brain tumors [13], [14] and lymphoid leukemias [15], MAGEA12 was up-regulated in bladder carcinoma [16], melanoma [17], breast cancer [18] and oral squamous cell carcinoma [19], whereas FCRL1 induction was observed in metastatic melanomas [21] and in different types of leukemias [20].
Differential HRM pattern of amplicons in GPM6B, MAGEA12, and FCRL1 differentiates HCC from normal liver
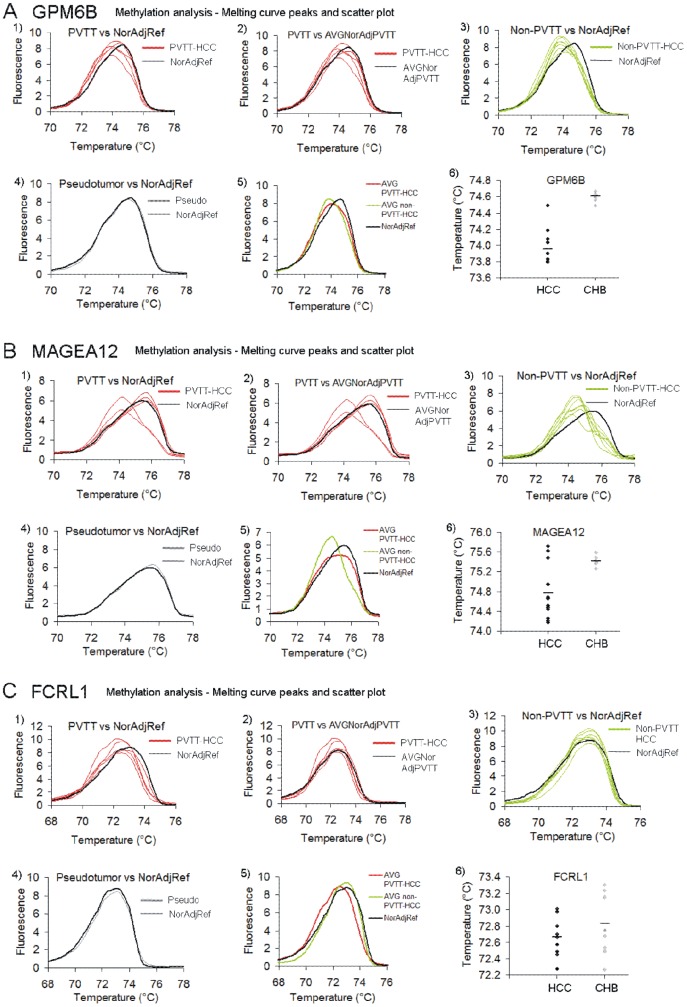
HRM analysis of tumors (n = 12) and adjacent normal tissues (n = 12) was performed. The melting curves were plotted and the temperature of a melting peak was calculated as described in the methods. As the methylated DNA amplicon melts at higher temperature than amplicons from unmethylated bisulfite converted DNA, samples of different methylation states can be differentiated by comparing the melting curve peaks. It is a common practice to compare tumors with pathologically normal adjacent tissue from the same patient. However, it may result in false negative results since so called normal liver can be already transformed molecularly and displayed altered DNA methylation patterns without exhibiting changes in histopathology, particularly in patients with highly invasive tumors. Five patients in the sample set were diagnosed with invasive HCC where portal vein tumor thrombus (PVTT) was detected. We assumed that normal adjacent liver can be already changed in these patients. We therefore created a normal reference curve (NorAdjRef) by averaging melting curves for all adjacent normal tissues excluding patients with PVTT. We suggest that such a reference curve could be further improved by including HRM results from a large sample of non-cancerous liver tissue and be used as a general standard for comparison for new cases particularly when invasion into adjacent tissue is suspected. Indeed, methylation of GPM6B probe in the tumor sample from patient 5 was the same as in its adjacent normal tissue, however it was different from NorAdjRef confirming the use of an averaged normal as a standard for comparison (Figure 2A, Figure 3A). Patients 1–6 with PVTT exhibited the same methylation state within FCRL1 probe when their tumors were compared to their matched adjacent normal tissue but again the HRM profiles were different from NorAdjRef (Figure 2C). Thus, by using an averaged normal standard we were able to call these samples as HCC, which would have been impossible if we used the adjacent tissue as a reference.
Figure 2. Methylation levels of GPM6B, MAGEA12 and FCRL1 promoters in HCC and non-cancerous tissue as measured by HRM.
(A–C) Melting curve peaks of GPM6B (A), MAGEA12 (B) and FCRL1 (C) amplicons in 12 tumor samples and 12 matched adjacent normal tissues. The peaks for HCC with PVTT (PVTT-HCC), HCC without PVTT (non-PVTT-HCC), and pseudotumor subjects were compared with the average of normal adjacent tissues from non-PVTT patients (NorAdjRef). In the second panel, melting peaks for tumor samples of HCC patients with PVTT were plotted together with the average of matched adjacent normal tissues from these patients, whereas the fifth panel shows the average of HCC with PVTT and HCC without PVTT in comparison with NorAdjRef. Temperature values for melting peaks in every patient are shown in scatter plots and in Table 3.
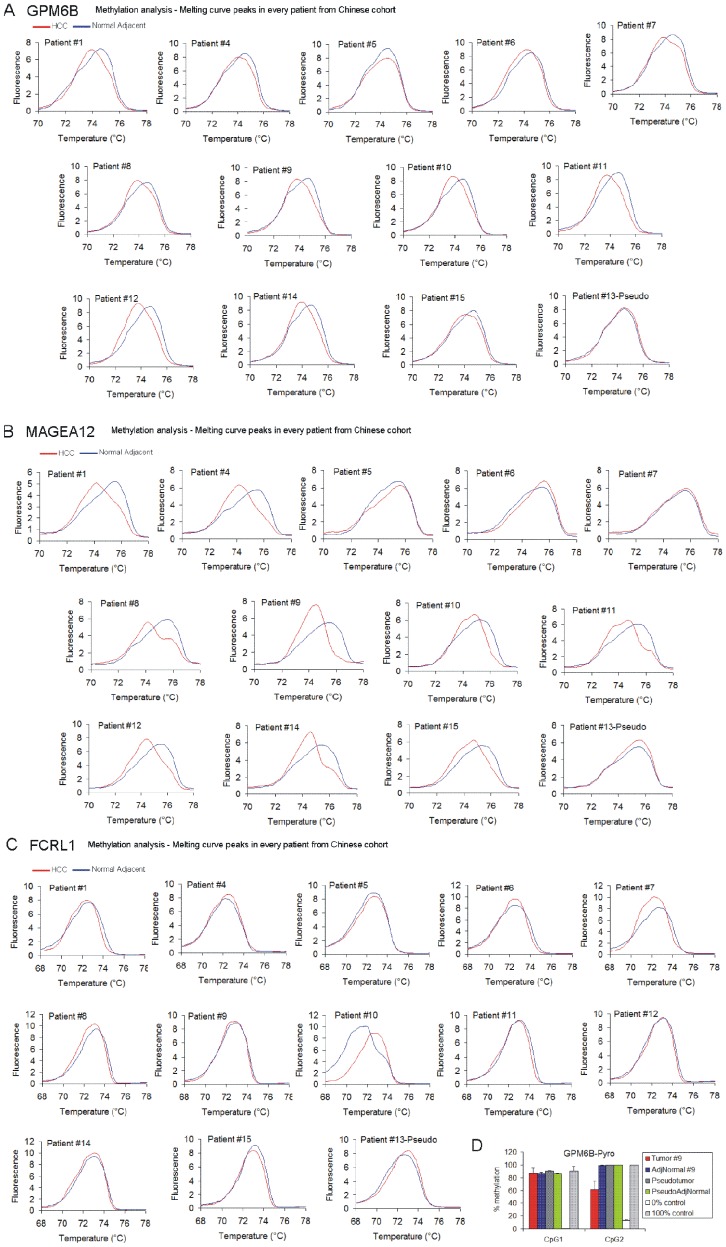
Figure 3. Methylation levels of GPM6B, MAGEA12 and FCRL1 promoters in individual HCC patients from the Chinese cohort as measured by HRM in comparison with the matched adjacent normal tissue from the same patient.
(A–C) Melting curve peaks of GPM6B (A), MAGEA12 (B) and FCRL1 (C) amplicons in tumor samples in comparison to matched adjacent normal tissue from the same patient. (D) Pyrosequencing of GPM6B region for patient 9 (HCC) and patient 13 (pseudotumor). The covered region overlaps with the sequence analyzed in HRM.
We have optimized three specific 150 bp DNA regions that could differentiate HCC from untransformed tissue using HRM. A region within GPM6B promoter was hypomethylated in tumors of all patients as compared with NorAdjRef (Figure 2A). All tumors were clearly separated and identified as HCC based on temperature values of melting peaks (Table 3, P = 3.97E-5, adjusted P = 1.17E-4, Mann-Whitney U test). Subjects with non-invasive HCC (non-PVTT) were more hypomethylated compared to invasive tumors as the difference in melting peaks between cancer and NorAdjRef indicates. Thus, the amplicon could differentiate also HCC without PVTT from HCC with PVTT, at least in the small number of cases examined here (Table 3, Figure 2A-charts 3 and 5, P = 0.03, adjusted P = 0.06, Mann-Whitney U test). HRM analysis of a region within MAGEA12 promoter differentiated all HCC without PVTT and 2 out of 5 HCC with PVTT from NorAdjRef (Figure 2B, Figure 3B, P = 1.7E-4, adjusted P = 5.1E-4, Mann-Whitney U test). Tumor samples from patients 5, 6, and 7 who were diagnosed with PVTT exhibited the same methylation level as their matched normal adjacent tissue (Figure 2B-chart 2, Figure 3B) and were not distinguished either from NorAdjRef (Figure 2B-chart 1). The average melting peak temperature value for the HCC without PVTT was lower than for the average HCC with PVTT indicating hypomethylation within MAGEA12 probe in non-invasive tumors (Table 3, Figure 2B-chart 5), although the change was not statistically significant in our set of samples (P = 0.64, Mann-Whitney U test). HRM for the FCRL1 probe differentiates HCC with PVTT from NorAdjRef (P = 2.5E-3, adjusted P = 7.5E-3, Mann-Whitney U test) but not HCC without PVTT from NorAdjRef (P = 0.11, Mann-Whitney U test) (Figure 2C-chart 1). The FCRL1 probe is hypomethylated in invasive compared to non-invasive tumors (Table 3, Figure 2C-chart 5, P = 2.5E-3, adjusted P = 7.5E-3, Mann-Whitney U test) and additionally differentiates these two kinds of tumors. Interestingly, although we nicely differentiate by HRM PVTT samples from NorAdjRef, there was no relevant difference when we compared FCRL1 methylation state in every PVTT tumor sample with the matched adjacent normal tissue from the same patient except patient 7 (Table 3, Figure 2C-chart2, Figure 3C). This suggests that the DNA methylation changes occurred already in the adjacent tissue in these livers. Patient 13 who was diagnosed with liver pseudotumor showed no difference in the HRM melting profile with its own adjacent tissue and NorAdjRef in all three probes (Table 3, Figure 2-charts 4, P>0.05). We confirmed this observation by pyrosequencing of GPM6B region for HCC subject 9 and pseudotumor patient as shown in Figure 3D.
Table 3. Temperature values of melting curve peaks of GPM6B (A), MAGEA12 (B) and FCRL1 (C) amplicons in tumor samples and normal liver tissues in Chinese (Ch) and Bangladeshi (B) cohorts as analyzed by HRM.
| GPM6B | MAGEA12 | FCRL1 | ||||||||||||||
| HCC | NorAdj/CHB | HCC | NorAdj/CHB | HCC | NorAdj/CHB | |||||||||||
| AVG | SD | AVG | SD | P | AVG | SD | AVG | SD | P | AVG | SD | AVG | SD | P | ||
| Ch | Patient 1 (PVTT) | 73.91 | 0.04 | 74.62 | 0.10 | <0.0001 | 74.21 | 0.13 | 75.41 | 0.21 | <0.0001 | 72.46 | 0.22 | 72.51 | 0.35 | 0.016 |
| Patient 4 (PVTT) | 74.04 | 0.05 | 74.59 | 0.15 | <0.0001 | 74.18 | 0.02 | 75.38 | 0.26 | <0.0001 | 72.49 | 0.17 | 72.27 | 0.19 | 0.018 | |
| Patient 5 (PVTT) | 74.49 | 0.01 | 74.49 | 0.01 | 0.018 | 75.62 | 0.01 | 75.40 | 0.15 | 0.166 | 72.49 | 0.18 | 72.54 | 0.20 | 0.018 | |
| Patient 6 (PVTT) | 74.19 | 0.13 | 74.56 | 0.11 | 0.0002 | 75.48 | 0.21 | 75.48 | 0.11 | 0.58 | 72.58 | 0.05 | 72.49 | 0.17 | 0.031 | |
| Patient 7 (PVTT) | 73.84 | 0.06 | 74.60 | 0.06 | <0.0001 | 75.72 | 0.03 | 75.60 | 0.05 | 0.06 | 72.28 | 0.27 | 72.68 | 0.00 | 0.0056 | |
| Patient 8 | 73.80 | 0.01 | 74.63 | 0.08 | <0.0001 | 74.26 | 0.20 | 75.43 | 0.29 | 0.0002 | 72.97 | 0.03 | 73.16 | 0.10 | <0.0001 | |
| Patient 9 | 73.83 | 0.14 | 74.63 | 0.08 | <0.0001 | 74.49 | 0.02 | 75.42 | 0.32 | 0.0006 | 72.58 | 0.54 | 72.75 | 0.68 | 0.084 | |
| Patient 10 | 73.80 | 0.01 | 74.63 | 0.01 | <0.0001 | 74.95 | 0.20 | 75.26 | 0.04 | 0.032 | 72.66 | 0.51 | 73.23 | 0.00 | 0.13 | |
| Patient 11 | 73.81 | 0.11 | 74.63 | 0.08 | <0.0001 | 74.70 | 0.02 | 75.40 | 0.24 | 0.0027 | 72.98 | 0.02 | 73.30 | 0.00 | 0.61 | |
| Patient 12 | 73.80 | 0.19 | 74.67 | 0.04 | <0.0001 | 74.45 | 0.07 | 75.43 | 0.20 | 0.0005 | 73.01 | 0.02 | 73.16 | 0.20 | 0.73 | |
| Patient 14 | 73.91 | 0.06 | 74.63 | 0.08 | <0.0001 | 74.52 | 0.03 | 75.40 | 0.24 | 0.0008 | 72.80 | 0.71 | 72.75 | 0.64 | 0.36 | |
| Patient 15 | 74.08 | 0.08 | 74.63 | 0.08 | <0.0001 | 74.66 | 0.03 | 75.37 | 0.19 | 0.0021 | 72.70 | 0.56 | 73.15 | 0.12 | 0.17 | |
| All HCC Average | 73.96 | 0.03 | 74.61 | 0.00 | <0.0001 | 74.77 | 0.03 | 75.41 | 0.19 | <0.0001 | 72.66 | 0.26 | 72.83 | 0.22 | 0.0028 | |
| Non-PVTT Average | 73.86 | 0.06 | 74.64 | 0.06 | <0.0001 | 74.57 | 0.06 | 75.39 | 0.21 | <0.0001 | 72.81 | 0.33 | 73.07 | 0.25 | 0.6 | |
| Pseudo 13 | 74.56 | 0.11 | 74.53 | 0.06 | 0.23 | 75.59 | 0.03 | 75.43 | 0.20 | 0.22 | 72.94 | 0.27 | 72.61 | 0.15 | 0.53 | |
| B | Patient 2 (HCC) | 74.09 | 0.12 | - | - | 0.47 | 73.15 | 0.06 | - | - | <0.0001 | 72.02 | 0.17 | - | - | 0.0086 |
| Patient 5 (HCC) | 73.74 | 0.07 | - | - | 0.03 | 73.22 | 0.06 | - | - | <0.0001 | 71.95 | 0.07 | - | - | 0.0002 | |
| Patient 6 (HCC) | 73.78 | 0.02 | - | - | 0.03 | 74.36 | 0.21 | - | - | 0.001 | 71.93 | 0.16 | - | - | 0.0028 | |
| Patient 9 (HCC) | 73.64 | 0.03 | - | - | 0.015 | 73.26 | 0.11 | - | - | <0.0001 | 72.10 | 0.11 | - | - | 0.0046 | |
| HCC Average | 73.81 | 0.00 | - | - | 0.0053 | 73.50 | 0.11 | - | - | <0.0001 | 72.00 | 0.05 | - | - | <0.0001 | |
| Patient 1 (CHB) | - | - | 74.12 | 0.17 | - | - | - | 75.74 | 0.18 | - | - | - | 72.13 | 0.08 | - | |
| Patient 3 (CHB) | - | - | 74.15 | 0.22 | - | - | - | 74.50 | 0.70 | - | - | - | 72.45 | 0.28 | - | |
| Patient 4 (CHB) | - | - | 74.19 | 0.27 | - | - | - | 75.56 | 0.03 | - | - | - | 72.25 | 0.01 | - | |
| Patient 8 (CHB) | - | - | 74.33 | 0.07 | - | - | - | 75.67 | 0.08 | - | - | - | 72.69 | 0.25 | - | |
| CHB Average | - | - | 74.20 | 0.18 | - | - | - | 75.36 | 0.10 | - | - | - | 72.38 | 0.01 | - | |
The value for every patient in Chinese cohort was compared with the average of normal adjacent tissue in 7 non-invasive patients (NorAdjRef, non-PVTT average). The value for every patient in Bangladeshi cohort was compared with the average of all CHB samples.
In summary, a combination of these three HRM optimized probes would differentiate tumors from non-tumors (using GPM6B probe and MAGEA12 in combination with GPM6B) and HCC with PVTT from non-PVTT HCC (using FCRL1 probe).
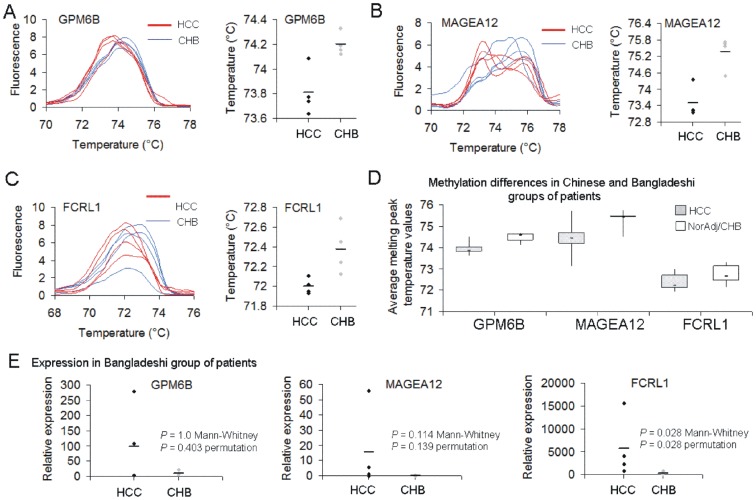
Validation of identified epigenetic biomarkers in Bangladeshi biopsy specimens from HCC and chronic hepatitis B (CHB) patients
The critical question in diagnosis of HCC is identifying early conversion of CHB to HCC. We therefore asked whether the identified probes could distinguish between HCC and CHB in an independent set of subjects. We tested 4 subjects with HCC and 4 subjects with CHB (Table 2 for clinical characteristics). The GPM6B probe which differentiated all HCC from NorAdjRef in the Shanghai group of patients also showed different melting patterns in HCC and CHB except one patient with HCC who was grouped together with CHB based on melting peak temperature (Table 3, Figure 4A, P = 0.028, adjusted P = 0.084, Mann-Whitney U test; P<0.0001, permutation test). Methylation state within MAGEA12 probe separated all HCC cases from CHB as indicated by lower melting temperature (Table 3, Figure 4B, P = 0.028, adjusted P = 0.084, Mann-Whitney U test; P = 0.02, permutation test). In HCC patients, we clearly observed two melting peaks indicating two populations of cells with low and high MAGEA12 methylation level. A probe within FCRL1 was hypomethylated in all tumors as compared with the average of all CHB patients (Table 3, Figure 4C, P = 0.028, adjusted P = 0.084, Mann-Whitney U test; P<0.0001, permutation test). When we grouped melting peak temperature values for all HCC samples and all adjacent normal and CHB samples, median value for each tested probe was lower in cancer than in normal indicating hypomethylation in cancer (Figure 4D). The differences were highly statistically significant for GPM6B and MAGEA12 probes (P<0.001, adjusted P<0.003, Mann-Whitney U test) as well as for FCRL1 but after excluding HCC samples without PVTT (P<0.01, adjusted P<0.03, Mann-Whitney U test). Expression of the tested genes as measured by QPCR in the Bangladeshi group was on average higher in HCC than in CHB subjects (see Figure 4E for exact values in every patient) with the most relevant changes observed for FCRL1 (Figure 4E, P = 0.028, adjusted P = 0.084, Mann-Whitney U test; P = 0.028, permutation test).
Figure 4. Methylation and expression levels of GPM6B, MAGEA12 and FCRL1 in HCC and chronic hepatitis B (CHB) patients as measured by HRM and QPCR, respectively.
Melting curve peaks of GPM6B (A), MAGEA12 (B) and FCRL1 (C) amplicons in 4 HCC and 4 CHB patients from Bangladesh. Temperature values for melting peaks in every patient are shown in scatter plots and in Table 3. (D) Box plots of the average melting peak temperature values across all the patients in both cohorts. Lower temperature value indicates lower methylation level within a probe. (E) Relative expression of the tested genes in HCC and CHB subjects as measured by QPCR and depicted as scatter plots. P values calculated using Mann-Whitney U test and permutation test are shown.
Discussion
Late onset of clinical symptoms of HCC accounts for late diagnosis and poor prognosis. As early detection of HCC improves cure rate from 5% to 80%, there is a need for development of diagnostic biomarkers [9]. Epigenetic alterations, especially changes in DNA methylation patterns, have been implicated in many types of human cancer and observed at initiation and progression stages [1], [2], [3]. Recent genome-wide approaches delineated cancer-specific changes in DNA methylation that open the door to the emerging field of DNA methylation markers for early detection, prognosis and response to therapy. For instance, DNA methylation within paired-like Homeodomain Transcription Factor 2 (PITX2) has been validated using paraffin-embedded tissues as a predictive marker of adjuvant tamoxifen therapy outcome in breast cancer patients [6]. Our recent studies revealed robust changes in the landscape of DNA methylation in HCC patients and showed that promoter specific hypomethylation is associated with activation of cancer-driving genes [4]. High frequency of DNA promoter hypomethylation was subsequently confirmed in another study on different sets of HCC samples [28] as well as in neuroblastoma samples [5]. These studies highlight the importance of promoter hypomethylation in cancer and the need to further explore the role of demethylation in cancer. One practical question is whether genes that are targets of hypomethylation in cancer can serve as markers that could improve early diagnosis and/or prognosis.
In the present study, we screened for potential DNA hypomethylation markers for liver cancer diagnosis among genes that we previously found to be hypomethylated in HCC patients. Our aim was to develop primers for specific probes within these genes that will differentiate HCC from normal liver, chronic hepatitis as well as PVTT invasive HCC using a simple PCR based method that could easily be translated into clinical settings. We have identified three specific DNA probes covering regions around 150 bp long as potential candidates for markers in HCC using a simple PCR-based HRM method. The probes correspond to three genes GPM6B, MAGEA12, and FCRL1 that were overexpressed and demethylated in our set of HCC patients (Figure 1B and 1C) and up-regulated in many other types of cancer (Figure 1D–F). Differential methylation of promoters of these genes was not previously reported in cancer although methylation-dependent regulation of MAGEA12 promoter was shown in several cancer cell lines [23].
As a reference in our analyses, we used the average of all adjacent normal tissues from HCC patients without PVTT (non-PVTT HCC) termed as a normal adjacent reference curve (NorAdjRef) rather than the adjacent to tumor liver sample of the same case. The rationale behind this approach is the many false negative results obtained when tumor samples are being compared with pathologically normal adjacent tissue from the same patient. We speculate that the normal tissue, especially in patients with already developed metastases, can be already transformed and bear altered DNA methylation patterns without changes in histology. In our study, it was the case for patient 5 who was identified based on HRM as a cancer subject only when GPM6B methylation in the tumor sample was compared with NorAdjRef (Figure 2A, Figure 3A). Similarly, FCRL1 methylation separated tumors of patients 1–6 from NorAdjRef but not from adjacent tissue from the same patient (Figure 2C). Our approach minimizes false negative results and increases specificity, which might facilitate detection of liver transformation early before the histopathological changes are visualized. Using an average normal curve provides an “objective” standard for comparison for unknown cases. Our results obtained from a limited set of subjects must be validated in a larger clinical study. Our data nevertheless point to the feasibility of this approach. It is particularly important to determine whether using an average non-cancer reference will indeed improve our ability to detect HCC prior to manifestation of histopathological changes.
The GPM6B probe differentiated all tested HCC samples either with or without PVTT from NorAdjRef (Table 3, Figure 2A). A probe within MAGEA12 should be used in combination with GPM6B in order to detect cancer as melting curves for tumors of a few patients with PVTT overlap with NorAdjRef curve (Table 3, Figure 2B). The third probe located within FCRL1 promoter is hypomethylated in invasive tumors and differentiates invasive from non-invasive cases (Table 3, Figure 2C). The combination of probes should detect all HCC cases and differentiate PVTT and non-PVTT HCC. All three probes distinguished HCC from CHB in Bangladeshi set of samples as compared with the average of CHB melting peaks (Table 3, Figure 4A–C). In both Chinese and Bangladeshi groups of patients, HCC was associated with hepatitis B virus (HBV) infection that is the major cause of primary liver cancer in these countries. The incidence of liver cancer in China and Bangladesh is rising together with HBV infection. The fact that we could validate cancer-specific hypomethylation of the identified probes in a different group of patients from a widely different ethnic background in a different geographical location points to the feasibility of using the identified probes as markers of HCC. High statistical relevance of the differences in melting temperature values (Figure 4D, P<0.001 for GPM6B and MAGEA12, P<0.01 for FCRL1 after excluding HCC without PVTT, Mann-Whitney U test) even with the small sample size provides justification for larger studies. Moreover, it will be of interest to test the identified probes in circulating tumor cells in blood as potentially non-invasive source of DNA. There is previous evidence for the feasibility of this approach in other cancers. A MAGEA gene expression test that provides information on multiple MAGEA genes in a single reaction detected circulating tumor cells in blood of melanoma, breast and colorectal patients [29]. Brennan et al. discovered methylation in ATAXIA TELANGIECTASIA MUTATED (ATM) intragenic loci in DNA from white blood cells as a potential marker of breast cancer risk [30]. Circulating methylated SEPTIN 9 (SEPT9) DNA and INTERMEDIATE FILAMENT FAMILY ORPHAN 1 (IFFO1) DNA in plasma were found to correlate with the occurrence of colorectal cancer [7] and ovarian cancer [8], respectively.
In contrary to most of the DNA methylation marker studies that focus on promoters that are hypermethylated in cancer, we unravel genes and specific probes within their promoters that are hypomethylated in cancer as compared with normal tissue. Using HRM, a straightforward method providing results in a short period of time, the identified probes are highly suitable for further testing for clinical applications and introduction into routine clinical tests after validation in larger studies.
Although our study is limited in the number of cases examined and did not have the necessary power to distinguish early and late stages of liver cancer, it is remarkable that even with a small sample size (cancer: n = 16, normal: n = 16) we were able to obtain highly significant results (Figure 4D). This positions these probes as excellent candidates for a larger clinical study. If proven successful early markers of HCC could be used to predict liver cancer in high-risk populations for developing HCC such as hepatitis B and C infection, type 2 diabetes mellitus, and alcohol-driven cirrhosis.
Supporting Information
Candidate genes for screening for epigenetic biomarkers of liver cancer.
(XLS)
Primer sequences and description of all the specific probes that were tested in HRM.
(DOC)
Acknowledgments
International Centre for Diarrhoeal Diseases Research, Bangladesh (icddr,b) acknowledges with gratitude the commitment of donors to the Centre's research efforts. MS is a fellow of the Canadian Institute for Advanced Research.
Funding Statement
This research was supported by grants from Ministere du Developpement Economique, de l'Innovation et de l'Exportation (MDEIE) program of the government of Quebec (No. 215004, to MS), the Canadian Institute of Health Research (No. MOP-42411, to MS) and International Centre for Diarrhoeal Diseases Research, Bangladesh (icddr,b). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jones PA, Baylin SB (2007) The epigenomics of cancer. Cell 128: 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Szyf M (2005) DNA methylation and demethylation as targets for anticancer therapy. Biochemistry (Mosc) 70: 533–549. [DOI] [PubMed] [Google Scholar]
- 3. Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, et al. (2001) Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet 10: 687–692. [DOI] [PubMed] [Google Scholar]
- 4. Stefanska B, Huang J, Bhattacharyya B, Suderman M, Hallett M, et al. (2011) Definition of the landscape of promoter DNA hypomethylation in liver cancer. Cancer Res 71: 5891–5903. [DOI] [PubMed] [Google Scholar]
- 5. Mayol G, Martin-Subero JI, Rios J, Queiros A, Kulis M, et al. (2012) DNA hypomethylation affects cancer-related biological functions and genes relevant in neuroblastoma pathogenesis. PLoS One 7: e48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martens JW, Margossian AL, Schmitt M, Foekens J, Harbeck N (2009) DNA methylation as a biomarker in breast cancer. Future Oncol 5: 1245–1256. [DOI] [PubMed] [Google Scholar]
- 7. deVos T, Tetzner R, Model F, Weiss G, Schuster M, et al. (2009) Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem 55: 1337–1346. [DOI] [PubMed] [Google Scholar]
- 8. Campan M, Moffitt M, Houshdaran S, Shen H, Widschwendter M, et al. (2011) Genome-scale screen for DNA methylation-based detection markers for ovarian cancer. PLoS One 6: e28141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takayama T (2011) Surgical treatment for hepatocellular carcinoma. Jpn J Clin Oncol 41: 447–454. [DOI] [PubMed] [Google Scholar]
- 10. Kristensen LS, Mikeska T, Krypuy M, Dobrovic A (2008) Sensitive Melting Analysis after Real Time- Methylation Specific PCR (SMART-MSP): high-throughput and probe-free quantitative DNA methylation detection. Nucleic Acids Res 36: e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drabek K, van de Peppel J, Eijken M, van Leeuwen JP (2011) GPM6B regulates osteoblast function and induction of mineralization by controlling cytoskeleton and matrix vesicle release. J Bone Miner Res 26: 2045–2051. [DOI] [PubMed] [Google Scholar]
- 12. Werner HB, Kramer-Albers EM, Strenzke N, Saher G, Tenzer S, et al. (2013) A critical role for the cholesterol-associated proteolipids PLP and M6B in myelination of the central nervous system. Glia 61: 567–586. [DOI] [PubMed] [Google Scholar]
- 13. Castells X, Garcia-Gomez JM, Navarro A, Acebes JJ, Godino O, et al. (2009) Automated brain tumor biopsy prediction using single-labeling cDNA microarrays-based gene expression profiling. Diagn Mol Pathol 18: 206–218. [DOI] [PubMed] [Google Scholar]
- 14. Castells X, Acebes JJ, Boluda S, Moreno-Torres A, Pujol J, et al. (2010) Development of a predictor for human brain tumors based on gene expression values obtained from two types of microarray technologies. OMICS 14: 157–164. [DOI] [PubMed] [Google Scholar]
- 15. Charfi C, Voisin V, Levros LC Jr, Edouard E, Rassart E (2011) Gene profiling of Graffi murine leukemia virus-induced lymphoid leukemias: identification of leukemia markers and Fmn2 as a potential oncogene. Blood 117: 1899–1910. [DOI] [PubMed] [Google Scholar]
- 16. Heidecker L, Brasseur F, Probst-Kepper M, Gueguen M, Boon T, et al. (2000) Cytolytic T lymphocytes raised against a human bladder carcinoma recognize an antigen encoded by gene MAGE-A12. J Immunol 164: 6041–6045. [DOI] [PubMed] [Google Scholar]
- 17. Gibbs P, Hutchins AM, Dorian KT, Vaughan HA, Davis ID, et al. (2000) MAGE-12 and MAGE-6 are frequently expressed in malignant melanoma. Melanoma Res 10: 259–264. [PubMed] [Google Scholar]
- 18. Otte M, Zafrakas M, Riethdorf L, Pichlmeier U, Loning T, et al. (2001) MAGE-A gene expression pattern in primary breast cancer. Cancer Res 61: 6682–6687. [PubMed] [Google Scholar]
- 19. Mollaoglu N, Vairaktaris E, Nkenke E, Neukam FW, Ries J (2008) Expression of MAGE-A12 in oral squamous cell carcinoma. Dis Markers 24: 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Du X, Nagata S, Ise T, Stetler-Stevenson M, Pastan I (2008) FCRL1 on chronic lymphocytic leukemia, hairy cell leukemia, and B-cell non-Hodgkin lymphoma as a target of immunotoxins. Blood 111: 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koh SS, Wei JP, Li X, Huang RR, Doan NB, et al. (2012) Differential gene expression profiling of primary cutaneous melanoma and sentinel lymph node metastases. Mod Pathol 25: 828–837. [DOI] [PubMed] [Google Scholar]
- 22. Wischnewski F, Friese O, Pantel K, Schwarzenbach H (2007) Methyl-CpG binding domain proteins and their involvement in the regulation of the MAGE-A1, MAGE-A2, MAGE-A3, and MAGE-A12 gene promoters. Mol Cancer Res 5: 749–759. [DOI] [PubMed] [Google Scholar]
- 23. Wischnewski F, Pantel K, Schwarzenbach H (2006) Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, -A2, -A3, and -A12 in human cancer cells. Mol Cancer Res 4: 339–349. [DOI] [PubMed] [Google Scholar]
- 24. Colella S, Shen L, Baggerly KA, Issa JP, Krahe R (2003) Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques 35: 146–150. [DOI] [PubMed] [Google Scholar]
- 25. Tost J, Gut IG (2007) DNA methylation analysis by pyrosequencing. Nat Protoc 2: 2265–2275. [DOI] [PubMed] [Google Scholar]
- 26. Kuo TC, Chang PY, Huang SF, Chou CK, Chao CC (2012) Knockdown of HURP inhibits the proliferation of hepacellular carcinoma cells via downregulation of gankyrin and accumulation of p53. Biochem Pharmacol 83: 758–768. [DOI] [PubMed] [Google Scholar]
- 27. Liao M, Tong P, Zhao J, Zhang Y, Li Z, et al. (2012) Prognostic value of matrix metalloproteinase-1/proteinase-activated receptor-1 signaling axis in hepatocellular carcinoma. Pathol Oncol Res 18: 397–403. [DOI] [PubMed] [Google Scholar]
- 28. Song MA, Tiirikainen M, Kwee S, Okimoto G, Yu H, et al. (2013) Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma. PLoS One 8: e55761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miyashiro I, Kuo C, Huynh K, Iida A, Morton D, et al. (2001) Molecular strategy for detecting metastatic cancers with use of multiple tumor-specific MAGE-A genes. Clin Chem 47: 505–512. [PubMed] [Google Scholar]
- 30. Brennan K, Garcia-Closas M, Orr N, Fletcher O, Jones M, et al. (2012) Intragenic ATM methylation in peripheral blood DNA as a biomarker of breast cancer risk. Cancer Res 72: 2304–2313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Candidate genes for screening for epigenetic biomarkers of liver cancer.
(XLS)
Primer sequences and description of all the specific probes that were tested in HRM.
(DOC)