Abstract
Plants use day-length information to coordinate flowering time with the appropriate season to maximize reproduction. In Arabidopsis, the long-day specific expression of CONSTANS (CO) protein is crucial for flowering induction. Although light signaling regulates CO protein stability, the mechanism by which CO is stabilized in the long-day afternoon has remained elusive. Here we demonstrate that FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1) protein stabilizes CO protein in the afternoon in long days. FKF1 interacts with CO through its LOV domain, and blue light enhances this interaction. In addition, FKF1 simultaneously removes CYCLING DOF FACTOR 1 (CDF1) that represses CO and FLOWERING LOCUS T (FT) transcription. Together with CO transcriptional regulation, FKF1 protein controls robust FT mRNA induction through multiple feedforward mechanisms that accurately control flowering timing.
Plants monitor seasonal day-length changes by sensing light conditions at a specific time of day to regulate flowering (1). In Arabidopsis, the day-length specific expression of FLOWERING LOCUS T (FT) protein is crucial for the proper timing of flowering (2–4). FT transcription is directly activated by CONSTANS (CO) transcription factor (5), which is stabilized only in the afternoon of long days (6). The regulation of long-day-specific CO protein expression is one of the most crucial mechanisms for photoperiodic flowering (2–4). Light signals perceived by phytochrome A (phyA) and cryptochrome (cry) photoreceptors stabilize CO protein in long days (6). However, the timing mechanism by which CO protein is stabilized specifically in the long-day afternoon was not well understood, as both phyA and cry proteins are expressed at similar levels throughout the day in long days (7). Previously we showed that FKF1 regulates CO transcription (8–10). Here we demonstrate that FKF1 also controls FT transcription by stabilizing CO protein and simultaneously degrading FT repressors in the long-day afternoon. Thus, our results indicate that FKF1 has multiple roles in photoperiodic flowering regulation.
As our mathematical gene-circuit model predicted that FKF1 protein would induce FT expression independently of CO transcriptional regulation (11), we examined the possibility of FKF1-dependent FT gene regulation. Since FKF1 degrades CDF1 transcription factor (9, 10), we hypothesized that if CDF1 acts as a FT transcriptional repressor, removing CDF1 protein might regulate FT expression. As FKF1 and CO proteins interact in yeast (12), a second possibility was that FKF1 regulates CO protein activity. To investigate additional roles of FKF1 protein in photoperiodic flowering, we tested these two possibilities.
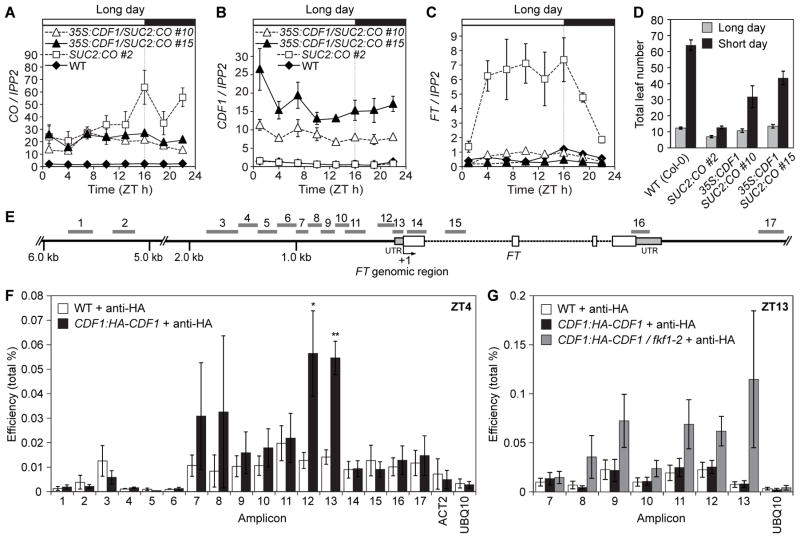
To test whether FKF1 regulates FT transcription through CDF1 protein stability regulation, we asked whether CDF1 directly regulates FT transcription. CDF1 mRNA was overexpressed in transgenic SUC2:CO-HA plants in which hemagglutinin (HA)-tagged CO was expressed by a phloem-specific SUCROSE-PROTON SYNPOTER2 (SUC2) promoter. Even though CO mRNA levels were similar in these lines (Fig. 1A, figs. S1 and S2), FT mRNA levels were reduced in the lines that had higher CDF1 expression (Fig. 1B and C and figs. S1 and S2). These plants showed later flowering phenotypes than the SUC2:CO-HA line (Fig. 1D). These results indicate that CDF1 protein represses FT transcription independently of CO transcription.
Fig. 1.
CDF1 represses FT transcription. (A to C) Gene expression analysis for CO (A), CDF1 (B), and FT (C) in wild type (WT), SUC2:CO-HA, and 35S:CDF1/SUC2:CO-HA plants grown in long days. IPP2 expression was used as a control. Experiments were repeated three times independently. (D) Flowering phenotypes of plants overexpressing CO and CDF1. Data are means ± SD of 16 plants. (E) Schematic drawing of the FT genomic region and the amplicon locations for ChIP experiments. (F and G) ChIP analysis with 10-day-old plants grown in long days. ACT2 and UBQ10 loci were used as controls. (F) WT and CDF1:HA-CDF1 plants were harvested in the morning (ZT4). Means ± SEM were calculated from seven independent replicates. **p<0.01, *p<0.05 in one-tail t-test. (G) WT, CDF1:HA-CDF1, and CDF1:HA-CDF1/fkf1-2 plants were harvested in the afternoon (ZT13). Data were calculated from five independent replicates.
We next examined whether CDF1 protein associates with the FT promoter. The amplicons of FT promoter regions 7, 8, 12, and 13 were enriched in CDF1:HA-CDF1 samples (9) compared with wild-type samples harvested at Zeitgeber time (=hours after light onset within a day) 4 (ZT4) (Fig. 1E and F), indicating that CDF1 binds to the FT promoter regions in the morning.
As both FKF1 and GIGANTEA (GI) proteins exist on the FT promoter (13) (Fig. 3G), we speculated that FKF1 could remove CDF1 on the FT promoter in the afternoon. We reasoned that more CDF1 proteins might remain associated with the FT promoter in the afternoon if FKF1 is absent. Our chromatin immunoprecipitation (ChIP) assay revealed that HA-CDF1 associated near the FT transcription start site in the afternoon only in the fkf1 mutant background (Fig. 1G).
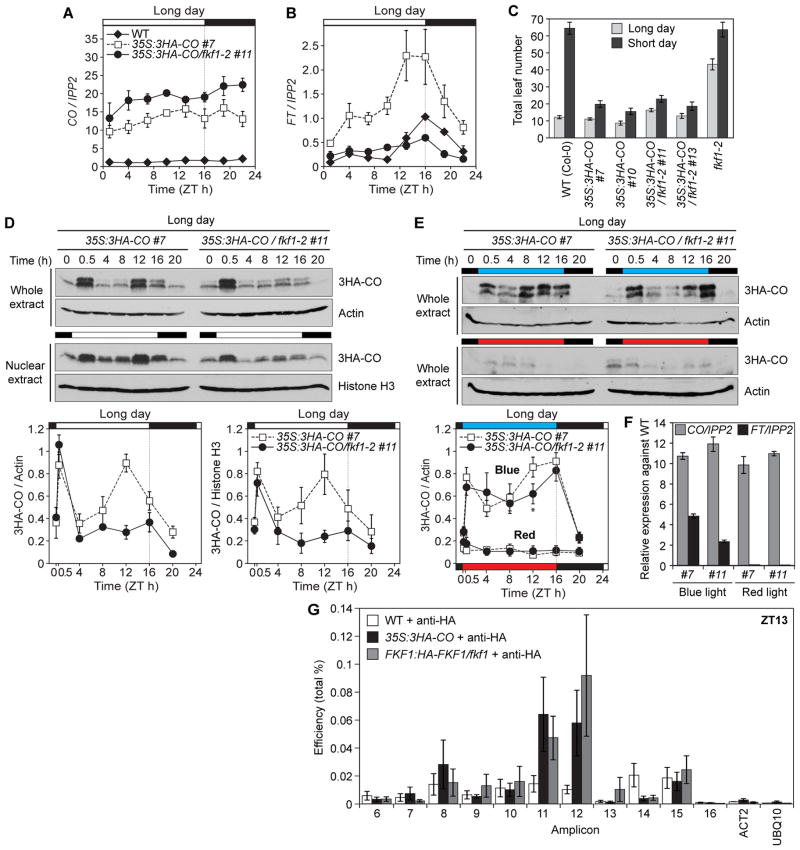
Fig. 3.
FKF1 regulates the light phase of CO protein expression. (A and B) Gene expression levels of CO (A) and FT (B) in WT, 35S:3HA-CO, and 35S:3HA-CO/fkf1-2 plants in long days. Experiments for (A, B, and D to F) were repeated three times independently. (C) Flowering time of CO overexpressors in WT and fkf1 mutant. Data are means ± SD of at least 13 plants. (D and E) FKF1 regulates CO protein stability. Actin and Histone H3 proteins were used as controls. Plants were grown for 10 days in long days (D) and transferred into blue and red light conditions on day 10 (E). (F) CO and FT mRNA levels in plants shown in (E). (G) Association of FKF1 and CO with FT promoter. Plants were grown for 10 days in long days and collected in the afternoon (ZT13). Means ± SEM were calculated from five independent replicates.
As FKF1 regulates the stability of CDF1 homologs that have overlapping function with CDF1 (14), we tested whether other CDF proteins also repress FT transcription. Since CO mRNA levels in the cdf1 cdf2 cdf3 cdf5 quadruple mutant are higher than in wild-type plants (14), we compared FT expression levels between the cdf quadruple mutant and the CO:HA-CO plant in which CO levels were elevated using the CO promoter. Even though CO mRNA levels in the quadruple mutant were lower than in the CO:HA-CO plant, FT mRNA levels in the mutant were higher than in the CO:HA-CO line (fig. S3). As the FT mRNA levels in the cdf1 single mutant resemble those in wild-type plants (9), our results indicate that these CDF proteins may redundantly repress FT transcription. Thus, FKF1 degrades CDF1 and possibly other CDF proteins to regulate FT expression.
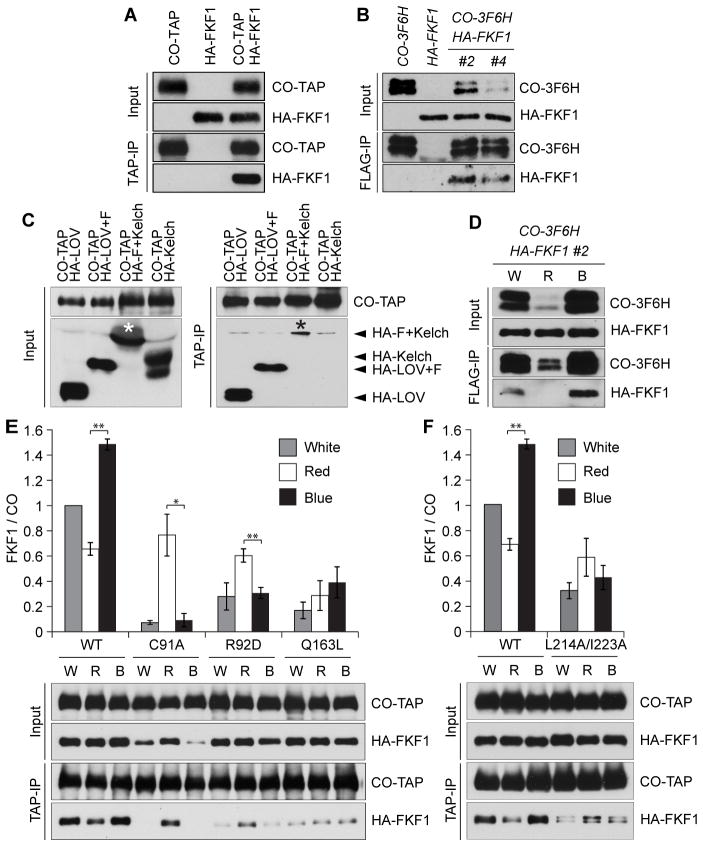
Next we examined whether there is an interaction between FKF1 and CO proteins. Using a Nicotiana benthamiana expression system (15), HA-FKF1 protein was co-immunoprecipitated with tandem affinity purification (TAP)-tagged CO (CO-TAP) (Fig. 2A). The FKF1-CO interaction was confirmed by reciprocal co-immunoprecipitation assays (fig. S4). We then analyzed the interaction in transgenic Arabidopsis plants expressing functional 3xFLAG and 6xHis-tagged CO (CO-3F6H) and HA-FKF1 (fig. S5). HA-FKF1 proteins were co-immunoprecipitated with CO-3F6H proteins (Fig. 2B), indicating that FKF1 and CO proteins can interact in vivo.
Fig. 2.
FKF1 interacts with CO in a blue-light enhanced manner. (A) The interaction between FKF1 and CO proteins in N. benthamiana. (B) In vivo FKF1-CO protein interaction. 10-day-old long-day grown plants were harvested at ZT13. (C) Mapping the CO binding site of FKF1. LOV+F and F+Kelch contain LOV and F-box domains, and F-box and Kelch repeat domains, respectively. Asterisks indicate HA-F+Kelch proteins. (D) Blue-light enhanced FKF1-CO interaction. Long-day grown plants were transferred to different light conditions at day 10 and harvested at ZT13. (E and F) FKF1 and CO interaction under different light conditions. Wild type and mutant forms of HA-FKF1 and CO-TAP were expressed in N. benthamiana. Quantified values are relative to the FKF1-CO interaction in white light for each combination. Means ± SEM were calculated from 3–6 independent samples. **p<0.01, *p<0.05 in one-tail t-test.
To identify the interaction site of FKF1 with CO, HA-tagged FKF1 domains (LOV, LOV+F, F+Kelch, and Kelch) were co-expressed with CO-TAP in N. benthamiana. HA-LOV and HA-LOV+F peptides interacted with CO-TAP protein (Fig. 2C). In addition, a small but reproducible amount of HA-F+Kelch peptide was co-immunoprecipitated with CO-TAP (Fig. 2C). These results suggest that the LOV domain is sufficient for CO-binding but the F-box region may also participate in this interaction.
As FKF1 and GI protein interaction occurs in a blue-light dependent manner (10), we investigated the blue-light dependency of the FKF1-CO protein interaction. More HA-FKF1 was co-immunoprecipitated with CO-3F6H under blue light than red light in vivo (Fig. 2D). However, this could be due to differences in CO protein levels under these conditions (Fig. 2D). As CO and FKF1 protein levels were similar in these conditions in N. benthamiana, we assessed the potential light dependency of the interaction. The amount of the FKF1-CO interaction was significantly greater in blue light than red light (Fig. 2E).
If the LOV domain is responsible for blue-light enhanced CO binding, we speculated that photochemically-blind LOV mutations (C91A, R92D, and Q163L) should alter the interaction (10). These LOV mutations weakened the interaction under blue and white light (Fig. 2E), indicating that blue light perceived by the LOV domain enhanced the interaction. As our results indicated the involvement of F-box with CO binding (Fig. 2C), we analyzed whether a functional F-box is important for binding. Two mutations (L214A/I223A), which eliminate the ability of F-box to bind to Arabidopsis Skp1-like (ASK1) proteins (16), were introduced to the FKF1 F-box. The F-box mutations also attenuated blue-light effects on the interaction (Fig. 2F), suggesting that the formation of the SCFFKF1 complex facilitates the FKF1-CO protein interaction.
To investigate the effect of the FKF1-CO interaction, we analyzed the effect of fkf1 mutation on CO activity. Since FKF1 affects CO transcription (8), we used lines in which CO mRNA is constitutively expressed (Fig. 3A and figs. S6A and S7). In the CO overexpressing lines, having the fkf1 mutation reduced FT mRNA levels (Fig. 3B and figs. S6B and S7). CO overexpressors in the fkf1 mutant flowered later than 35S:3HA-CO plants in long days (Fig. 3C). We observed a similar fkf1 effect on FT mRNA expression when CO mRNA levels were elevated in the tissues where CO is expressed (fig. S8).
The timing of FKF1 protein expression under light coincides with CO protein stabilization (6, 8). As the fkf1 mutation reduced CO activity (Fig. 3B), we speculated that FKF1 may stabilize CO protein. We analyzed the diurnal CO protein profiles in fkf1. CO protein peaks at ZT0.5 and ZT12 in long days in 35S:CO plants (6). Our 35S:3HA-CO lines showed a similar daily CO protein pattern, although there were doublet bands corresponding to the 3HA-CO proteins (Fig. 3D and fig. S6C) as also observed previously (17). In fkf1, CO protein fell to low levels in the afternoon (ZT12-16), though the ZT0.5 CO peak was still present (Fig. 3D and fig. S6C). We found FKF1 in both cytosolic and nuclear fractions (fig. S9). The profile of CO abundance in the nucleus was very similar to that in the whole protein extracts (Fig. 3D and fig. S6C), demonstrating that FKF1 stabilizes CO protein in both cytoplasm and nucleus. In short days, CO levels were extremely low (6) (fig. S10A), and there were no obvious difference in CO protein expression (fig. S10B) between wild-type and fkf1 backgrounds. These findings imply that rhythmic FKF1 expression may determine the timing of stabilizing CO in long days.
Blue light stabilizes CO protein in long days (6), and our data suggest that blue light enhances the FKF1-CO interaction. We analyzed the daily CO protein profiles under blue- and red-light conditions. CO protein was stable under blue light. The CO protein level was slightly but significantly lower in fkf1 background at ZT12 (Fig. 3E), indicating that FKF1’s contribution is restricted to the afternoon. The CO protein profiles in blue light resemble those in the white-light grown phyB mutant (6), suggesting that without phyB-dependent destabilization of CO, blue light perceived by cryptochromes can stabilize CO protein throughout the day. In addition, the fkf1 mutation reduced FT mRNA levels without changing CO mRNA levels under blue light (Fig. 3F). Under red light, CO protein was unstable in all CO overexpressors (Fig. 3E). This is most likely due to the activation of phyB signaling, which degrades CO protein (6). Together with the previous observation (6), these results suggest that the balance between blue and red light, which work antagonistically, determines CO protein levels.
As FKF1 stabilizes CO protein even in the nucleus (Fig. 3D), we postulated that FKF1 and CO interactions might also occur on the FT promoter. We found that both FKF1 and CO proteins associated near the FT transcription start site (13) (Fig. 3G), where CORE (CO-responsive element) sequences exist (18). These results indicate that FKF1 may interact with CO protein on the FT promoter.
Our results imply that FKF1 has at least three different roles in the regulation of FT expression (fig. S11). One, FKF1 regulates CO transcription by degrading CDF proteins (9, 10, 14). Two, as CDF proteins are also FT repressors, the same FKF1-mediated degradation mechanism may also regulate FT transcription. Three, FKF1 is involved in the stabilization of CO protein in the long-day afternoon, increasing the expression of FT.
We refined our previous model (11) by including these biochemical functions of FKF1 (15). The new mathematical model focuses on the regulation of CO and FT mRNA and CO protein (Fig. 4A). FKF1 and GI modulate the CDF1 protein, which represses CO and FT (Fig. 4A) and represents the functions of all CDF proteins in the model. Falling CDF1 protein levels relieve repression of CO at the end of long days in the simulated wild type, leading to the characteristic shoulder in the mRNA profile around ZT13 (Fig. 4B and fig. S12). The simulated fkf1 mutant reduces CO transcript levels by 59% at ZT13 (Fig. 4B). In the model, CO is stabilized by FKF1 in a light-dependent manner (fig. S12). The simulated fkf1 mutant both fails to degrade CDF1 and fails to stabilize CO (fig. S13), reducing the amount of FT over one cycle (FTAREA) by ~78% of wild-type levels in long days (Fig. 4C), similar to the FT levels in fkf1 (8). In addition, the model recreated the altered FT profiles of CO overexpression lines shown in Fig.1 and 3 (figs. S14 and S15). These results indicate that the two FKF1-dependent mechanisms described are sufficient to explain the observed CO and FT mRNA profiles.
Fig. 4.
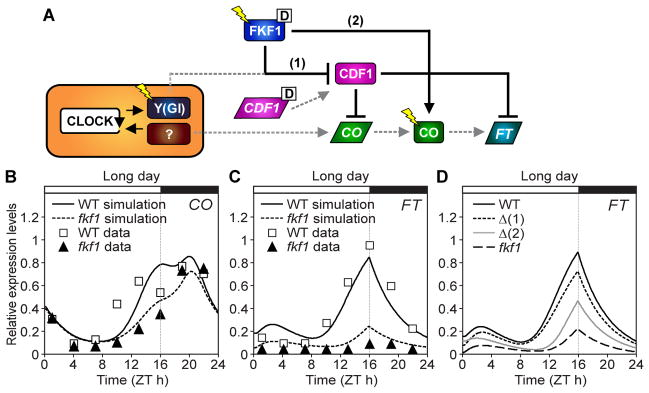
Simulation of the photoperiod sensor. (A) Schematic of the new model incorporating the roles of CDF1 and FKF1 in regulating FT. Parallelograms, rectangles, and flashes represent mRNAs, proteins, and light inputs, respectively. “D” indicates components input as experimental data (15) (fig. S19). Solid lines indicate FKF1 dependent mechanisms. The rest of interactions are shown in dotted lines. (B and C) Simulations of CO (B) and FT (C) mRNA are plotted against experimental data (8) for wild type and fkf1 mutants in long days. (D) Effects of partial loss of FKF1 function in FT induction. Δ(1) and Δ(2) means that either FKF1 function (1) or (2) in the diagram shown in (A) was removed to simulate FT patterns.
Using the model, we estimated the importance of each FKF1 function under long-day conditions, by simulating partial loss-of-function mutants that are not accessible genetically. If FKF1 could not degrade CDF1 protein but stabilized CO protein [mutant designated Δ(1)], the simulated decreased by ~22% from the wild-type levels (Fig. 4D). If FKF1 degraded CDF1 but could not stabilize CO protein [mutant Δ(2)], the simulated FTAREA decreased by ~51% (Fig. 4D). Although the exact balance varies among model versions (fig. S16), both mechanisms were required to explain the very low levels of FT observed in full fkf1 mutants (fig. S13H).
Our results suggest that FKF1 regulates FT expression through a multiple-feedforward motif (Fig. 4A) that can enhance signal detection (19). Photoperiod sensing depends upon correctly-timed operation of this motif. Our data indicate that the circadian clock conveys time information for stabilizing CO protein through the timing of FKF1 expression. We therefore investigated the impact of removing circadian control from these two components, via constitutive expression of CO and FKF1, while GI and CDF1 mRNA expression remained rhythmic. Our model predicted that this would stabilize CO protein (fig. S17A) and alter the FT mRNA profile, with a near-linear rise during the daytime, and also increase FT expression levels depending on FKF1 protein levels (fig. S17B). To verify our model predictions, we analyzed CO protein and FT mRNA expression in 35S:3HA-CO/35S:HA-FKF1 lines (fig. S18). Constitutive FKF1 protein expression stabilized CO protein even during the early part of the day (fig. S18). In addition, FT expression in these lines increased linearly throughout the day and FT levels were higher in the line with a higher FKF1 level (fig. S18). These results validated our prediction and indicate the importance of circadian regulation of FKF1 expression for day-length-dependent CO protein stabilization.
The FKF1 photoperiod sensor uses multiple, partially redundant switches to allow strong activation in long days. As the sun rises higher in the sky each day when spring approaches, plants can sense the increased intensity in the blue-light range of the spectrum each afternoon through multiple photoreceptors, including FKF1. The complexity of this mechanism in even a temperate species like Arabidopsis suggests that it has the flexibility to regulate successful reproduction in a wide range of environments.
Supplementary Material
Acknowledgments
We thank S. Harmer, K. Torii, D. Nusinow, S. Ito, and H. Kinmonth-Schultz for critial reading of the manscript and G. Coupland for providing us cdf quadruple mutant. Y.H.S. is partly supported by Next Generation Biogreen 21 Program grant (SSAC, PJ008109). R.W.S. was supported by BBSRC (BB/F59011/1, BB/F005237/1). SynthSys is partly supported by BBSRC and EPSRC (BB/G019621). This work was supported by NIH grant (GM079712) to T.I.
Footnotes
References and Notes
- 1.Thomas B, Vince-Prue D. Photoperiodism in plants. 2. Academic Press; San Diego: 1997. [Google Scholar]
- 2.Kobayashi Y, Weigel D. Genes Dev. 2007;21:2371. doi: 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- 3.de Montaigu A, Toth R, Coupland G. Trends Genet. 2010;26:296. doi: 10.1016/j.tig.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Amasino R. Plant J. 2010;61:1001. doi: 10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- 5.Samach A, et al. Science. 2000;288:1613. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- 6.Valverde F, et al. Science. 2004;303:1003. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 7.Mockler T, et al. Proc Natl Acad Sci U S A. 2003;100:2140. doi: 10.1073/pnas.0437826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. Nature. 2003;426:302. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- 9.Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. Science. 2005;309:293. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- 10.Sawa M, Nusinow DA, Kay SA, Imaizumi T. Science. 2007;318:261. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salazar JD, et al. Cell. 2009;139:1170. doi: 10.1016/j.cell.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 12.Fukamatsu Y, et al. Plant Cell Physiol. 2005;46:1340. doi: 10.1093/pcp/pci144. [DOI] [PubMed] [Google Scholar]
- 13.Sawa M, Kay SA. Proc Natl Acad Sci U S A. 2011;108:11698. doi: 10.1073/pnas.1106771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fornara F, et al. Dev Cell. 2009;17:75. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 15.See details in materials and methods that are available as supporting material on Science Online.
- 16.Han LQ, Mason M, Risseeuw EP, Crosby WL, Somers DE. Plant J. 2004;40:291. doi: 10.1111/j.1365-313X.2004.02207.x. [DOI] [PubMed] [Google Scholar]
- 17.Zuo Z, Liu H, Liu B, Liu X, Lin C. Curr Biol. 2011;21:841. doi: 10.1016/j.cub.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tiwari SB, et al. New Phytol. 2010;187:57. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- 19.Alon U. Nat Rev Genet. 2007;8:450. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.