Abstract
KChIPs enhance functional expression of Kv4 channels by binding to an N-terminal regulatory region located in the first 40 amino acids of Kv4.2 that we call the Functional Expression Regulating N-terminal (FERN) domain. Mutating two residues in the FERN domain to alanines, W8A and F11A, disrupts KChIP binding and regulation of Kv4.2 without eliminating the FERN domain’s control of basal expression level or regulation by DPP6. When Kv4.2(W8A,F11A) is co-expressed with wild type Kv4.2 and KChIP3 subunits, a dominant negative effect is seen where the current expression is reduced to levels normally seen without KChIP addition. The dominant negative effect correlates with heteromultimeric channels remaining on intracellular membranes despite KChIP binding to non-mutant Kv4.2 subunits. In contrast, the deletion mutant Kv4.2(Δ1–40), eliminating both KChIP binding and the FERN domain, has no dominant negative effect even though the maximal conductance level is 5x lower than seen with KChIP3. The 5x increased expression seen with KChIP integration into the channel is fully apparent even when a reduced number of KChIP subunits are incorporated as long as all FERN domains are bound. Our results support the hypothesis that KChIPs enhances Kv4.2 functional expression by a 1:1 suppression of the N-terminal FERN domain and by producing additional positive regulatory effects on functional channel expression.
Introduction
The transient A type potassium current, ISA, is formed from Kv4 family alpha subunits, as well as KChIP (K Channel Interacting Proteins: KChIP1–4) and DPL (Dipeptidyl peptidase like Proteins: DPP6, DPP10) auxiliary subunits (Jerng et al. 2004a). In heterologous expression systems, functional expression of Kv4 proteins is poor with the channel largely located in intracellu-lar membrane compartments (Kunjilwar et al. 2004). Co-expression with KChIPs or DPLs greatly enhances the expressed functional current (An et al. 2000, Morohashi et al. 2002, Shibata et al. 2003, Nadal et al. 2003, Jerng et al. 2004b, Foeger et al. 2010, Seikel & Trimmer 2009). A number of different effects have been attributed to auxiliary subunits to account for this increase in functional current. Our studies here concern the molecular requirements for KChIP auxiliary subunit proteins to modulate the functional expression of channels formed from Kv4.2 alpha sub-units.
KChIPs are members of the large neuronal calcium sensor (NCS) family of four EF-hand calcium binding proteins (Braunewell & Gundelfinger 1999). Four different KChIP genes have been identified in mammals with a related NCS protein, frequenin, also showing functional regulation of Kv4 channels (An et al. 2000, Morohashi et al. 2002, Nakamura et al. 2001). KChIPs were originally identified as Kv4 interacting proteins based on yeast 2-hybrid screens with the N-terminus of Kv4 proteins (An et al. 2000). Functional expression studies subsequently showed that KChIP proteins could regulate current expression levels as well as the gating properties of the channels in the surface membrane. Later studies have supported a forward trafficking role for KChIPs, releasing trapped channels onto the cell surface, as well as important roles for KChIPs in driving channel assembly, stabilizing channel protein complexes, and slowing the turnover of channels on the cell surface (Foeger et al. 2010, Bahring et al. 2001, Kunjilwar et al. 2004).
DPL proteins are structurally similar to the Dipeptidyl peptidase proteins, except they lack enzymatic activity (Jerng et al. 2004a). Two family members DPP6 (also called DPPX or DPL1) and DPP10 (also called DPPY and DPL2) have been shown to be auxiliary subunits of Kv4 channels (Jerng et al. 2004b, Nadal et al. 2003). Unlike KChIP proteins, DPLs are single transmembrane proteins with the majority of the protein contained in the large extracellular C-terminus. Despite these structural differences, both DPLs and KChIPs produce similar changes in the surface expression of Kv4 alpha subunits.
Deletions in the first 40 amino acids of the Kv4 alpha subunit N-terminus significantly increases the functional expression of Kv4.2 channels and eliminates KChIP regulation (Bahring et al. 2001). Based on its dual role in controlling basal expression level and regulation by KChIPs, we have named this 40 amino acid regulatory region the FERN domain, for Functional Expression Regulating N-terminal domain. Structural studies were performed to characterize how KChIPs interact with the tetrameric N-terminus of Kv4 channels (Wang et al. 2007, Pioletti et al. 2006). The structural models show a 4:4 stoichiometry of KChIP:Kv4.2 subunits, with KChIPs interacting in at least two distinct sites: Site 1 is at the N-terminus of Kv4.2 within the FERN domain and Site 2 is at the T1-T1 assembly interface. Although these structural models were made based on a tetrameric soluble N-terminal fragment of the Kv4 channel, this proposed stoichiometry is also supported by biochemical analysis of purification of Kv4-KChIP channel complexes (Kim et al. 2004a). At Site 1, a hydrophobic pocket within the KChIP protein binds Kv4.2 N-terminus which can be disrupted by the double point mutation Kv4.2(W8A,F11A) (Zhou et al. 2004). Although the W8A, F11A mutation disrupts KChIP binding to the FERN domain, Kv4.2(W8A,F11A) is also trapped on intracellular membranes, suggesting that these residues are not required for the intracellular retention function of the FERN domain (Zhou et al. 2004).
Here we report that Kv4.2 subunits containing a FERN domain but not bound to KChIPs act as a dominant negative suppressing the expression of KChIP bound Kv4.2 subunits. This dominant negative effect is lost when the FERN domain is deleted suggesting that the key factor is not the reduced number of KChIPs incorporated into the channel, but rather the number of unbound FERN domains present in the channel. We conclude that an important function of KChIPs in promoting channel expression is to directly bind and suppress the function of the FERN domain. Finally, in addition to suppression of the FERN domain function, KChIP3 was also found to have additional positive effects that further boost Kv4 channel functional expression by 5x. Interestingly, these positive effects occur even when a reduced number of KChIP3 subunits are bound to the channel as long as no unbound FERN domains are present.
Methods
DNA subcloning
Kv4.2, Kv4.2(W8A,F11A) (originally named Kv4.2ΔKCB), KChIP3, HA-KChIP3, KChIP1, (KChIP1)Kv4.2 cDNA expression constructs were described previously (Zhou et al. 2004, Kunjilwar et al. 2004, Strang et al. 2003). The DPP6 construct used was the long splice variant as described previously (Jerng et al. 2004b). Kv4.2(EGFP) was made by PCR to replace the Kv4.2 stop codon with a Sal1 site in frame with a Sal1 site in the N-terminal poly-linker of pEGFP-N1 (Clontech), introducing 2 amino acids (Ser, Thr) at the Sal1 site. Epitope tagging was performed by PCR modification to the N-terminal sequence. Kv4.2(Δ1–40) was constructed by PCR to put rKv4.2 AA 41-end in frame after a Kozak and Initiator Met sequence. All constructs were confirmed by DNA sequencing.
Cell Culture
CHO-K1 cells were obtained from ATCC and maintained in DMEM medium supplemented with 10% FBS, 40 μg/ml L-proline, 100 U/ml penicillin and 100 U/ml Streptomycin. COS7 cells were obtained from ATCC and maintained in DMEM medium supplemented with 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin. Transfection protocols for CHO-K1 and COS7 cells and titration of cDNA levels for optimal expression of Kv4.2 and KChIP3 have been described previously (Zhou et al. 2004, Kunjilwar et al. 2004). Red-fluorescent protein cDNA (pCMV-DsRed2) was used as a filler to keep the total cDNA amount fixed and to identify transfected cells for recordings. Recordings were performed at 18–26 h after transfection.
Immunocytochemistry
Immunocytochemistry was performed in transfected COS7 cells as described previously (Zhou et al. 2004). Kv4.2(EGFP) was visualized by green fluorescence. KChIP3 was visualized by staining with KChIP3 Ab FL214 (Santa Cruz, 1 ug/ml) followed by Goat anti-Rb Alexa594 (Molecular Probes, 1:200). Kv4.2(W8A,F11A) was visualized by anti-Kv4.2 (Chemicon, 5 μg/ml) and goat anti-Rb Alexa488 (Molecular Probes, 1:200). For double staining of Kv4.2(W8A,F11A) and KChIP3, we used an HA-tagged KChIP3 and stained with anti-HA (Santa Cruz, 1 μg/ml) and detected with goat anti-mouse Alexa 594 (Molecular Probes, 1:200). For immunostaining of DPP6, we used a myc-tagged DPP6 stained with anti-myc (9E10, Santa Cruz, 1 μg/ml) and detected with goat anti-mouse Alexa 594 (Molecular Probes, 1:200). For imaging the surface membrane, cells were treated with cholera toxin B-Alexa 594 (CT-B-594) (Vibrant Lipid Raft Labeling System, Invitrogen) at 4 °C using the manufacturer’s protocol, followed by fixation and immunofluorescence. CT-B-594 binds to the surface ganglioside GM-1. For imaging internal membrane, cells were briefly treated with brefeldin A BIODIPY 558/568 (rBrefA) (Invitrogen) as described previously, followed by fixation and immunofluorescence (Deng et al. 1995). For imaging the cytoplasm, cells were transfected with EGFP. Images were collected as z-stacks using an Olympus FV300 or Zeiss LSM510 and displayed as projection images. All transfections were independently replicated at least 3 times. All scale bars are 20 μm.
Immunoprecipitation
Immunoprecipitation was performed on cell lysates from transfected COS7 cells as described previously (Zhou et al. 2004). The antibodies used were: Control- goat IgG (ICN), Goat anti 4.2 antibody (N-15, Santa Cruz), goat anti-myc antibody (A-14G, Santa Cruz) and goat anti HA antibody (Y-11G, Santa Cruz) at a final concentration of 10 μg/ml of each. Reported experiments were all independently replicated at least 3 times.
Western Blot
Western blots were performed as described previously (Zhou et al. 2004). Primary antibodies used were anti Kv4.2 antibody (AB5630 at 1:200–1000, Chemicon) and anti-KChIP3 (FL214 at 1 μg/ml, Santa Cruz). Bound antibodies were detected by Goat anti-Rb IgG (Pierce) at 1:10,000 and visualized by chemiluminescence (Pierce). Lane “L” is a fraction of the lysate run as a loading control. Based on the original volume this lane has 1/5 as much of the original sample as was run in the IP lanes, assuming 100% IP efficiency. Antibody used in the immunoprecipitation “Crl IgG”, “Kv4.2-Ab”, “HA-Ab”, “myc-Ab” is indicated for each lane.
Electrophysiology
Ionic currents were recorded with an Axopatch-200B amplifier (Axon Instruments) using whole-cell voltage clamp configuration as described previously (Zhou et al. 2004, Kunjilwar et al. 2004). Current stimulus protocols and data collection were controlled by pClamp software (Axon Instruments). Pipettes contained (in mM): 140 KCl, 1 MgCl2, 1 EGTA, 10 HEPES (pH 7.4) and 0.133 CaCl2. The calculated free Ca2+ concentration was~ 10 nM at pH 7.4. The bath solution was (in mM): 2 KCl, 138 NaCl, 1.8 CaCl2, 1 MgCl2, 10 HEPES (pH 7.4). All the recordings were done at room temperature (25+2°C).
Modeling
Modeling assumed that the expressed subunit composition matched the cDNA composition. With 2 types of Kv4.2 subunits in the transfection, with “U” being the mole fraction of non-KChIP binding subunit (for Unbound), and “B” being KChIP Bound subunits, then the formulas for the different channel compositions are: 4B:0U = (1-U)4; 3B:1U = 4*(1-U)3*U; 2B:2U = 6*(1-U)2*U2; 1B:3U = 4*(1-U)*U3; 0B:4U = U4. The fractional current predicted was then modeled with a current of 1 for full KChIP effect or a current of 0.033 (fractional expression for Kv4.2 alone is 1/30 of the KChIP level). The subunit compositions giving a current level of 1 were based on the different Models: 4 KChIPs only – 4B:0U; 3 or 4 KChIPs – 4B:0U, 3B:1U; 2, 3 or 4 KChIPs - 4B:0U, 3B:1U, 2B:2U; and 1 or more KChIPs - 4B:0U, 3B:1U, 2B:2U, 1B:3U.
Data Analysis
Data analysis was performed using Clampfit (Axon Instruments) and Origin software (OriginLab). Pooled data are expressed as Mean + SEM. N’s for all summary data are 6–15 independent recordings. Statistical comparisons between groups of data were carried out with the two-tailed Student’s t test for paired or unpaired data or 1 way ANOVA using Origin and Analyze-it for Excel (Analyse-it Software). Tests of significance were performed based on standard hypothesis testing approaches using t-tests with a significance level of 0.01. Significance at 0.01 level indicated by (*); NSD – Not Significantly Different.
Results
The Kv4.2(W8A,F11A) mutation selectively eliminates KChIP but not DPP6 modu-lation
We have previously reported that mutagenesis of W8A and F11A in Kv4.2 (Kv4.2(W8A,F11A)) eliminates KChIP binding, trafficking, and functional modulation of the channel (Zhou et al. 2004). We have hypothesized that the Kv4.2(W8A,F11A) mutation produces a highly selective loss in KChIP modulation, by disruption of KChIP binding, rather than some other mechanism, since other properties of the channel, recorded in the absence of KChIPs, appeared to be essentially normal (Zhou et al. 2004). In order to further test this hypothesis, we sought to compare the ability of a different Kv4.2 auxiliary subunit, DPP6, to modulate the mutant Kv4.2(W8A,F11A) alpha subunit protein.
Kv4.2 or Kv4.2(W8A,F11A) cDNAs were transfected into CHO-K1 cells with and without co-expression of KChIP3 or DPP6 auxiliary subunits. After overnight expression, transfected cells were recorded by whole cell voltage-clamp and the A type current expression level and functional properties measured. As described previously, the functional current level expressed by the Kv4.2(W8A,F11A) alpha subunit is not altered by the co-expression of KChIP3, leaving the expression level approximately 30 times lower than wild type Kv4.2 channels co-expressed with KChIP3, and not significantly different from Kv4.2 expressed in the absence of auxiliary subunit proteins (Fig. 1A,B) (Zhou et al. 2004). In addition, the functional properties of channels formed when Kv4.2(W8A,F11A) is co- expressed with KChIP3 are distinct from those of Kv4.2 expressed with KChIP3, with a faster multi-exponential inactivation and slower recovery from inactivation (data not shown).
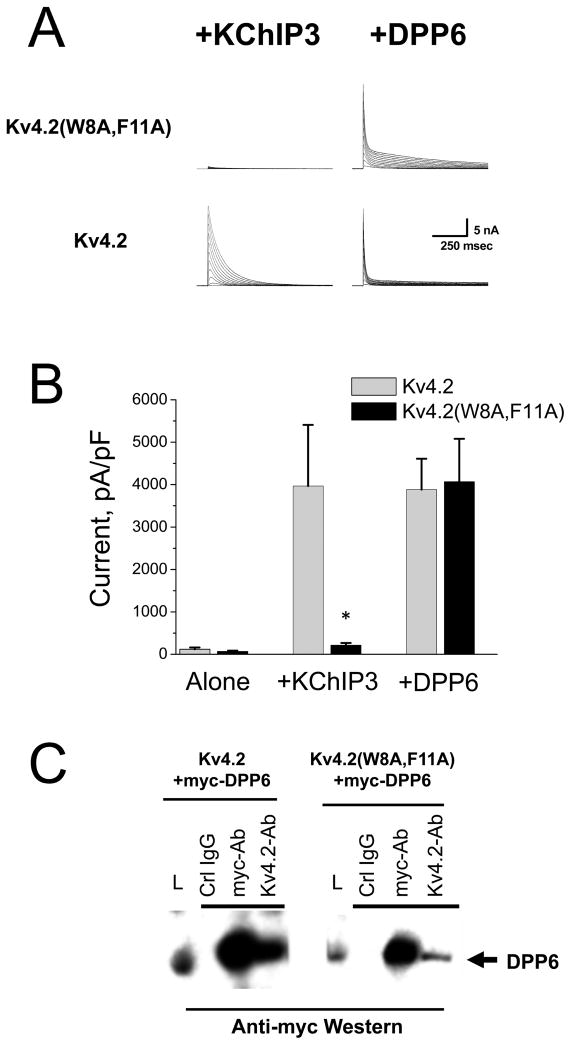
Figure 1. Kv4.2(W8A,F11A) mutant selectively disrupts regulation by KChIP3 but not DPP6.
A) Expressed currents for CHO-K1 cells transfected with the combination of subunits indicated. Kv4.2 expression is enhanced by both KChIP3 and DPP6. Kv4.2(W8A,F11A) expression is only enhanced by DPP6, not KChIP3. B) Summary data for currents in response to a depolarizing voltage step to +50 mV recorded by whole-cell voltage clamp from CHO-K1 cells. Kv4.2 and Kv4.2(W8A,F11A) expressed alone produce low levels of A type K current that are not significantly different. Co-transfection with KChIP3 cDNA significantly boosts the functional expression of Kv4.2 by 30 fold with no significant change in the functional expression of Kv4.2(W8A,F11A). Kv4.2(W8A,F11A), however, is regulated by another auxiliary subunit protein DPP6 with no significant difference to Kv4.2. C) Both Kv4.2 and Kv4.2(W8A,F11A) subunits show co-assembly with DPP6 in co-immunoprecipitation studies. Myc-tagged DPP6 was co-transfected with either Kv4.2 or Kv4.2(W8A,F11A). Lysate loading control lane (L) contains 1/5 of the amount of material used in each IP reaction. Antibodies used in precipitations shown above the lane. Anti-myc western to detect DPP6 shows that precipitation of either wild type or mutant Kv4.2 subunits co-precipitates DPP6 (Kv4.2-Ab lanes).
In contrast to the lack of modulation by KChIP3, DPP6 robustly modulates the functional expression of Kv4.2(W8A,F11A). Fig. 1A shows the dramatic increase in functional expression of Kv4.2(W8A,F11A) by DPP6. Compared to wild type Kv4.2, the peak current level generated by co-expression of DPP6 with Kv4.2(W8A,F11A) is not significantly different from wild type Kv4.2 (Fig. 1B). Also evident in these experiments is a slight change in the inactivation time course for the mutant compared to wild type Kv4.2 co-expressed with DPP6 (Fig. 1A). This effect is likely due to the mutation slightly destabilizing the Kv4.2 N-type fast inactivation process (Gebauer et al. 2004).
Unlike KChIPs, which do not co-immunoprecipitate the Kv4.2(W8A,F11A) channel, DPP6 is able to co-assemble with the mutant. In Fig. 1C, we co-expressed a myc-tagged DPP6 construct with Kv4.2 or Kv4.2(W8A,F11A) and tested for co-assembly by co-immunoprecipitation. We find that precipitation of myc-DPP6 co-precipitates Kv4.2 as well as Kv4.2(W8A,F11A) alpha subunits.
Finally, we examined if the co-expression with DPP6 produces similar changes in the subcellular distribution of Kv4.2(W8A,F11A) and Kv4.2 subunits. For these experiments we followed three distinct subcellular compartments in COS-7 cells, which are large flat cells that allow excellent visualization of internal membrane systems, based on labeling with selective reagents: the surface membrane- cholera toxin B-Alexa 594 (CT-B-594) staining at 4 °C (Fig. S1A), Internal Membranes- staining with Brefeldin A BIODIPY 558/568 (rBrefA) (Fig. S1B); Cytoplasm-EGFP expression (Fig. S1C).
Expressed alone, KChIP3 has a uniform distribution expected for a soluble protein (Fig. S2A), whereas DPP6 is localized to membranes, including the surface membrane (Fig. S2B). Expression of Kv4.2(EGFP) alone shows that the majority of the fluorescent protein co-localizes with rBrefA (Fig. S2C) and not CT-B-594 (Fig. S2D), and thus is found in internal membranes. If instead we co-express Kv4.2(EGFP) with KChIP3, the channel protein now redistributes out of the internal membrane systems and shows extensive co-localization with the surface marker CT-B-594 (Fig. S2E). Identical results are obtained using antibody staining with the expression of a wild type non-fluorescence Kv4.2 construct in these cells (Zhou et al. 2004). Finally, a similar redistribution of Kv4.2(EGFP) onto the surface membrane is produced by DPP6 co-expression (Fig S2F).
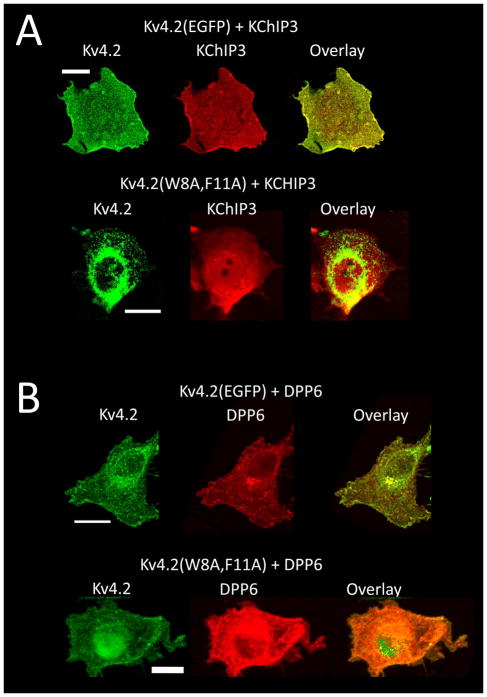
We hypothesized that the differences in functional expression of Kv4.2(W8A,F11A) when co-expressed with KChIP or DPP6 auxiliary subunits were due to differential co-assembly, rather than differential trafficking of the co-assembled channel. However, KChIPs are reported to have additional interaction sites on the channel besides the Site 1 that we have mutated here (Kim et al. 2004b, Scannevin et al. 2004). It is possible that these alternative interactions are disrupted during solubilization, so we compared the subcellular localization of Kv4.2(W8A,F11A) with KChIP and DPP6 auxiliary subunits when they are co-expressed in COS-7 cells. Previous studies have shown that KChIP proteins alter the membrane trafficking of Kv4.2 channels allowing them to redistribute onto the cell surface (Fig. 2A) (O’Callaghan et al. 2003, Flowerdew & Burgoyne 2009, Shibata et al. 2003, Kunjilwar et al. 2004, Bahring et al. 2001, Zhou et al. 2004). Fig. 2A shows that Kv4.2(EGFP) movement onto the cell surface is accompanied by a matching redistribution of KChIP3. In contrast when Kv4.2(W8A,F11A) is co-expressed with KChIP3, Kv4.2(W8A,F11A) is trapped in internal membranes while KChIP3 remains cytoplasmic. With DPP6 co-expression, on the other hand, we see extensive labeling of both Kv4.2(EGFP) and Kv4.2(W8A,F11A) on the surface membrane with strong co-localization with DPP6 (Fig. 2B). We conclude that there is no inherent defect in the the Kv4.2(W8A,F11A) channel that prevents achieving a high level of functional expression. Rather this mutation is highly selective for disrupting KChIP binding and functional regulation, leaving DPP6 modulation unaffected, with only a small change in channel gating properties.
Figure 2. Differential effects on subcellular distributions when Kv4.2 or Kv4.2(W8A,F11A) are co-expressed with KChIP3 or DPP6.
Projection images of confocal stacks. A) Im-munofluorescence studies on COS7 cells transfected with KChIP3 and Kv4.2(EGFP) or Kv4.2(W8A,F11A). Kv4.2(EGFP) and KChIP3 redistribute together onto the surface membrane compartment, whereas co-expressed Kv4.2(W8A,F11A) and KChIP3 remain in separate subcel-lular compartments: Kv4.2(W8A,F11A) –intracellular membranes, KChIP3- cytoplasm. B) Im-munofluorescence studies on COS7 cells transfected with myc-DPP6 and Kv4.2(EGFP) or Kv4.2(W8A,F11A). Following co-expression with DPP6, both Kv4.2(EGFP) and Kv4.2(W8A,F11A) redistribute onto the surface membrane compartment and extensively co-localize with DPP6. Scale bars are 20 μm.
Mixing of Kv4.2(W8A,F11A) with wild type subunits produces dominant negative actions in the presence of KChIP3
Our ability to produce a Kv4.2 subunit that is essentially normal other than failing to bind to KChIPs provides us with an opportunity to express channels with substoichiometric numbers of KChIP binding sites by varying the Kv4.2 to Kv4.2(W8A,F11A) cDNA ratio used in the trans-fection. By expressing such subunit mixtures in the presence of KChIPs, we can ask if channels incorporating a substoichiometric number of KChIP proteins are expressed normally on the cell surface.
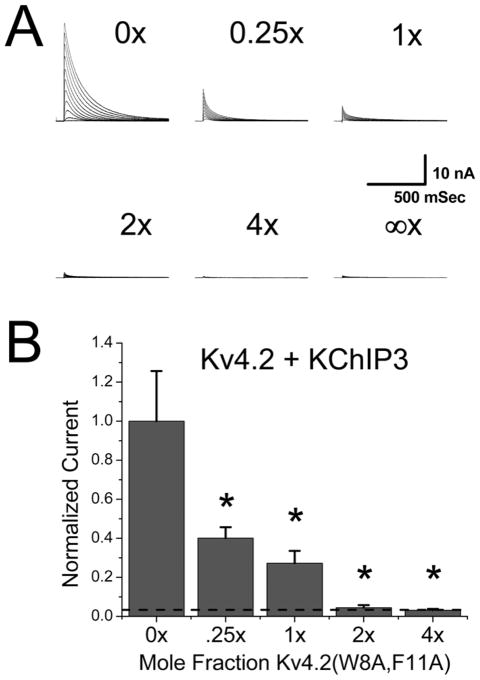
To test the functional effects of varying KChIP incorporation into channels, the ratio of wild type Kv4.2 to Kv4.2(W8A,F11A) cDNA in the transfection mix was varied. As can be seen in Fig. 3A, increasing the ratio of Kv4.2(W8A,F11A) in the channel expression mix produces a dramatic suppression of functional A-current expression. Fig. 3B summarizes the results which clearly show that addition of even 25% as much Kv4.2(W8A,F11A) as Kv4.2 cDNA suppresses functional A-current expression by over 50%. These constructs only differ by two amino acids, and, as discussed previously, in the presence of DPP6 they express equally well. We conclude, therefore, that Kv4.2(W8A,F11A) has a strong dominant negative effect in the presence of Kv4.2 + KChIP3 even at low expression ratios as might expected if all four subunits must bind KChIPs to increase functional current expression.
Figure 3. Kv4.2(W8A,F11A) dominantly suppresses KChIP3 enhancement of Kv4.2 current.
Titration experiment to test for effects of mixing Kv4.2(W8A,F11A) into the expression of Kv4.2 plus KChIP3. Kv4.2 and KChIP3 cDNA levels were fixed to produce maximal expression in CHO-K1 cells. A variable amount of Kv4.2(W8A,F11A) cDNA was added into the transfec-tion mix and the total cDNA level kept constant by mixing in a variable amount of pCMV-DsRed2 reporter gene cDNA. A) Representative current traces for CHO-K1 cells transfected with Kv4.2 plus KChIP3 and Kv4.2(W8A,F11A) at the mole ratio indicated relative to Kv4.2. Infinite ratio indicates expression level seen for Kv4.2(W8A,F11A) plus KChIP3 without Kv4.2 cDNA added. B) Summary data where molar ratio of Kv4.2(W8A,F11A) cDNA added relative to wild type Kv4.2 cDNA is compared to the fraction of expressed A-current as measured by peak current at +50 mV. Dashed line shows expected current expression level for Kv4.2 alone without KChIP3. Functional A-current levels are suppressed by Kv4.2(W8A,F11A) co-expression in a dose dependent manner. One tailed t-tests show that expression is significantly reduced at all doses of Kv4.2(W8A,F11A) compared to expression without Kv4.2(W8A,F11A) addition. Expression at high levels of Kv4.2(W8A,F11A) was not significantly different from Kv4.2 expression without KChIP3.
Kv4.2(W8A,F11A) mutant traps wild type channels in intracellular membranes
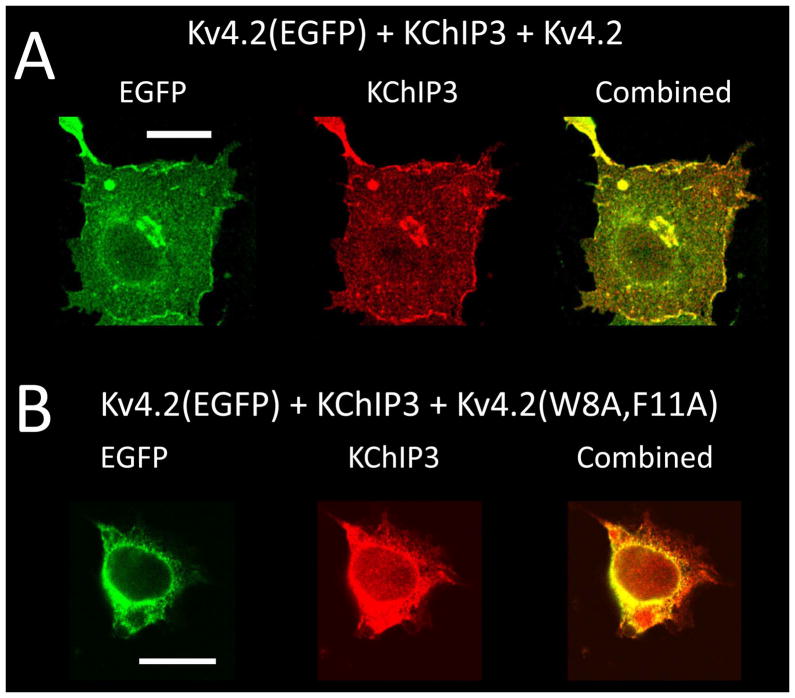
We next wanted to determine if the dominant negative effects of Kv4.2(W8A,F11A) result from the retention of Kv4.2 subunits on intracellular membranes even if they are bound to KChIPs. We therefore tested the effects of adding a non-fluorescent channel construct to the co-localization and trafficking of Kv4.2(EGFP) and KChIP3. In Fig. 4A, we performed a triple transfection of Kv4.2 with Kv4.2(EGFP) and KChIP3, using an equal amount of Kv4.2 and Kv4.2(EGFP) cDNA in the transfection. The presence of the non-fluorescent subunit has no effect on either the trafficking of fluorescent protein out of the internal membrane systems or the co-localization of KChIP3 and Kv4.2(EGFP). In Fig. 4B, we performed the same experiment only now substitution Kv4.2(W8A,F11A) for Kv4.2. In this case, we see that the wild type Kv4.2(EGFP) protein remains trapped in internal membrane despite the redistribution and co-localization of KChIP3 into the same membrane compartments. These results show that Kv4.2(W8A,F11A) causes an accumulation of wild type channels in intracellular membrane compartments despite KChIP3 recruitment to this subcellular compartment.
Figure 4. Kv4.2(W8A,F11A) changes the subcellular distribution of EGFP tagged wild type subunits co-expressed with KChIP3.
Channel constructs expressed in COS7 cells. Kv4.2(EGFP) co-expressed with KChIP3 and either Kv4.2 or Kv4.2(W8A,F11A), followed by fixation and immunofluorescence staining for KChIP3. Kv4.2(EGFP) localized by green fluorescence (EGFP image), and KChIP3 localized by red immunofluorescence. Projection images of confocal stacks shown. A) Mixing of wild type Kv4.2 into the Kv4.2(EGFP) plus KChIP3 expression mix does not affect the redistribution or co-localization of Kv4.2(EGFP) and KChIP3 onto the cell surface. B) In contrast, mixing Kv4.2(W8A,F11A) into the Kv4.2(EGFP) plus KChIP3 expression mix prevents the redistribution of Kv4.2(EGFP) protein from the internal membrane system, even though there is considerable co-localization of KChIP3 with the channel on internal membranes. Scale bars are 20 μm.
KChIP binding to channels containing Kv4.2(W8A,F11A) subunits
To test if KChIPs are able to bind stably to Kv4.2 subunits in heteromultimeric channels that also containing subunits with the Kv4.2(W8A,F11A) mutation, we performed co-immunoprecipitation experiments. To identify mutant and normal Kv4.2 subunits on Western blots, we again expressed wild type Kv4.2 as an EGFP fusion which runs at a higher molecular weight than Kv4.2(W8A,F11A). For these experiments, we expressed KChIP3 as an HA-tagged construct so we could use different antibodies to precipitate and detect the same protein. Fig. 5A shows, as expected, that Kv4.2(EGFP) efficiently co-precipitates with HA-KChIP3. In Fig. 5B, we see that if we mix in a non-fluorescent wild type Kv4.2 subunit into the transfection with Kv4.2(EGFP) plus KChIP3, then immunoprecipitation with anti-HA antibody pulls down both Kv4.2(EGFP) and Kv4.2. Likewise, anti-Kv4.2 antibody efficiently pulls down KChIP3 as expected. Interestingly, the results are virtually identical even if Kv4.2(W8A,F11A) cDNA is added to the expression mix, Fig. 5C. We see no loss of KChIP co-precipitation with anti-Kv4.2 antibodies and importantly, we see that both Kv4.2(EGFP) and Kv4.2(W8A,F11A) are pulled down by precipitation with anti-HA antibody. We have previously shown that Kv4.2(W8A,F11A) does not co-immunoprecipitate with HA-KChIP3 (Zhou et al. 2004); therefore, the co-immunoprecipitation we observe here must be due to pull down of heteromultimeric channels containing both Kv4.2(EGFP) and Kv4.2(W8A,F11A) that have stably incorporated KChIP3. Based on these results, we conclude that the functional defect produced by Kv4.2(W8A,F11A) is due to defective trafficking of channels with substoichiometric incorporation of KChIP subunits.
Figure 5. Kv4.2(W8A,F11A) co-expression does not prevent normal association of KChIP3 with wild type subunits.
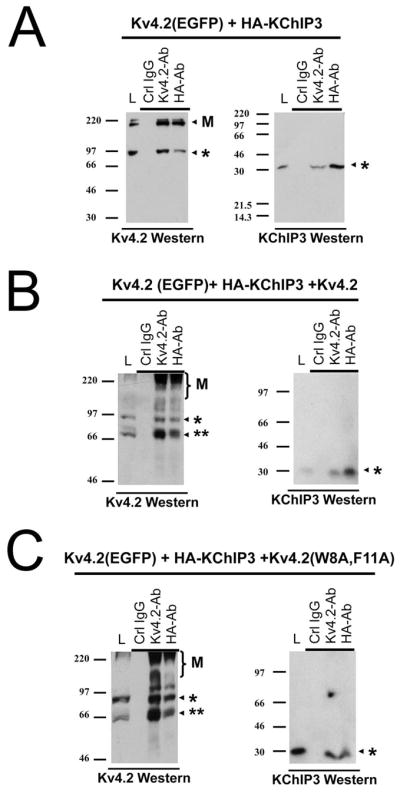
Co-precipitation experiments for Kv4.2(EGFP) co-expressed with HA tagged KChIP3 and mixed with either wild type or Kv4.2(W8A,F11A) mutant subunits. Trans-fections used in the experiments indicated above the Western blots. Antibodies used in the Western blots to detect proteins co-precipitated under the conditions indicated are listed below the Western. Lysate loading control lane (L) contains 1/5 of the amount of material used in each IP reaction. Note: Kv4.2 proteins typically show some multimeric laddering in Western blots, visible at high molecular weights (Indicated by “M”). Co-ippt. conditions shown above the appropriate lane. Single asterisk indicates location of Kv4.2(EGFP) or HA-KChIP3 respectively. Double asterisk indicates location of Kv4.2 or Kv4.2(W8A,F11A) respectively. A) Kv4.2(EGFP) is precipitated by anti-HA antibodies indicating it forms stable complexes with KChIP3. Complementary KChIP3 Westerns show co-IP of KChIP with anti-Kv4.2 antibodies. B) Co-expression with Kv4.2 in the mix results in the co-ippt. of the lower molecular weight Kv4.2 along with Kv4.2(EGFP) when precipitated by anti-HA antibodies. Likewise KChIP3 is co-ppt when Kv4.2-Ab is used. C) Co-expression with Kv4.2(W8A,F11A) in the mix also shows the efficient co-ppt. of both Kv4.2(W8A,F11A) and Kv4.2(EGFP) by either anti-Kv4.2 or anti-HA. KChIP3 co-ppt. by anti-Kv4.2 antibodies is also preserved under these conditions
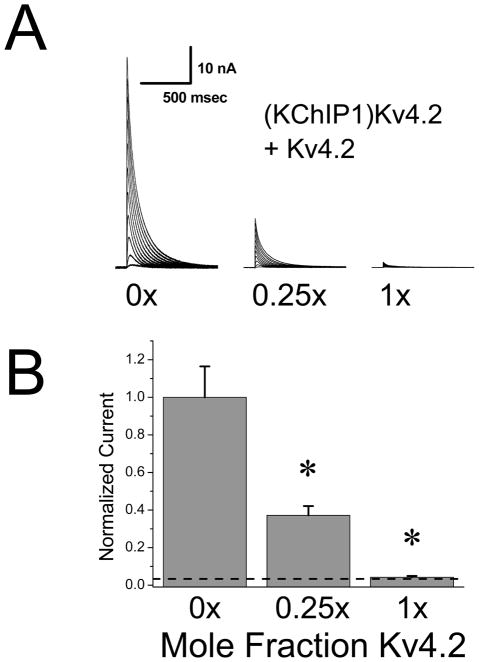
Dominant Negative effects of wild type Kv4.2 subunits not bound to KChIPs
Our hypothesis that substoichiometric incorporation of KChIP into channels fails to promote surface expression makes an interesting prediction that wild type Kv4.2 subunits should also have a dominant negative effect if they are incorporated into a channel without binding to KChIP. In our previous studies, we have shown that KChIP1 and KChIP3 produce a similar regulation of Kv4.2 channel functional expression, and the KChIP1 regulation is also disrupted in the Kv4.2(W8A,F11A) mutant. We further showed that a fusion of KChIP1 at the N-terminus of Kv4.2, making (KChIP1)Kv4.2, fully reproduces the trafficking and gating changes normally seen following KChIP1 co-expression (Zhou et al. 2004). This construct then allows us to express Kv4.2 subunits with a fixed 1:1 KChIP stoichiometry, and to test the hypothesis that Kv4.2 without KChIP is a dominant negative construct. To perform these experiments, we expressed (KChIP1)Kv4.2 and varied the amount of wild type Kv4.2 subunit added to the transfection mix. As usual, the amount of a control CMV promoter construct (pCMV-DsRed2) was varied to maintain a constant cDNA and promoter load in the transfections. We then recorded from cells and measured the levels of functional current expressed dependent upon the ratio of cDNA for wild type Kv4.2 compared to the fusion construct (KChIP1)Kv4.2 (Fig. 6A). Similar to our results with Kv4.2(W8A,F11A) we find that expression of (KChIP1)Kv4.2 is very sensitive to addition of even small amounts of non-KChIP bound wild type subunit. With a cDNA ratio 25% for Kv4.2 compared to (KChIP1)Kv4.2, we find that the functional current expression is suppressed by over 60%. At equimolar cDNA ratios (1:1) the functional current expression is essentially back down to levels seen without KChIP addition. We also confirmed that the KChIP binding mutant Kv4.2(W8A,F11A) suppresses expression of (KChIP1)Kv4.2 similarly to wild type Kv4.2 suggesting that the W8A,F11A mutations are not producing any additional effects in this assay (data not shown). Summary results for these experiments are presented in Fig. 6B.
Figure 6. Wild type Kv4.2 subunits dominantly suppress KChIP1 enhancement of Kv4.2 expression.
Titration experiment testing for the suppression of (KChIP1)Kv4.2 expression by Kv4.2 wild type subunits. Fusion construct (KChIP1)Kv4.2 has enhanced functional expression similar to co-expression of KChIP1 and Kv4.2 ((Zhou et al. 2004)). A variable amount of Kv4.2 cDNA was added into the trasfection mix and the total cDNA level kept constant by mixing in a variable amount of pCMV-DsRed2 reporter gene cDNA. A) Currents expressed relative to the molar ratio of Kv4.2 to (KChIP1)Kv4.2 cDNA added to the transfection mix. B) Summary results comparing the molar ratio of Kv4.2 cDNA added relative to (KChIP1)Kv4.2 cDNA to the fraction of expressed A-current as measured by peak current at +50 mV. Addition of Kv4.2 cDNA significantly reduces the peak current expression. Dashed line indicates Kv4.2 current expression level without KChIP1 addition. Kv4.2 addition significantly suppresses (KChIP1)Kv4.2 functional expression back to the level seen without KChIP1 being present. At 1x level, the current expression level is not significantly different from Kv4.2 alone.
Kv4.2 Subunits lacking FERN Domain are not Dominant Negative
Our results thus far show that Kv4.2 channels with less than 4 KChIPs do not express well. There are three likely models to explain such a result: 1) Each FERN domain must be independently suppressed by its own KChIP. 2) A non-FERN site is present on each channel that must be independently suppressed by its own KChIP. Or, 3) Four KChIPs must be bound to the channel to produce an independent positive signal that is needed for efficient functional expression of Kv4.2 channels. If Option 1 is true, then we predict that subunits lacking both KChIP binding and the FERN domain will no longer show dominant negative suppression of Kv4.2 functional expression. On the other hand, if Options 2 or 3 are true, then we should still see a dominant negative effect due to reduced integration of KChIPs into the channel. We therefore constructed a deletion construct for Kv4.2 that lacks the first 40 residues and thus is missing both the FERN domain and the KChIP binding motif, Kv4.2(Δ1–40) (Bahring et al. 2001, Foeger et al. 2010).
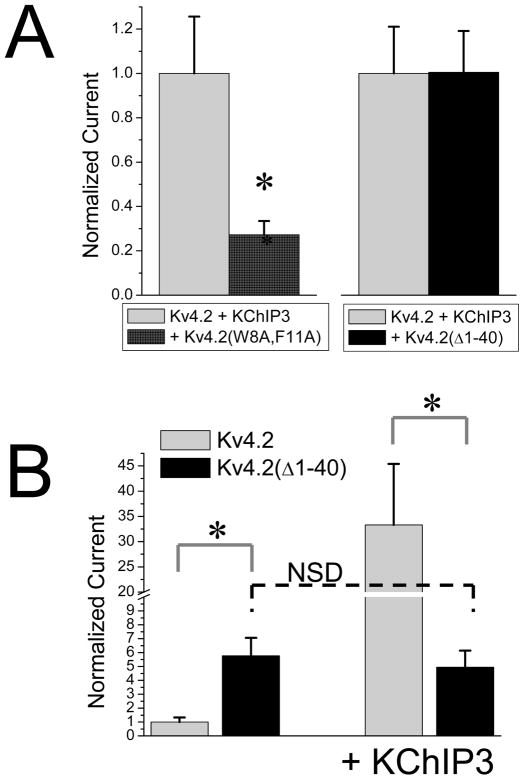
We tested for dominant negative effects of Kv4.2(Δ1–40) by co-expressing this subunit with Kv4.2 and KChIP3. Fig. 7A shows the effects of co-transfecting equimolar amounts of either Kv4.2(W8A,F11A) or Kv4.2(Δ1–40) cDNA with Kv4.2 and KChIP3. As described previously, Kv4.2(W8A,F11A) profoundly suppresses the expression of functional A-current; however, the Kv4.2(Δ1–40) subunit does not. In fact, there is no significant effect of Kv4.2(Δ1–40) in this assay, showing that reduced incorporation of KChIP3 into the channel is only a problem if a FERN domain that has not bound a KChIP is present in the channel.
Figure 7. Kv4.2 Subunits lacking FERN Domain do not have Dominant Negative effects on functional expression.
Dominant negative effects of Kv4.2(W8A,F11A) were compared to those of Kv4.2(Δ1–40) that lacks both the KChIP binding motif and the FERN domain. A) Addition of isomolar Kv4.2(W8A,F11A) significantly suppresses Kv4.2 + KChIP3 functional expression as described previously. In contrast, the non-KChIP3 binding construct Kv4.2(Δ1–40) shows no significant suppression of current. B) Deletion of the N-terminus of Kv4.2 shows a significant enhancement of functional expression due to the removal of the FERN domain. However, when KChIP3 is added to the expression mix, Kv4.2(Δ1–40) functional expression is around 5 fold lower than Kv4.2, and not significantly different from Kv4.2(Δ1–40) expressed without KChIP3. Asterisk- significant difference at the 0.01 level. N.S.D.- not significantly different.
To determine whether the only role for KChIP3 in regulating functional expression is to suppress the FERN domain, we compared the level of functional expression produced by Kv4.2(Δ1–40) to Kv4.2 co-expressed with KChIP3 (Fig. 7B). As expected, due to the removal of the FERN domain, the functional expression of Kv4.2(Δ1–40) is much greater than Kv4.2 expressed alone, increasing by 6 fold. As expected, due to removal of the N-terminal KChIP binding motif, Kv4.2(Δ1–40) expression level is insensitive to the co-expression of KChIP 3(Bahring et al. 2001, Foeger et al. 2010). Interestingly, however, the expression level produced by co-expressing Kv4.2 with KChIP3 is 5x fold greater than Kv4.2(Δ1–40) strongly suggesting that KChIP3 co-assembly with wild type Kv4.2 subunits produces positive effects on functional expression that are missing in the expression of Kv4.2(Δ1–40). Because Kv4.2(Δ1–40) does not show dominant negative effects when mixed with Kv4.2 plus KChIP3, such positive signals must be produced even with reduced levels of KChIP3 incorporated into the channels.
Discussion
In this study, we have shown that Kv4.2 subunits that have not bound KChIPs dominantly suppress the enhanced expression of Kv4.2 subunits that are KChIP bound. The dominant negative effect correlates with the retention of KChIP bound Kv4.2 subunits in intracellular membranes. When the initial 40 amino acids of the Kv4.2 protein are deleted the dominant negative effects are lost, despite the fact that the N-terminal KChIP binding motif is removed. We have named this 40 amino acid region of the N-terminus responsible for the dominant negative effect the FERN domain. Our results suggest that a failure of KChIPs to bind to the FERN domain is responsible for producing the domain negative effects we have observed.
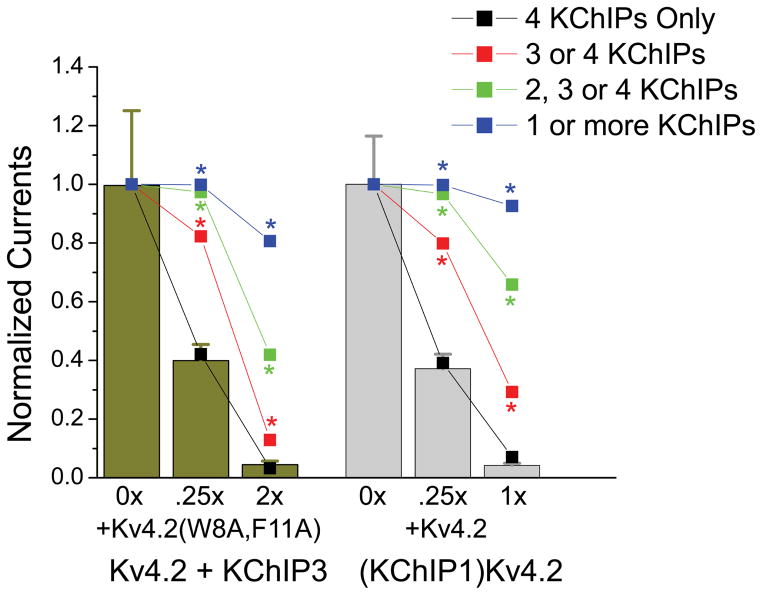
To estimate how completely FERN domains must be bound up by KChIPs to promote enhanced functional expression, we constructed a simple quantitative model and compared the predictions of this model to our results in the experiments shown in Figs 3 and 6 (Modeling described in Experimental Procedures). For our model we assumed random binomial, tetrameric assembly of mutant and wild type Kv4.2 subunits into channels in proportion to the amount of cDNA used in the transfection. We then used this model to predict the expected changes in functional channel expression if minimally 1, 2, 3, or 4 KChIPs must be bound to the channel to enhance functional expression. The results of this analysis are shown plotted against the observed data in Fig. 8, and clearly show that the observed expression levels are indistinguishable from what would be expected if channels must incorporate 4 KChIPs into the channel in order to have enhanced functional expression. Our results suggest that if even one Kv4.2 subunit does not bind KChIPs and thus exposes its FERN domain, functional expression of the channel is fully suppressed.
Figure 8. Binomial assembly modeling shows that titration experiment results are predicted if 4 KChIP proteins must be bound to the channel to enhanced functional expression.
Results from our titration experiments presented in Figs. 3 and 6 were compared to model predictions based on the need to incorporate 1, 2, 3, or 4 KChIPs into a channel in order to enhance functional expression. Results show that only the model where 4 KChIPs must be bound to enhance functional expression provides an accurate fit with predictions under all conditions tested that are not significantly different from our data. This result is consistent with a single unbound FERN domain fully suppressing enhanced channel functional expression. All other model predictions are significantly different from the measured data under the conditions indicated by the asterisks.
Because KChIPs bind to the FERN domain, it is tempting to speculate that at least some of the ability of KChIPs to traffic channels to the cell surface is due to the physical covering up of this regulatory region. Alternatively, however, the KChIP binding may recruit some additional proteins into this same regulatory region to completely suppress FERN domain function and promote functional surface expression. Scannevin et al. have suggested that the hydrophobic residues of the Kv4.2 FERN domain are aggregating in the absence of KChIPs, producing insoluble aggregates that are incapable of trafficking to the cell surface (Scannevin et al. 2004). In this case, KChIPs would be expected to occlude aggregation of the FERN domain. Other studies suggest that KChIPs produce forward trafficking signals and affect channel subunit stability and turnover from the surface membrane (Foeger et al. 2010, Bahring et al. 2001, Kunjilwar et al. 2004). We clearly see that KChIPs produce a redistribution of Kv4.2 channels within COS-7 cells only when all FERN domains are bound with KChIPs. We do not see evident aggregation of Kv4.2 into large aggresomes in the absence of KChIP binding (see Fig. S2C,D), as might be predicted from the Scannevin model; however, we cannot rule out smaller aggregates. Our results strongly point towards the importance of an interplay between the FERN domain and KChIPs, however, future studies will be needed to distinguish between the physical aggregation model of Scannevin et al and more typical trafficking models.
Our studies show that an important function of KChIPs is mediated through its regulation of FERN domain function; however, we have also noted a second effect of KChIP3 that acts to increase functional expression 5x higher than seen following removal of the FERN domain. This second effect does not require incorporation of 4 KChIPs into the channel, since it still occurs when Kv4.2(Δ1–40) is co-expressed with Kv4.2 and KChIP3. The mechanism for this positive effect of KChIPs is an important subject of future research, but could include direct KChIP effects on channel assembly or stability, or lifetime (Kunjilwar et al. 2004, Cui et al. 2008, Foeger et al. 2010, Shibata et al. 2003).
Finally, we have shown that DPP6 is able to modulate the surface expression of channels that are insensitive to KChIP proteins. This result suggests that DPLs have a separate mechanism available to override the retention effects produced by the FERN domains. Although DPLs are proposed to interact with Kv4 channels in the peri-transmembrane region (Ren et al. 2005), whether DPP6 also interacts directly with the FERN domain through a secondary binding site or promotes enhanced cell surface expression by another mechanism remains to be seen (Seikel & Trimmer 2009, Foeger et al. 2012, Cotella et al. 2012, Cotella et al. 2010). Other studies comparing DPP6 and KChIP regulation of Kv4.2 channels in HEK cells have suggested a unique role of KChIPs in stabilizing total channel protein in the cell, whereas DPP6 primarily shifts the protein onto the cell surface (Foeger et al. 2012). However, knockdown of DPP6 in cerebellar granule neurons and hippocampal pyramidal neurons shows a dramatic loss of both Kv4.2 and KChIP3 when DPP6 is lost (Nadin & Pfaffinger 2010). It therefore remains to be seen how these different regulatory systems actual work together to control normal channel functional expression in native neurons.
Supplementary Material
Acknowledgments
This work was supported by Grants R01NS31583, R01 GM090029 and P01NS37444 from the National Institutes of Health and the Cores of the Baylor College of Medicine Mental Retardation Research Center Grant HD24064.
The abbreviations used are
- EGFP
enhanced green fluorescent protein
- CHO
Chinese hamster ovary
Footnotes
The authors have no conflicts of interest to declare.
References
- An WF, Bowlby MR, Betty M, et al. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- Bahring R, Dannenberg J, Peters HC, Leicher T, Pongs O, Isbrandt D. Conserved Kv4 N-terminal domain critical for effects of Kv channel-interacting protein 2.2 on channel expression and gating. J Biol Chem. 2001;276:23888–23894. doi: 10.1074/jbc.M101320200. [DOI] [PubMed] [Google Scholar]
- Braunewell KH, Gundelfinger ED. Intracellular neuronal calcium sensor proteins: a family of EF-hand calcium-binding proteins in search of a function. Cell Tissue Res. 1999;295:1–12. doi: 10.1007/s004410051207. [DOI] [PubMed] [Google Scholar]
- Cotella D, Radicke S, Bortoluzzi A, Ravens U, Wettwer E, Santoro C, Sblattero D. Impaired glycosylation blocks DPP10 cell surface expression and alters the electrophysiology of Ito channel complex. Pflug Arch Eur J Phy. 2010;460:87–97. doi: 10.1007/s00424-010-0824-2. [DOI] [PubMed] [Google Scholar]
- Cotella D, Radicke S, Cipriani V, et al. N-glycosylation of the mammalian dipeptidyl aminopeptidase-like protein 10 (DPP10) regulates trafficking and interaction with Kv4 channels. Int J Biochem Cell Biol. 2012;44:876–885. doi: 10.1016/j.biocel.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Cui YY, Liang P, Wang KW. Enhanced trafficking of tetrameric Kv4.3 channels by KChIP1 clamping. Neurochem Res. 2008;33:2078–2084. doi: 10.1007/s11064-008-9688-7. [DOI] [PubMed] [Google Scholar]
- Deng Y, Bennink JR, Kang HC, Haugland RP, Yewdell JW. Fluorescent conjugates of brefeldin A selectively stain the endoplasmic reticulum and Golgi complex of living cells. J Histochem Cytochem. 1995;43:907–915. doi: 10.1177/43.9.7543914. [DOI] [PubMed] [Google Scholar]
- Flowerdew SE, Burgoyne RD. A VAMP7/Vti1a SNARE complex distinguishes a non-conventional traffic route to the cell surface used by KChIP1 and Kv4 potassium channels. Biochem J. 2009;418:529–540. doi: 10.1042/BJ20081736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foeger NC, Marionneau C, Nerbonne JM. Co-assembly of Kv4 {alpha} subunits with K+ channel-interacting protein 2 stabilizes protein expression and promotes surface retention of channel complexes. J Biol Chem. 2010;285:33413–33422. doi: 10.1074/jbc.M110.145185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foeger NC, Norris AJ, Wren LM, Nerbonne JM. Augmentation of Kv4.2-encoded currents by accessory dipeptidyl peptidase 6 and 10 subunits reflects selective cell surface Kv4.2 protein stabilization. J Biol Chem. 2012;287:9640–9650. doi: 10.1074/jbc.M111.324574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer M, Isbrandt D, Sauter K, Callsen B, Nolting A, Pongs O, Bahring R. N-type inactivation features of Kv4.2 channel gating. Biophys J. 2004;86:210–223. doi: 10.1016/S0006-3495(04)74097-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerng HH, Pfaffinger PJ, Covarrubias M. Molecular physiology and modulation of somatodendritic A-type potassium channels. Mol Cell Neurosci. 2004a;27:343–369. doi: 10.1016/j.mcn.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Jerng HH, Qian Y, Pfaffinger PJ. Modulation of Kv4.2 Channel Expression and Gating by Dipeptidyl Peptidase 10 (DPP10) Biophys J. 2004b;87:2380–2396. doi: 10.1529/biophysj.104.042358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim LA, Furst J, Butler MH, Xu S, Grigorieff N, Goldstein SA. Ito Channels Are Octomeric Complexes with Four Subunits of Each Kv4.2 and K+ Channel-interacting Protein 2. J Biol Chem. 2004a;279:5549–5554. doi: 10.1074/jbc.M311332200. [DOI] [PubMed] [Google Scholar]
- Kim LA, Furst J, Gutierrez D, Butler MH, Xu S, Goldstein SA, Grigorieff N. Three-Dimensional Structure of I(to); Kv4.2-KChIP2 Ion Channels by Electron Microscopy at 21 A Resolution. Neuron. 2004b;41:513–519. doi: 10.1016/s0896-6273(04)00050-9. [DOI] [PubMed] [Google Scholar]
- Kunjilwar K, Strang C, DeRubeis D, Pfaffinger PJ. KChIP3 rescues the functional expression of Shal channel tetramerization mutants. J Biol Chem. 2004;279:54542–54551. doi: 10.1074/jbc.M409721200. [DOI] [PubMed] [Google Scholar]
- Morohashi Y, Hatano N, Ohya S, Takikawa R, Watabiki T, Takasugi N, Imaizumi Y, Tomita T, Iwatsubo T. Molecular Cloning and Characterization of CALP/KChIP4, a Novel EF-hand Protein Interacting with Presenilin 2 and Voltage-gated Potassium Channel Subunit Kv4. J Biol Chem. 2002;277:14965–14975. doi: 10.1074/jbc.M200897200. [DOI] [PubMed] [Google Scholar]
- Nadal MS, Ozaita A, Amarillo Y, et al. The CD26-related dipeptidyl aminopeptidase-like protein DPPX is a critical component of neuronal A-type K+ channels. Neuron. 2003;37:449–461. doi: 10.1016/s0896-6273(02)01185-6. [DOI] [PubMed] [Google Scholar]
- Nadin BM, Pfaffinger PJ. Dipeptidyl peptidase-like protein 6 is required for normal electrophysiological properties of cerebellar granule cells. J Neurosci. 2010;30:8551–8565. doi: 10.1523/JNEUROSCI.5489-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TY, Pountney DJ, Ozaita A, Nandi S, Ueda S, Rudy B, Coetzee WA. A role for frequenin, a Ca2+-binding protein, as a regulator of Kv4 K+-currents. Proc Natl Acad Sci U S A. 2001;98:12808–12813. doi: 10.1073/pnas.221168498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Callaghan DW, Hasdemir B, Leighton M, Burgoyne RD. Residues within the myristoylation motif determine intracellular targeting of the neuronal Ca2+ sensor protein KChIP1 to post-ER transport vesicles and traffic of Kv4 K+ channels. J Cell Sci. 2003;116:4833–4845. doi: 10.1242/jcs.00803. [DOI] [PubMed] [Google Scholar]
- Pioletti M, Findeisen F, Hura GL, Minor DL., Jr Three-dimensional structure of the KChIP1-Kv4.3 T1 complex reveals a cross-shaped octamer. Nat Struct Mol Biol. 2006;13:987–995. doi: 10.1038/nsmb1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Hayashi Y, Yoshimura N, Takimoto K. Transmembrane interaction mediates complex formation between peptidase homologues and Kv4 channels. Mol Cell Neurosci. 2005;29:320–332. doi: 10.1016/j.mcn.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Scannevin RH, Wang K, Jow F, et al. Two N-Terminal Domains of Kv4 K(+) Channels Regulate Binding to and Modulation by KChIP1. Neuron. 2004;41:587–598. doi: 10.1016/s0896-6273(04)00049-2. [DOI] [PubMed] [Google Scholar]
- Seikel E, Trimmer JS. Convergent modulation of Kv4.2 channel alpha subunits by structurally distinct DPPX and KChIP auxiliary subunits. Biochemistry. 2009;48:5721–5730. doi: 10.1021/bi802316m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata R, Misonou H, Campomanes CR, et al. A fundamental role for KChIPs in determining the molecular properties and trafficking of Kv4.2 potassium channels. J Biol Chem. 2003;278:36445–36454. doi: 10.1074/jbc.M306142200. [DOI] [PubMed] [Google Scholar]
- Strang C, Kunjilwar K, DeRubeis D, Peterson D, Pfaffinger PJ. The role of Zn2+ in Shal voltage-gated potassium channel formation. J Biol Chem. 2003;278:31361–31371. doi: 10.1074/jbc.M304268200. [DOI] [PubMed] [Google Scholar]
- Wang H, Yan Y, Liu Q, et al. Structural basis for modulation of Kv4 K+ channels by auxiliary KChIP subunits. Nat Neurosci. 2007;10:32–39. doi: 10.1038/nn1822. [DOI] [PubMed] [Google Scholar]
- Zhou W, Qian Y, Kunjilwar K, Pfaffinger PJ, Choe S. Structural Insights into the Functional Interaction of KChIP1 with Shal-Type K(+) Channels. Neuron. 2004;41:573–586. doi: 10.1016/s0896-6273(04)00045-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.