Discovery and Origin of Cellular Senescence
Classical work by Hayflick and Moorhead1 uncovered more than 50 y ago the biological and evolutionarily conserved phenomenon of cellular senescence. The authors demonstrated that primary fibroblasts exhibit only a finite proliferative capacity in culture, before they exit the cell cycle in a state known as replicative senescence (“Hayflick phenomenon”).1 This type of senescence is caused by progressive telomeric erosion associated with the accumulation of DNA damage. Alternatively, several stress factors, including hypermitogenic stimuli like oncogenic Ras, reactive oxygen species and cytotoxic drugs that cause DNA damage, as well as mitotic spindle dysfunction and aneuploidy can trigger an accelerated antiproliferative response, known as stress-induced, premature senescence (SIPS).2
(Patho)biological Role of Senescence
Senescence comes along as a (patho)biologically relevant, but two-edged, cellular stress response. On one hand, a protective and tumor-suppressive role of cellular senescence has been demonstrated at the pre-malignant level.3 On the other hand, senescence exhibits detrimental effects at the cellular and organ level, including proliferative exhaustion of progenitor and stem cells or promotion of inflammatory processes linked to the so-called senescence-associated secretory phenotype (SASP) (Fig. 1).4 Interestingly, elimination of accumulating senescent cells in vivo in the mouse effectively delays aging-associated disorders, thereby corroborating the causative link between cellular senescence and tissue dysfunction in age-related phenotypes.5 Thus, cellular senescence seems to be ultimately connected to health and lifespan regulation during organismal aging.
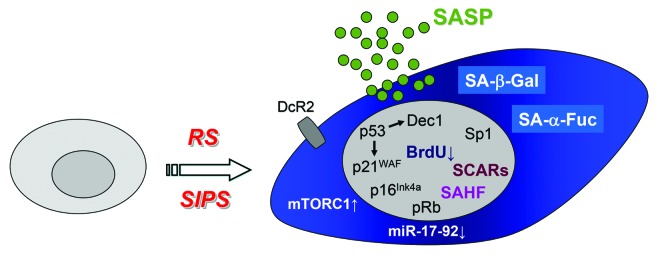
Figure 1. Characteristic features of senescent cells. Cells undergoing replicative senescence (RS) or stress-induced premature senescence (SIPS) are distinguished by an enlarged and flattened morphology and several molecular and subcellular changes, including activation of tumor suppressor pathways (p53-p21WAF, p53-Dec1, p16Ink4a), chromatin alterations (DNA-SCARS, DNA segments with chromatin alterations reinforcing senescence; SAHF, senescence-associated heterochromatic foci) and activation of certain transcription factors (Sp1) as well as production of secreted factors (SASP, senescence-associated secretory phenotype). Moreover, cell surface expression of decoy receptor 2 (DcR2) typically increases during senescence. Lastly, the lysosomal compartment expands considerably in cells undergoing cellular senescence. Here, as demonstrated by Hildebrand et al.,8 the classical and widely used senescence-associated β-galactosidase activity (SA-β-Gal)7 is now joined by a novel and more robust lysosomal (bio)marker for cellular senescence, SA-α-Fuc (senescence-associated α-fucosidase activity).
Characteristics and Markers of Senescent Cells
In order to detect senescent cells in culture, and clearly more challenging under in vivo and in situ conditions, several markers with varying robustness are being used beyond the typical flattened cellular morphology and increased cell surface (Fig. 1). Overall, proliferative arrest, apoptosis resistance and altered gene expression and miRNA profiles (Fig. 1) represent general features of senescent cells.2 At the nuclear architecture level, formation of DNA-SCARS (DNA segments with chromatin alterations reinforcing senescence) and senescence-associated heterochromatic foci (SAHF)4 mirror the repression of proliferative genes and therefore the lack of DNA synthesis as detected by a lack of BrdU incorporation and G1 arrest. At the molecular checkpoint level, the p53-p21WAF1 pathway and the tumor suppressor p16INK4a, as well as rapamycin-sensitive mTOR (mammalian Target of Rapamycin) signaling take over synergistic roles in the induction and maintenance of the senescent phenotype (Fig. 1).2,6 Lastly, at the biochemical and enzymatic level, a considerable expansion of the lysosomal compartment and, hence, increased granularity is typically observed in cells undergoing senescence. That is demonstrated by an increased senescence-associated β-galactosidase activity (SA-β-Gal; measured at pH 6), the classical marker that is widely used to detect senescent cells.7
Senescence-Associated Lysosomal α-L-Fucosidase (SA-α-Fuc): A new and Robust Senescence Marker
Several of the markers described above may work in a cell type- and senescence stimulus-dependent manner and are therefore not always reliable. For example, p16INK4a increases during replicative aging. However, p16INK4a is either not expressed or inactivated in certain tumor cells, which, however, are still prone to SIPS due to an intact p53 pathway. Moreover, solely determining SA-β-Gal activity can potentially result in both wrong positive results (increased SA-β-Gal activity due to hyper-dense cell cultures), or wrong negative results, where SA-β-Gal is not or only weakly induced and therefore not sensitively enough detected upon cellular senescence. Taking in particular the latter observation into account, Hildebrand et al. investigated comprehensively several lysosomal hydrolases, besides SA-β-Gal, for their suitability as senescence markers.8 Interestingly, the authors identified α-L-fucosidase, a glycosidase involved in the metabolism of certain glycolipids and glycoproteins, as a novel and promising biomarker for cellular senescence. Hildebrand et al. tested senescence-associated α-fucosidase activity (SA-α-Fuc) in various senescence models in cell culture, including replicative and oncogene-induced senescence, as well as SIPS. Unequivocally, both at the transcriptional and the enzymatic level SA-α-Fuc turned out as an at least equivalent, and in most cases an even more reliable, marker for the detection of cellular senescence as compared with SA-β-Gal.
Taken together, SA-α-Fuc represents a convenient, sensitive and robust senescence marker in cell culture experiments employing both rodent and human senescence models. Further characterization of SA-α-Fuc expression and activity under stringent in vivo and in situ settings could include the detection of rather sparsely occurring senescent cells at the cancer stem and progenitor level, as well as the analysis of senescent cells during tissue aging.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25318
References
- 1.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Collado M, Serrano M. The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer. 2006;6:472–6. doi: 10.1038/nrc1884. [DOI] [PubMed] [Google Scholar]
- 3.Kang TW, Yevsa T, Woller N, Hoenicke L, Wuestefeld T, Dauch D, et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–51. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- 4.Rodier F, Campisi J. Four faces of cellular senescence. J Cell Biol. 2011;192:547–56. doi: 10.1083/jcb.201009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–6. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demidenko ZN, Zubova SG, Bukreeva EI, Pospelov VA, Pospelova TV, Blagosklonny MV. Rapamycin decelerates cellular senescence. Cell Cycle. 2009;8:1888–95. doi: 10.4161/cc.8.12.8606. [DOI] [PubMed] [Google Scholar]
- 7.Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hildebrand DG, Lehle S, Borst A, Haferkamp S, Essmann F, Schulze-Osthoff K. α-Fucosidase as a novel convenient biomarker for cellular senescence. Cell Cycle. 2013;12 doi: 10.4161/cc.24944. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]


