Abstract
The acquisition of massive but localized chromosome translocations, a phenomenon termed chromothripsis, has received widespread attention since its discovery over a year ago. Until recently, chromothripsis was believed to originate from a single catastrophic event, but the molecular mechanisms leading to this event are yet to be uncovered. Because a thorough interpretation of the data are missing, the phenomenon itself has wrongly acquired the status of a mechanism used to justify many kinds of complex rearrangements. Although the assumption that all translocations in chromothripsis originate from a single event has met with criticism, satisfactory explanations for the intense but localized nature of this phenomenon are still missing. Here, we show why the data used to describe massive catastrophic rearrangements are incompatible with a model comprising a single event only and propose a molecular mechanism in which a combination of known cellular pathways accounts for chromothripsis. Instead of a single traumatic event, the protection of undamaged chromosomes by telomeres can limit repetitive breakage-fusion-bridge events to a single chromosome arm. Ultimately, common properties of chromosomal instability, such as aneuploidy and centromere fission, might establish the complex genetic pattern observed in this genomic state.
Keywords: chromothripsis, breakage-fusion-bridge, chromosomal instability, centromere fission, mitosis, telomere
Introduction
The phenomenon of localized but massive DNA fragmentation was described first as an acquisition of rearrangements through a single catastrophic event1 and was subsequently termed chromothripsis.2 Whereas chromothripsis was used initially to describe a considerable number—dozens or more—of translocations limited to a single chromosome,1 the term has since been applied to a wide variety of chromosomal alterations irrespective of the involvement of one or more chromosomes or the number of detected alterations.3 Currently, chromothripsis is used to describe chromosomal rearrangements affecting between one and a dozen or more chromosomes in tumors or cell lines,1,3,4 in the germ line5,6 or in chronic inflammation.7 Even though the term chromothripsis is being applied to a continuously expanding group of complex, otherwise inexplicable, chromosome aberrations, efforts to provide a mechanistic basis for the phenomenon have met with little success. Since a satisfactory elucidation is lacking, the phenomenon itself has wrongly acquired the role of mechanism3,5; considerable importance has been attributed to massive chromosome fragmentation, but chromothripsis appears to be a genetic state at best.
Chromothripsis was thoroughly reviewed shortly after its first description,8 but the first critical considerations regarding its proposed underlying mechanisms have been published only recently.9,10 According to the initial description, numerous simultaneous breaks shatter a single chromosome, forming dozens of fragments that are subsequently reassembled in a random fashion by non-homologous end joining (NHEJ). However, the single event nature of chromothripsis has met with criticism,10 because it is inferred from a undocumented computer model verified only for a minimal subset of samples.1 Other authors indeed consider the possibility that chromosome pulverization might be a multi-step process,9,11 in agreement with the classical stepwise model of tumor development.12
Notwithstanding recent reviews, which warn for genetic and experimental factors that might skew interpretation,13 the status of chromothripsis as a novel phenomenon has remained unchallenged. Here, we argue that part of the genetic alterations attributed to chromothripsis, including examples from the initial description, are brought about by a reiteration of breakage-fusion-bridge (BFB) cycles. Possibly, the entire spectrum genome instability cannot be accounted for by simple genetics, and several explanations tailored to individual examples might be needed. Previously described mechanisms, however, can account for many aspects of chromosome pulverization.
Incompatibilities in a Single Event Model
Although the percentage of tumors that can be attributed to chromothripsis is very low (2–3%), the proposed catastrophe leading to pulverization of individual chromosomes can be considered spectacular. Other phenomena that generate substantial DNA damage in a short time have been described, for example, apoptosis14 or meiotic recombination,15 but breaks in these events are spread over the entire chromosome complement. Thus, the most remarkable feature of chromothripsis is that a large number of translocations affect a small number of chromosomes. The limited number of chromosomes typically involved in a single sample might have contributed to the hypothesis that a massive number of breaks are acquired simultaneously.
A Monte Carlo simulation—a mathematical algorithm based on random sampling—is used to corroborate the formation of massive breakage in a single event.1 This computer model predicts an escalation of distinct copy number (CNum) states during a sequential accumulation of translocations. Since the observed ratio of translocations and CNum states falls outside the predicted range for repetitive breakage, the assumption of a single catastrophic event is made. The problem is that a purely qualitative attribute (sequencing of a translocation) is compared with a quantitative measurement (CNum). Massive sequencing potentially identifies a translocation present only in a single cell, whereas CNum analysis provides a global value for the entire sample. Thus, loss of a chromosomal region form part of the cell population, a consequence of heterogeneous growth,16 does not impede sequence-based identification of the fusion point, but will alter the CNum of the segments involved. Subsequent scoring of translocations present only in a subset of cells might produce a gross overestimation of the total number of translocations per cell. CNum analysis indeed indicates that genetic regions undergoing chromothripsis are subject to heterogeneity, and are inherited by a subset of cells instead of the whole population (see below).
Notwithstanding the problems with a computer model, the comparison of fusion points to CNum and heterozygosity status is an essential step that helps to understand the behavior of a cell population (Fig. 1). In contrast to the predictions made by the Monte Carlo simulation, thorough scrutiny of the data seems to reveal that several features of chromothripsis are incompatible with a single round of breakage and rejoining. According to the initial model, fragments that are not included in the reassembled chromosome are lost to the cell. In contrast, all fragments except the future telomeres must be fused at both ends when reincorporated in the reassembled chromosome; a single round of breakage and ligation also means that intra-chromosomal fragments can only recombine with other intra-chromosomal fragments, and all of these must be present at the same CNum (Fig. 1A and B). DNA segments lost to the cell, or incorporated in a second chromosome structure, might be present in lower CNum, but only if they behave independently. In contrast, translocations spanning segments that have different CNum can be formed only after multiple rounds of breakage and fusion (Fig. 1C); as a minimum, these translocations need an event to produce the translocation and an independent event to generate CNum deviations. In some samples attributed to chromothripsis, however, the combination of CNum analysis and massive sequencing shows translocations between fragments that originate from the same chromosome but have acquired different CNum (Fig. 2), invalidating models based on a single event.
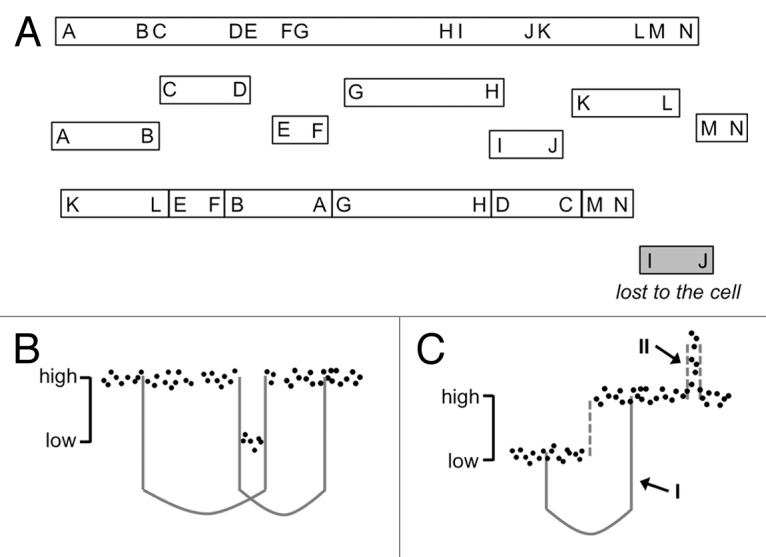
Figure 1. Possibilities and impossibilities of a single event model. (A) The hitherto proposed mechanism leading to chromothripsis comprises a single round of breakage and fusion, in which any fragment evading reassembly is excluded from the population. In the proposed pathway, clonal expansion of the reassembled chromosome leads to a unique and integer (“high” or “low”) copy number for each segment. (B) A single catastrophic event can produce interleaved translocations in the reassembled chromosome (left, for example A-G and H-D), and eliminate segments to produce two unique copy number states. (C) A single event cannot cause translocations between segments that bear different copy numbers (I) or non-integer CNum states that exceed the ploidy of chromothripsis-associated chromosomes (II). The latter two phenomena require differential transmission of chromosomal segments within the cell population and are probably linked to tumor heterogeneity.
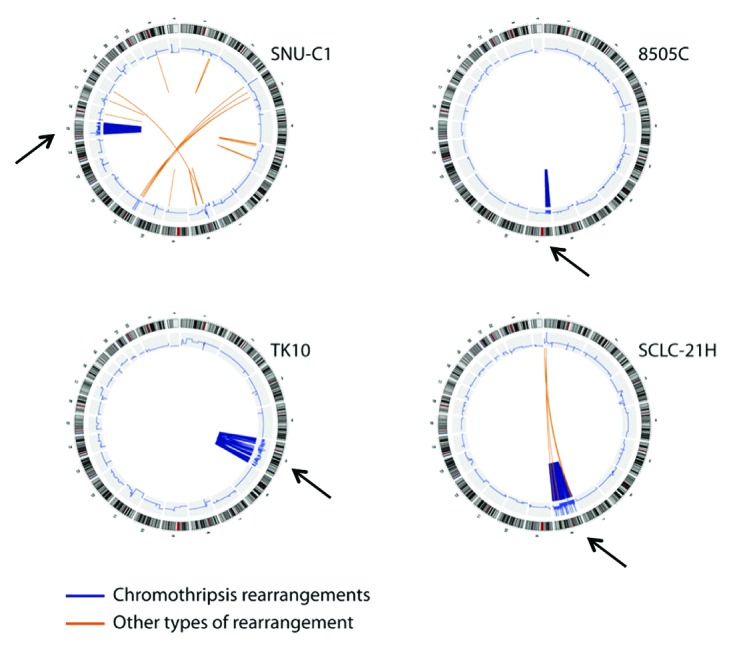
Figure 2. Whole-genome analysis in chromothripsis. Reprinted from Stephens et al. Cell 2011; 144:27–40, with permission from Elsevier. Circos plots of cell lines used for the original description of chromothripsis. Chromosome banding is shown in the outer circle and CNum analysis in the inner circle. Translocations are depicted inside the inner circle. Please note the localized CNum heterogeneity in the affected chromosomes only (arrows), evident from an increased signal noise as compared with unaffected chromosomal regions.
CNum analysis not only assists the interpretation of individual translocations, but can also provide information on the behavior of the whole cell population. Although the details of the Monte Carlo simulations are not published, a model in which translocations are caused in multiple successive events apparently would result in a wide range of CNum states, whereas a single event would limit the possibilities of CNum variation1 and instead result in an oscillation between two states.13 The “simple” mathematics of a single breakage and fusion provide the same conclusion (Fig. 1); individual segments are either included with peers or excluded from the reformed chromosome. What is considered a unique CNum state, however, could be a matter of debate; whereas the CNum of most chromosomal segments faithfully follows the ploidy determined by spectral karyotyping, the CNum in regions of chromothripsis deviate considerably from ploidy status and no longer are found as integer values (Figs. 2 and 3). Such deviations from integer CNum states can only occur in a heterogeneous population, in which part of the cells have gained or lost a particular marker. It has been recognized that intra-tumor variation could be an important factor to establish genetic patterns that resemble chromothripsis,13 but mathematical methods to quantify heterogeneity are still awaiting development. Recent techniques, however, have made it possible to carry out CNum analysis on individual cells.17 When applied to cell lines that show chromosomal instability (CIN), these experiments showed that individual cells acquire additional copies of small regions, revealing marked cell-to-cell differences and heterogeneity. Although analysis of single cells17 identified multiple CNum differences between cell pairs, the high numbers of translocations apparently found in chromothripsis were never reached. Probably, massive sequencing has identified the sum of many translocations, each of which represent a small proportion of the entire sample. In conclusion, CNum alterations in small genomic regions acquired in individual cells do not necessarily represent the entire cell population.
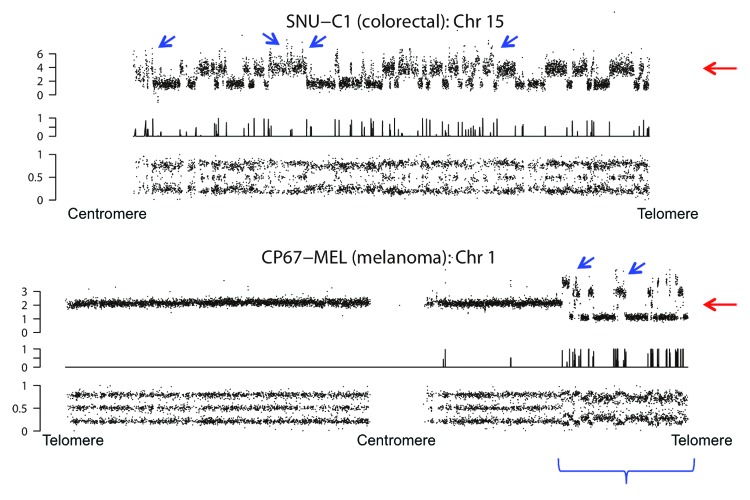
Figure 3. Analysis of single chromosomes. Reprinted from Stephens et al. Cell 2011; 144:27–40, with permission from Elsevier. Comparison of CNum (upper part), predicted breakpoints (middle part) and allelic balance (lower part) for two chromosomes undergoing chromothripsis. Red arrows indicate whole chromosome CNum; blue arrows show examples of regions with higher CNum than the whole chromosome. Please note the increased noise in CNum analysis and allelic balance (blue accolade) for regions strongly affected by chromothripsis. The genomic affected regions extend up to the telomere, but no involvement of other chromosome arms was evident (Fig. 2, SNU-C1). The short arm of chromosome 15 comprises rDNA repeats and is not analyzed.
The non-integer CNum states corresponding to regions of chromothripsis, evident from increased noise in CNum analyses (Figs. 2 and 3), indicate that these genomic segments in particular suffer from sample heterogeneity. In addition, the CNum of individual small segments frequently exceeds ploidy of the corresponding chromosome; the CNum of several small regions reaches eight or more, whereas karyotyping shows a few additional copies of the affected chromosome (Fig. 3). In combination, these data show that regions of chromothripsis acquire a large variety of unique CNum states. The combination of a lower than expected number of translocations and higher than expected variation in CNum states would probably classify the data as a product of sequential events in the Monte Carlo simulation. Apart from the CNum, the allelic balance ratios also indicate that chromothripsis is associated with heterogeneity; whereas allelic ratios follow the limitations imposed by ploidy in normal segments, intermediate values are observed frequently for markers located in chromothriptic segments (Fig. 3). Since sample heterogeneity requires differential retention of translocations—single-cell analysis demonstrates that accumulation of heterogeneity is a stepwise process17—signal noise in data concerning the entire cell population contradicts models based on a single event. In conclusion, population-wide methods of genetic analysis favor a multi-step process in which translocations accumulate sequentially.
Alternative Explanations
Even though chromothripsis has received criticism in earlier reviews,9,10,13 few alternative explanations for the observed data have been proposed. An open question is how the data from sequencing and CNum analysis can be unified. To answer this question, it is important to know the nature of the data; techniques with the potential to detect alterations in individual cells (massive sequencing and karyotyping) are compared with population-wide analyses (CNum and allelic ratio). Even though a multi-step model is more probable, it not only has to elucidate the generation of breaks that lead to translocations, but also support the observed CNum variation and sample heterogeneity. Finally, a comprehensive model has to include a mechanism that prevents breakage of unaffected chromosomal regions.
Since a single round of breakage and rejoining cannot account for all chromosomal alterations detected, the hypothesis has been expanded with an exacerbation of breakage by replication defects.18 Apparently, sequestration of lagging metaphase chromosomes in micronuclei leads to a delay in the replication of the DNA contained within. This delay might then provoke a damage response, restricted to the micronucleus, which supposedly limits chromothripsis to a few chromosomes. Since micronucleus formation and delayed replication take place in a single cell cycle, the micronucleus model still does not explain why regions of chromothripsis acquire non-integer CNum. In conclusion, a model than spans multiple divisions, thereby triggering the differential propagation of breakage products, is a prerequisite for the emergence of non-integer CNum and sample heterogeneity.
Chromothripsis Correlates with CIN and BFB
The Monte Carlo simulations agree with a single event only if CNum states closely follow ploidy; the examples shown above reveal that most of the data cannot be interpreted in this way. Instead, CNum analysis and heterozygosity indicate that multiple sequential events sculpt the genetic landscape in chromothripsis. This, however, leaves the question open as to what might be the underlying mechanism. Several of the cell lines used for the initial description, for example TK1019 and 8505C,20 were originally established over two decades ago and show signs of chromosomal instability (CIN). A subclass of CIN might thus account for the accumulation of breaks in a limited region of the genome.
Whereas CIN is mostly known for the gains and losses of whole chromosomes, it also includes chromosome breakage and structural alterations.21 The hypothesis that structural and numerical alterations are associated is supported by the samples of chromothripsis, as many of these present intra- and interchromosomal translocations together with copy number alterations.1 One feature of CIN in particular, formation of dicentric chromosomes and the cyclic process known as breakage-fusion-bridge (BFB),22,23 seems a likely cause of the break distribution observed in chromothripsis. Although dicentric chromosomes were considered briefly when massive but localized damage was first described,1 their relation to the observed data and to a single catastrophic event has remained elusive. A later review considered chromothripsis and BFB separate phenomena.13 Nonetheless, BFB fits with the observed data, because breakage occurs at a random position between the two centromeres when the kinetochores of dicentric chromosomes segregate to different spindle poles.23 Chromosomal regions not enclosed between the two centromeres, effectively most of the genome, will not undergo this type of mitotic spindle-induced breakage.
The tension generated by the mitotic spindle not only produces breakage of dicentric chromosomes, it also segregates the two DNA fragments into different daughter cells. BFB thus provides a mechanism for the enrichment of a chromosome segment in a proportion of daughter cells, and subsequent differences in growth or survival of these daughter cells might limit the transmission of additional copies to a proportion of the cell population. Differential transmission of chromosome segments in BFB, in multiple sequential events, thus causes heterogeneity and explains why individual markers appear in a non-integer CNum. Since amplification of small chromosome regions is a common consequence of ongoing BFB cycles,24,25 BFB might also explain the sharp increase in the CNum of small regions, even exceeding ploidy of the corresponding chromosome arm (Fig. 3, arrows). The remarkable size limitation of the amplified segments might be explained by the finding that a previously damaged chromosomal region remains susceptible for renewed breakage after fusion of DNA ends.26
Combination of the main characteristics of BFB, random breakage of dicentrics and inheritance of the fragments by daughter cells, has important consequences for the genomic landscape in chromothripsis. Random break distribution, albeit restricted to a single dicentric chromosome, means that each round of division can position a rupture in a new genomic location; each mitosis thus contributes to the acquisition of additional CNum states by the cell population. Since BFB is a cyclic process that repeats itself from one cell division to another, exponential growth of cancer cells implies that CNum states accumulate at an exponential rate as well. Importantly, the two daughter cells that inherit a chromosome fragment behave independently and therefore produce unrelated new breaks in the subsequent BFB cycle (Fig. 4, left). The analysis of translocations in single cells17 corroborates the cell-autonomous and individual nature of breakage. A theoretical doubling of breakage sites in each division means that the typical number of translocations in a chromothripsis sample,1 between 50 and 100, might already be acquired in the sixth or seventh generation of daughter cells after BFB has first started.
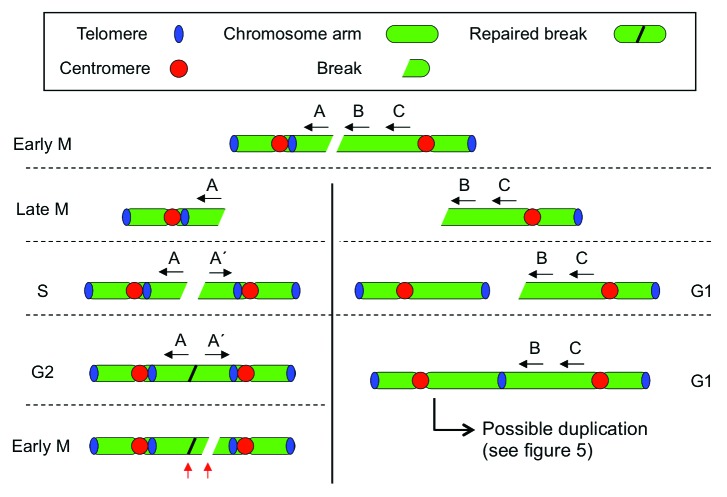
Figure 4. BFB and chromothripsis. Mitotic breakage of a dicentric chromosome, a common product of centromere fission in CIN cells,23 gives rise to two fragments that are promptly segregated by the dividing cell. The fragments thus formed may be repaired in G2 phase after replication (left) and give rise to an inverted repeat that increases the CNum of the region involved but no additional chromosome in spectral karyotyping. In the subsequent division, breakage might occur at another position on the same chromosome arm (red arrows). Alternatively, the breakage product may again capture a telomere (right) and propagate BFB to a healthy chromosome. If a telomere is captured in G1, the fusion product is duplicated in the subsequent S phase and the situation depicted in Figure 5 occurs. In both cases, new dicentric chromosomes are formed and BFB continues. Differential survival of the two daughter cells may result in a non-integer CNum for each product. Horizontal dotted lines indicate cell cycle transitions, the vertical line indicates separated daughter cells.
Limiting BFB to Single Chromosomes
A remarkable feature of chromothripsis is the localization of multiple breaks to single chromosomes or chromosome arms.1,3,27 Whereas some examples undergo translocations between regions of chromothripsis and other chromosomes, other cell lines confine breakage without apparent spreading.1 This observation seems to contradict the general view of BFB, thought to propagate rather than limit chromosomal instability. BFB is thought to propagate breakage because the segregation of chromosome segments in mitosis results in daughter cells containing a reactive DNA end subject to renewed fusion (Fig. 4).21 NHEJ is the preferred repair pathway for breaks generated in mitosis,28 as is the case for BFB, because homologous recombination is suppressed during this part of the cell cycle;29 massive sequencing indicated that non-homologous end joining (NHEJ) indeed is the predominant break repair mechanism in chromothripsis.1
A central role for chromosome fusion in BFB is ascribed to telomeres.30 Telomere structure depends on components of the NHEJ repair machinery31 and can be substrate for DNA end-to-end fusions.32 To convert a telomere to a suitable NHEJ substrate, however, its function must be compromised by genetic inactivation of one or more structural proteins,33 and the actual DNA-joining steps of NHEJ are largely repressed in normal telomeres. Although the molecular and functional analysis of telomeres indicates a dual role in fusion and protection, a survey of clinical samples revealed the paucity of chromosome end-to-end fusions between in carcinomas, responsible for less than 1% of karyotype aberrations.23 NHEJ apparently is sufficiently suppressed in telomeres to delay repair until a more suitable substrate becomes available, since the majority of breaks in carcinomas are processed through self-ligation after replication (Fig. 4, left),23 or form neoacrocentric chromosomes when they acquire a new telomere through the alternative recombination (ALT) pathway.34 Intermediate replication of a chromosome segment, followed by self-ligation, results in an antiparallel orientation of fusion products, evidence of which is abundant in samples of chromothripsis.1 Thus, alternative pathways of repairing a reactive DNA end compete with telomere capture in BFB, and pathways that prevent the involvement of healthy chromosomes seem dominant. Suppression of NHEJ in telomeres might be the key mechanism in the limitation of chromothripsis to single chromosomes.
Although NHEJ is largely suppressed, telomeres occasionally fuse with the isolated breaks generated in BFB (Fig. 4, right), illustrated further by the capture of chromosome arms in clinical samples.23 Several of the chromothripsis samples, too, show repeated translocations between two or more chromosomes.1 Other samples, however, reveal perfect limitation to just one chromosome arm; another mechanism therefore explains confinement of breakage. This latter group includes samples in which two copies of the same affected chromosome are present, suggesting that duplication protects against capture of healthy chromosomes. The presence of two copies of the same chromosome has far-reaching implications for BFB, because their breakage in mitosis produces a pair of reactive ends, instead of a single “half-break,” that form the preferred substrate for NHEJ instead of the semi-protected telomeres.35,36 The doubling of chromothriptic chromosomes thus is an important step to establish a metastable genome (Fig. 5). This metastable state may persist over many divisions and promote further breakage of the same region, or can be brought to an end by alternative competing repair pathways such as ALT telomere acquisition.34 Finally, breakage of each copy in a different but adjacent location greatly facilitates the formation of tandem translocations or internal deletions. In conclusion, the genetic makeup of cell lines undergoing chromothripsis is compatible with a multi-step model involving BFB, in which telomeres are partially shielded from NHEJ-dependent end fusion.
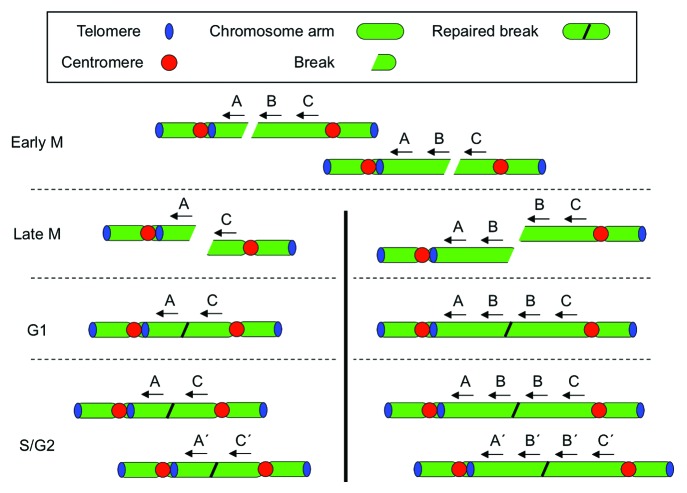
Figure 5. Formation of tandem breaks by aneuploidy. Spectral karyotyping indicates the presence of multiple copies of chromosomes affected by chromothripsis, in addition to the corresponding normal chromosomes.1 Breakage of two dicentric chromosomes in a single round of BFB generates two reactive ends that form the preferred substrate for subsequent fusion by NHEJ. In contrast to an isolated fragment formed after breakage of a single dicentric chromosome (Fig. 4, left), these two fragments can be fused in G1 without intermittent replication but are doubled again in the subsequent S phase. This gives rise to a metastable situation, in which the same chromosomes break over and over again. In addition, the presence of two copies facilitates the formation of internal deletions (left) and tandem duplications (right). Horizontal dotted lines indicate cell cycle transitions; the vertical line indicates separated daughter cells.
Starting BFB in Chromothripsis
A final question that remains to be answered is how BFB is started in chromothripsis if telomeres are largely protected. The nature of dicentric chromosomes leads to distribution of breaks between the two centromeres, since the mitotic spindle exerts pulling forces on these structures.22 Fusion of two chromosomes therefore generates breaks in arms originally part of each chromosome. In a considerable proportion of cases, chromothripsis is restricted to a single chromosome arm and reaches up to the telomere (Fig. 3, lower panel), but no evidence for fusion to a second chromosome was uncovered; this observation undoubtedly has contributed to the single-event model. In the multi-step model supported by the CNum analysis, such restriction means that a single arm is limited by the two centromeres of a dicentric chromosome from the start of BFB. Again, a particular feature of CIN, in this case centromere fission,21,37 provides a possible explanation. Centromere fission—breakage of a chromosome in or adjacent to the centromere—is closely associated with karyotype evolution in CIN tumors23 and can be brought about by mitotic spindle defects such as merotelic attachments37,38 or by cohesion fatigue leading to unscheduled sister chromatid separation.39 Depending on the selection of a fusion partner, the fragments formed by centromere fission—chromosome arms limited by a protective telomere on one extreme and an unprotected half-break on the other extreme—can fuse to the telomere of a receptor chromosome, self-ligate after replication, or acquire a new telomere through the ALT pathway.23 The behavior of centromeric breaks thus shows remarkable parallels to the genetic alterations in chromothripsis.
Importantly, the samples used to describe chromothripsis themselves reveal evidence of centromere fission; not only is chromothripsis regularly limited to a single chromosome arm without evidence for breaks in other chromosomes, also the CNum differences between arms that acquire translocations and adjoining arms show that the centromere is a breakage hotspot.1 In addition, cell lines used to identify chromothripsis show direct signs of centromere fission; whereas the original TK10 cells had a t(3;5) translocation,19 a t(3;9) is identified in the current karyotype.1 A likely scenario includes a dicentric t(3;5) intermediate that broke again and formed the current der(5) chromosomes. Since the chromosome arm that initiates this process (from chromosome 3) is located outside the two centromeres in the dicentric intermediate, it does not suffer breakage itself and goes undetected in massive sequencing.
Although several examples of chromothripsis show a typical limitation of breakage to single chromosome arms, other samples suffer from interchromosomal translocations. These indicate that two arms from different chromosomes are affected, and breakage along the chromosome arms can start BFB cycles in agreement with these examples.24,25 In some cases, translocations accumulate in a sharply defined region along the chromosome arm.1 Possibly, a fragile site40 flanks such regions and provokes BFB in these cases of chromothripsis. In conclusion, whereas the protective effect of telomeres likely is responsible for the limitation of BFB, its initiation might depend on a different mechanism of chromosome breakage.
Concluding Remarks
Somewhat more than a year after its initial description, chromothripsis has wrongly acquired the status of mechanism, although the underlying pathways have yet to be discovered. Due to the lack of explanations, the single-event model based solely on computer simulation has remained unchallenged even though it has met with criticism.10 Whereas the single event model based on computer simulation is not supported by the observed data, a sequential model explains most if not all features of chromothripsis. The classical features of CIN themselves might not justify the massive number of breaks, but the close relation between BFB and CIN21,23 provides a plausible basis for the major part of the data observed. Particular phenomena associated with CIN, for example, centromere fission, might at least partially explain initiation of BFB, and acquisition of additional copies of chromothriptic chromosomes helps to prevent spreading of BFB by providing substrates for NHEJ. Finally, the dicentric chromosomes associated with BFB are highly unstable; the low percentage of chromothripsis in cancer cell lines might therefore reflect the instability of dicentric chromosomes instead of a separate carcinogenic mechanism.
A hitherto ignored feature of the chromothriptic genome regions, heterogeneity, might be brought about by the differential survival of daughter cells that previously inherited different combinations of BFB products. Importantly, the presence of two metastable chromosome populations in separate sets of daughter cells is compatible with the supposed, but neither true nor complete, “two-state” mode of genetic inheritance. In conclusion, a combination of BFB and tumor heterogeneity provides a likely explanation for the genetic signature that characterizes chromothripsis. Possibly, a combination of established pathways might clarify novel complex genetic patterns in the future, eliminating the need for additional terminology.
Acknowledgments
The authors thank Catherine Mark for editorial assistance. This work is financed by grants PS09/00572 (Fondo de Investigación en Salud) and BFU2009-08395E, (Ministerio de Economía y Competitividad) to K.v.W., and grants S2010/BMD2502 (Comunidad Autónoma de Madrid) and SAF2010-21205 (Ministerio de Economía y Competitividad) to C.M.A. K.v.W. is supported by a JAE-doc fellowship from the Spanish National Research Council (CSIC). The Department of Immunology and Oncology was founded and is supported by the CSIC and by Pfizer.
Glossary
Abbreviations:
- ALT
alternative pathway of telomere maintenance
- BFB
breakage-fusion-bridge
- CIN
chromosomal instability
- CNum
copy number
- NHEJ
non-homologous end joining
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25266
References
- 1.Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyerson M, Pellman D. Cancer genomes evolve by pulverizing single chromosomes. Cell. 2011;144:9–10. doi: 10.1016/j.cell.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 3.Kloosterman WP, Hoogstraat M, Paling O, Tavakoli-Yaraki M, Renkens I, Vermaat JS, et al. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol. 2011;12:R103. doi: 10.1186/gb-2011-12-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu C, Wyatt AW, McPherson A, Lin D, McConeghy BJ, Mo F, et al. Poly-gene fusion transcripts and chromothripsis in prostate cancer. Genes Chromosomes Cancer. 2012;51:1144–53. doi: 10.1002/gcc.21999. [DOI] [PubMed] [Google Scholar]
- 5.Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet. 2011;20:1916–24. doi: 10.1093/hmg/ddr073. [DOI] [PubMed] [Google Scholar]
- 6.Chiang C, Jacobsen JC, Ernst C, Hanscom C, Heilbut A, Blumenthal I, et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet. 2012;44(S1):390–7, S1. doi: 10.1038/ng.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lapuk AV, Wu C, Wyatt AW, McPherson A, McConeghy BJ, Brahmbhatt S, et al. From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer. J Pathol. 2012;227:286–97. doi: 10.1002/path.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maher CA, Wilson RK. Chromothripsis and human disease: piecing together the shattering process. Cell. 2012;148:29–32. doi: 10.1016/j.cell.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer. 2012;12:663–70. doi: 10.1038/nrc3352. [DOI] [PubMed] [Google Scholar]
- 10.Righolt C, Mai S. Shattered and stitched chromosomes-chromothripsis and chromoanasynthesis-manifestations of a new chromosome crisis? Genes Chromosomes Cancer. 2012;51:975–81. doi: 10.1002/gcc.21981. [DOI] [PubMed] [Google Scholar]
- 11.Rausch T, Jones DT, Zapatka M, Stütz AM, Zichner T, Weischenfeldt J, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 13.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–36. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 14.Walker PR, Sikorska M. Endonuclease activities, chromatin structure, and DNA degradation in apoptosis. Biochem Cell Biol. 1994;72:615–23. doi: 10.1139/o94-081. [DOI] [PubMed] [Google Scholar]
- 15.Youds JL, Boulton SJ. The choice in meiosis - defining the factors that influence crossover or non-crossover formation. J Cell Sci. 2011;124:501–13. doi: 10.1242/jcs.074427. [DOI] [PubMed] [Google Scholar]
- 16.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–6. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–8. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bear A, Clayman RV, Elbers J, Limas C, Wang N, Stone K, et al. Characterization of two human cell lines (TK-10, TK-164) of renal cell cancer. Cancer Res. 1987;47:3856–62. [PubMed] [Google Scholar]
- 20.Ito T, Seyama T, Mizuno T, Tsuyama N, Hayashi T, Hayashi Y, et al. Unique association of p53 mutations with undifferentiated but not with differentiated carcinomas of the thyroid gland. Cancer Res. 1992;52:1369–71. [PubMed] [Google Scholar]
- 21.Martínez-A C, van Wely KH. Are aneuploidy and chromosome breakage caused by a CINgle mechanism? Cell Cycle. 2010;9:2275–80. doi: 10.4161/cc.9.12.11865. [DOI] [PubMed] [Google Scholar]
- 22.McClintock B. The Stability of Broken Ends of Chromosomes in Zea Mays. Genetics. 1941;26:234–82. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martínez-A C, van Wely KH. Centromere fission, not telomere erosion, triggers chromosomal instability in human carcinomas. Carcinogenesis. 2011;32:796–803. doi: 10.1093/carcin/bgr069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciullo M, Debily MA, Rozier L, Autiero M, Billault A, Mayau V, et al. Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum Mol Genet. 2002;11:2887–94. doi: 10.1093/hmg/11.23.2887. [DOI] [PubMed] [Google Scholar]
- 25.Lo AW, Sabatier L, Fouladi B, Pottier G, Ricoul M, Murnane JP. DNA amplification by breakage/fusion/bridge cycles initiated by spontaneous telomere loss in a human cancer cell line. Neoplasia. 2002;4:531–8. doi: 10.1038/sj.neo.7900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pobiega S, Marcand S. Dicentric breakage at telomere fusions. Genes Dev. 2010;24:720–33. doi: 10.1101/gad.571510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu P, Erez A, Nagamani SC, Dhar SU, Kołodziejska KE, Dharmadhikari AV, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guerrero AA, Martínez-A C, van Wely KH. Merotelic attachments and non-homologous end joining are the basis of chromosomal instability. Cell Div. 2010;5:13. doi: 10.1186/1747-1028-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothkamm K, Krüger I, Thompson LH, Löbrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–15. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung AL, Deng W. Telomere dysfunction, genome instability and cancer. Front Biosci. 2008;13:2075–90. doi: 10.2741/2825. [DOI] [PubMed] [Google Scholar]
- 31.Boulton SJ, Jackson SP. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–28. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samper E, Goytisolo FA, Slijepcevic P, van Buul PP, Blasco MA. Mammalian Ku86 protein prevents telomeric fusions independently of the length of TTAGGG repeats and the G-strand overhang. EMBO Rep. 2000;1:244–52. doi: 10.1093/embo-reports/kvd051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–7. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gagos S, Chiourea M, Christodoulidou A, Apostolou E, Raftopoulou C, Deustch S, et al. Pericentromeric instability and spontaneous emergence of human neoacrocentric and minute chromosomes in the alternative pathway of telomere lengthening. Cancer Res. 2008;68:8146–55. doi: 10.1158/0008-5472.CAN-08-0945. [DOI] [PubMed] [Google Scholar]
- 35.Agarwal S, Tafel AA, Kanaar R. DNA double-strand break repair and chromosome translocations. DNA Repair (Amst) 2006;5:1075–81. doi: 10.1016/j.dnarep.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 36.Bernstein C, Bernstein H, Payne CM, Garewal H. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis. Mutat Res. 2002;511:145–78. doi: 10.1016/S1383-5742(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 37.Guerrero AA, Gamero MC, Trachana V, Fütterer A, Pacios-Bras C, Díaz-Concha NP, et al. Centromere-localized breaks indicate the generation of DNA damage by the mitotic spindle. Proc Natl Acad Sci USA. 2010;107:4159–64. doi: 10.1073/pnas.0912143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci. 2003;116:4213–25. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- 39.Daum JR, Potapova TA, Sivakumar S, Daniel JJ, Flynn JN, Rankin S, et al. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr Biol. 2011;21:1018–24. doi: 10.1016/j.cub.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barlow JH, Faryabi RB, Callén E, Wong N, Malhowski A, Chen HT, et al. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152:620–32. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]