Abstract
It is now clear that animal genomes are predominantly non-protein-coding, and that these sequences encode a wide array of RNA transcripts and other regulatory elements that are fundamental to the development of complex life. We have previously argued that the proportion of an animal genome that is non-protein-coding DNA (ncDNA) correlates well with its apparent biological complexity. Here we extend on that work and, using data from a total of 1,627 prokaryotic and 153 eukaryotic complete and annotated genomes, show that the proportion of ncDNA per haploid genome is significantly positively correlated with a previously published proxy of biological complexity, the number of distinct cell types. This is in contrast to the amount of the genome that encodes proteins, which we show is essentially unchanged across Metazoa. Furthermore, using a total of 179 RNA-seq data sets from nematode (47), fruit fly (72), zebrafish (20) and human (42), we show, consistent with other recent reports, that the vast majority of ncDNA in animals is transcribed. This includes more than 60 human loci previously considered “gene deserts,” many of which are expressed tissue-specifically and associated with previously reported GWAS SNPs. These results suggest that ncDNA, and the ncRNAs encoded within it, may be intimately involved in the evolution, maintenance and development of complex life.
Keywords: noncoding RNA, non-coding DNA, evolution, complexity, lincRNA, small RNA
Background
Until relatively recently, it was widely accepted that at least 95% of the mammalian genome was non-functional or evolutionarily redundant. Historically, this notion appears to stem largely from two unexpected findings of the 1960s and 1970s: (1) that the majority of most complex metazoan genomes are composed of repetitive elements of diverse origin; and (2) that unlike prokaryotic genomes, eukaryotic genomes contain large amounts of DNA between protein-coding exons (i.e., introns) and between protein-coding genes themselves (i.e., intergenic gene deserts) relegating protein-coding sequences to ~3% of the genome. Despite work from the first half of the 20th century from McClintock and others that showed that repetitive elements could act as control elements, which were specifically activated at particular developmental time points,1,2 and early suggestions that introns and intergenic spaces could in-principle house suites of regulatory elements,3 relatively little research focused on non-genic elements.
Intriguingly, there is renewed and increasing support for the view that non-protein-coding regions may be of particular evolutionary importance. Indeed, one of the primary conclusions of the ENCODE project is that, at varying levels of significance, a large proportion of the human genome is functional.4-8 Additionally, extending on McClintock’s work, it is now clear that the transposon-derived and other repetitive elements that make up nearly 50% of the mammalian genome have been co-opted into functional roles in a variety of cellular and developmental contexts. For example, they are expressed in a variety of different tissues and conditions,9 are transcribed tissue-specifically10 and can serve as alternative upstream promoters that show specific and regulated deposition of particular epigenetic mark.10,11 They can also be site-specifically methylated9 and can function as binding sites of CTCF, a protein known as the “master weaver” of the genome, that frequently serves an epigenetic boundary element.12,13 Indeed, a B1 SINE element subtype, B1-X35S, which is present in more than 14,000 copies in the mouse genome, mediates epigenetic insulation by binding of the transcription factors dioxin receptor (AHR) and SLUG (SNAI2).13 Repetitive elements are also associated with the biology of reproduction,14 have been causally associated with the evolution of pregnancy in placental mammals15and show high levels exaptation by exonization in all mammals.16
It is also now clear that the metazoan genome produces a wide array of RNA species, most of which are derived from the tracts of intergenic and intronic DNA that have no protein-coding capacity. Even for well-described protein-coding genes, it is now well-accepted that that any given locus produces a complex array of overlapping transcripts with different splicing patterns, including antisense transcripts, many of which are further processed and capped,17 variously described as an interlaced architecture,18 and islands of protein-coding information in sea of cis- and trans-acting regulatory RNA information.19 Transcription in eukaryotes, including complex animals, moreover, extends far beyond traditional protein-coding genes, mostly as long non-protein-coding RNAs, including the well-described lincRNAs,20-22 polyadenylated long non-coding RNAs, which are frequently oriented bidirectional to coding genes,23 and a host of nuclear-specific transcripts with an unknown function.18 Furthermore, a recent study that combined tiling arrays with high-throughput RNA sequencing revealed extensive previously undetected transcription around the p53 and HOX genes and throughout unannotated intergenic regions.24 These long ncRNAs are further complemented by an ever-increasing catalog of small RNAs, including microRNAs and other PIWI- and Argonaute-assocaited RNAs,25,26 small RNAs derived from transcription start sites (e.g., tiRNAs)27,28 and splice sites (spliRNAs)27 as well as species derived from structural or housekeeping RNAs (e.g., snoRNA-derived RNAs and tRNA-derived RNAs).29-32
We have previously argued that, in contrast to gene number, the proportion of genomic DNA that is non-protein-coding (ncDNA) shows a strong correlation with apparent biological complexity.33,34 In light of the increasing evidence that non-protein-coding sequences encode functional elements (e.g., repeat elements co-opted into regulatory roles or ncRNAs), this relationship warrants further investigation. Here, we extend our previous analysis to more than 1,500 prokaryotic species and 150 multicellular organisms. Consistent with our previous work, our analysis is focused on haploid genome composition, thus removing the confounding factor of ploidy or the contaminating DNA of prey, which are likely to be the primary cause of the large genome sizes attributed to lungfish and amoeba, respectively (reviewed in ref. 34). To attempt to further remove any ambiguity associated with the phrase “biological complexity,” the analysis described below uses a previously published metric for organismal complexity,35 which is itself based on a number of previous studies that concluded complexity is best approximated by the number of different cell types.36-39 We also perform an RNA-seq meta-analysis across four multicellular organisms and find, consistent with expectation, that the majority of the non-protein-coding regions of these genomes are transcribed. Taken together, these data suggest that there is a close relationship between the expansion of ncDNA in higher organisms and organismal complexity.
Results
Protein-coding gene sets show little variation over long evolutionary time frames
One of the biggest surprises to come from the recent genome-sequencing projects is the apparent lack of correlation between the number of protein-coding genes and biological complexity, which is sometimes referred to as the G-value paradox.40 To investigate this relationship, we collected genome annotation data from 1,627 prokaryotic species and 153 multicellular organisms (see “Materials and Methods” and Table 1; Table S1), and then partnered this with a recently published metric of 73 organisms that uses the number of distinct cell types as a proxy for organismal complexity35 (Table S1). Consistent with previous reports, we find that gene number and biological complexity in multicellular animals are not correlated. For example, the fruit fly Drosophila melanogaster, which has at least 64 distinct cell types (Table S1), has 16,000 protein-coding genes,41 while the considerably less complex Caenorhabditis elegans has only 28 distinct cell types35 (Table S1). Indeed, within the eukaryotic lineage, the species with highest number of annotated genes are unicellular amoeba and protists: Paramecium tetraurelia has nearly 40,000 protein-coding genes,42 and Trypanosoma cruzi and Tetrahymena thermophila are predicted to have 22,57043 and 27,000,44 respectively (Table S1). In multicellular animals, most have ~20,000 genes including the fish Takifugu rubripes,45 chicken (Gallus gallus),46 mouse (Mus musculus),47 gorilla (Gorilla gorilla)48 and humans47,49 (Table S1). A systematic examination of the number of genes across multicellular animals revealed that there is no significant difference in gene number between vertebrates, which have a minimum 100 distinct cell types, and invertebrates, with ~50 cell types (Table 1; Table S1, two-tailed p = 0.756, Mann-Whitney U test), consistent with the hypothesis that gene number does not scale with organismal complexity.
Table 1. Gene number across fungi, protists and Metazoa.
| Taxa (number of species) | Minimum | Median | Mean | Maximum |
|---|---|---|---|---|
| Unicellular fungi (15) |
1,996 |
6,516 |
6,489 |
13,286 |
| Multicelluar fungi (45) |
5,120 |
10,980 |
11,414 |
20,548 |
| Protists (23) |
3,396 |
8,920 |
12,418 |
39,642 |
| Invertebrates (26) |
10,685 |
19,870 |
20,773 |
33,925 |
| Vertebrates (24) | 13,756 | 19,924 | 21,398 | 38,612 |
Next, we investigated levels of homology between animal protein-coding gene sets to assess if the relative constancy in gene number was indicative of similarly equivalent proteomes. We systematically queried the literature and Homologene,50 and found that for eight representative metazoan species, the majority of genes were homologous (Table 2). For example, 63% and 85.7% of Amphimedon queenslandica (basal marine sponge) and Mus musculus (common mouse) genes have identifiable orthologs (Table 2), indicating that the core protein-coding componentry of complex animals may have been present since the dawn of multicellularity and, despite lineage-specific expansions of particular gene families and innovations, has not changed appreciably despite the development of more complex body plans. This is consistent with the universal genome hypothesis, which posits that an ancestral and basal genome that encodes all major developmental programs essential for various phyla of Metazoa emerged in a unicellular or a primitive multicellular organism shortly before the Cambrian period.51 Interesetingly, A. queenslandica expresses not only members of the Wnt and TGF-β signaling pathway, but also the Notch-Delta signaling system and a proneural basic helix loop helix (bHLH) gene that resembles the conserved molecular mechanisms of primary neurogenesis in bilaterians.52,53
Table 2. Gene homology in Metazoa.
| Species | Number of protein-coding genes | Number of protein-coding genes with homologs (%) | Source |
|---|---|---|---|
|
A. queenslandica |
30,060 |
18,693 (62.2) |
ref. 53 |
|
D. pulex |
30,907 |
19,641 (63.5) |
ref. 74 |
|
C. elegans |
20,132 |
8,678 (43.1) |
* |
|
T. urticae |
18,414 |
11,805 (64.1) |
ref. 75 |
|
D. melanogaster |
13,827 |
9,282 (67.1) |
* |
|
D. rerio |
26,690 |
21,084 (79.0) |
* |
|
T. guttata |
18,581 |
14,527 (78.2) |
ref. 76 |
| M. musculus | 25,388 | 21,766 (85.7) | * |
Values obtained from HomoloGene Release 65.50
Gene number and homology, however, may belie a complexity contained with the protein-coding repertoire of a genome that is only evident when examined at the level of encoded amino acids. It has been recently argued that examining the total number of amino acids encoded by all transcript isoforms shows a positive correlation with biological complexity.35 This analysis, however, yields a poor correlation (R2 = 0.1333) and is biased by the relative depth of the annotated proteome for a given species. For example, in this previous work, human was assessed to have a proteome more than 106 amino acids larger than mouse, which, given that these species have nearly an identical number of cell types (see ref. 35; Table S1), is likely to reflect differences in depth of polling, not biology. For example, the EMBL-EBI Proteomics Identification Database (PRIDE)54 reports 4,457 experimental entries for Homo sapiens, but only 773 for Mus musculus.
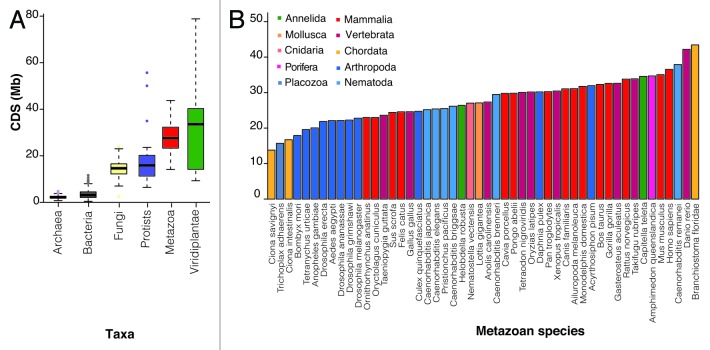
To attempt to accurately assess how much of the genome is protein-coding, we directly examined the number of non-redundant bases that are ever engaged in a coding sequence (CDS), which will capture the genomic regions associated with all annotated splice isoforms, across 1,627, prokaryotic and 153 eukaryotic complete and annotated genomes (see “Materials and Methods”; Table S1). Investigation of 10 prior mouse and 12 human genome builds revealed that the number of annotated coding bases has changed relatively little in recent years (Fig. S1), suggesting that this metric is likely to be both robust and representative of the biology of the system. Consistent with gene number annotations, we observe increases in Mb of coding sequence between prokaryotes and eukaryotes, but no subsequent correlation with organismal complexity among multicellular eukaryotes (Fig. 1A). We observed that within the Metazoa there is generally little variation in the overall amount of the genome devoted to CDS, and that there is no relationship between Mb of CDS and biological complexity (Fig. 1B).
Figure 1. Protein-coding sequence (CDS) across taxa and a subset of metazoan species. (A) Total protein-coding sequence (CDS) across major taxa. (B) CDS across well-annotated metazoan species. Note that among metazoan there is little divergence in the amount of total amount of genomic sequence devoted to generating protein-coding genes.
Biological complexity and the nc/tg ratio
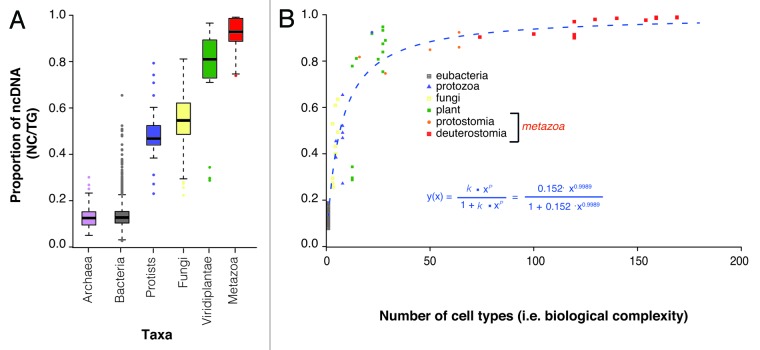
We have previously shown that there is a correlation between biological complexity and the amount of the genome that is non-protein-coding,33,34 calculated by taking all genomic bases that are only ever non-protein-coding and dividing by total halploid genome size (nc/tg).34 Here, we extended our prior work to the 1,627 prokaryotic and 153 eukaryotic genomes described above and found a clear correlation between the nc/tg ratio and increasing complex taxonomic groups (p < 2.2e-1.6, Kruskal-Wallis test, Fig. 2A). The range of nc/tg values is considerable, with the averages for archaea and bacteria being nearly identical (two-tailed p = 0.359, Mann-Whitney U test) at 0.130 and 0.136, respectively, and extending to ~0.98 in the Metazoa. The average value for each taxa is minimally influenced by data points outside the first or third quartiles. For example, there are less than 50 bacterial species, of the more than 1,500 surveyed, with nc/tg values greater than the maximum of the third quartile, 0.25, and the majority of these are species in evolutionary transition. This includes Mycobacterium leprae, which has an nc/tg value of 0.50, which is driven by the loss of functional protein-coding genes due to its endosymbiotic lifestyle.55
Figure 2. Non-protein-coding DNA content across taxa and its association with organismal complexity. (A) The proportion of non-protin-coding DNA per total haploid genome (nc/tg ratio) across taxa. (B) The nc/tg ratio values as a function of the distinct number of cell types, a proxy of biological complexity. The best fit curve, modified Hill’s equation, which itself is a logistic function, is given in blue text.
In contrast to gene number or bases of coding sequence, we observed statistically significant nc/tg ratio differences within and between multicellular taxonomic groups. For example unicellular and multicellular fungi have average nc/tg ratios of 0.399 and 0.585 (two-tailed p = 6.7e-08, Mann-Whitney U test); invertebrates and vertebrates have average values of 0.873 and 0.976 (two-tailed, p = 1.5e-10, Mann-Whitney U test); and mammals show the highest the average nc/tg ratio of 0.989, which is significantly higher than the vertebrate average (one-tailed, p = 7.6e-07, Mann-Whitney U test).
To further refine the association of nc/tg ratio values and organismal complexity, we investigated the 73 species with a previously defined number of cell types.35 Examining these species revealed a positive correlation between the nc/tg ratio and organismal complexity (Fig. 2B, Spearman correlation coefficient r = 0.952, p value < 0.0001). We found that the distribution of values was well described by a modified Hill’s equation56 (which is itself a modified logistic function, see “Discussion”), in the form y = Kxn/(1+Kxn) where K = 0.15219 ± 0.02272 with a p value < 0.0001 and n = 0.99888 ± 0.06943 with a p value < 0.0001 (Fig. 2B). This distribution is consistent with patterns observed in complex information systems theory, in which the amount of encoded information approaches an asymptote defined by the maximum allowable entropy (see “Discussion”).
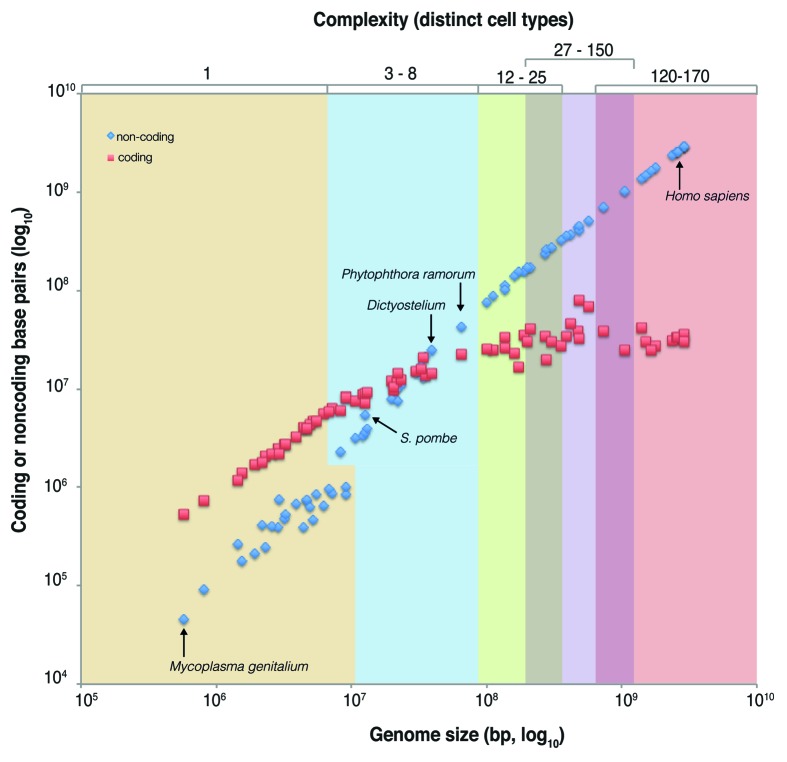
To investigate the relationship between both coding and non-protein-coding sequence and organismal complexity simultaneously, we examined their contributions to the total genome size of species with defined cell numbers (Fig. 3). The data show that increasing genome size in prokaryotes is associated with a corresponding, and nearly exponential, increase in protein-coding bases, which extends to basal multicellular eukaryotes, but then asymptotes for all complex multicellular animals (Fig. 3, red data points). This is consistent with the data reported above, showing that both gene number and the amount of CDS is relatively static across multicellular animal genomes. In contrast, the amount of the genome that is non-protein-coding grows exponentially in correlation with biological complexity (Fig. 3, blue data points). Intriguingly, the intersection of the coding and non-protein-coding series occurs at data points associated with simple multicellular organisms, i.e., between the data points for S. pombe and Dictyostelium, which supports the hypothesis that elements associated and embedded within ncDNA may facilitate increased organismal complexity.
Figure 3. The relationship between biological complexity and genome composition. In this plot, the 73 organisms with a previously defined number of distinct cell types (e.g., relative biological complexity, see Table S1; ref. 35) are shown as pairs of data points, with one depicting total protein-coding sequence bases (red) and one total non-protein-coding bases (blue) which cumulatively give the total genome size (x-axis). Non-protein-coding sequence increases exponentially with the number of distinct cell types, while protein-coding sequence is asymptotic. Note that the intersection of the protein-coding and non-protein-coding data sets occurs among simple multicellular organisms.
The extent of genomic transcription in four animals
We have previously postulated that one of the primary roles of ncDNA may be to produce regulatory RNAs, many of which may act in both cis- and trans- to modulate epigenetic states and control protein-coding gene expression.19,34,57,58 To assess if animal genomes are indeed all widely transcribed, we performed a meta-analysis of RNA-Seq data sets from four organisms: Caenorhabditis elegans (47 data sets), Drosophila melanogaster (72 data sets), Danio reiro (20 data sets) and Homo sapiens (42 data sets, Table 3; Table S2). By combining data sets from multiple sources, we were able to achieve high levels of coverage. For example, the total sequencing depth of the C. elegans RNA-seq data was equivalent to 668-fold genomic coverage (Table 3). The majority of the data sets examined were polyA+ enriched, i.e., they represented RNA sequencing data from polyadenylated and canonical mRNAs, and were therefore amenable to de novo transcript assembly using the Tophat-Cufflinks pipeline.59 Each data set was individually mapped with Tophat and assembled into de novo transcripts with Cufflinks and then merged (see “Materials and Methods”; assembled transcriptomes are available for visualization). This revealed that even though our analysis was limited to polyA+ transcripts, at least 80% of the C. elegans and ~60% of the Drosophila, D. rerio and H. sapiens genomes were transcribed, the vast majority of which is non-protein coding.
Table 3. RNA-seq coverage in four metazoan species.
| Species | Genome size (Mb) | CDS (Mb, % of genome) | RNA-seq data sets queried | Total RNA-seq (Gb, fold genome coverage) | Exonic coverage (Mb, % of genome)* | Transcriptomic coverage (% of genome)§ |
|---|---|---|---|---|---|---|
|
C. elegans |
100.28 |
25.40 (25.33) |
47 |
67.05 (668) |
45.38 (45.25) |
80.40 (80.17) |
|
D. melanogaster |
162.37 |
22.78 (14.03) |
72 |
103.92 (640) |
49.61 (30.56) |
98.75 (60.82) |
|
D. rerio |
1409.77 |
42.19 (2.99) |
20 |
154.47 (109) |
142.21 (10.09) |
894.72 (63.47) |
| H. sapiens | 2897.32 | 36.53 (1.26) | 42 | 353.69 (122) | 798.31 (27.554) | 1631.10 (56.29) 2034.94 (70.24)1 2253.78 (77.78)2 |
Exonic coverage is limited to regions defined as such in de-novo transcriptome builds derived from the Tophat-Cufflinks pipeline,59 as described in the “Materials and Methods.” §Transciptomic coverage includes both processed exons and the introns that join them. For H. sapiens, two additional coverage values are given.(1) RNA-seq coverage of all mapped tags that fall into a cluster of at least 16 mapped reads, and (2) all mapped tags across all data sets.
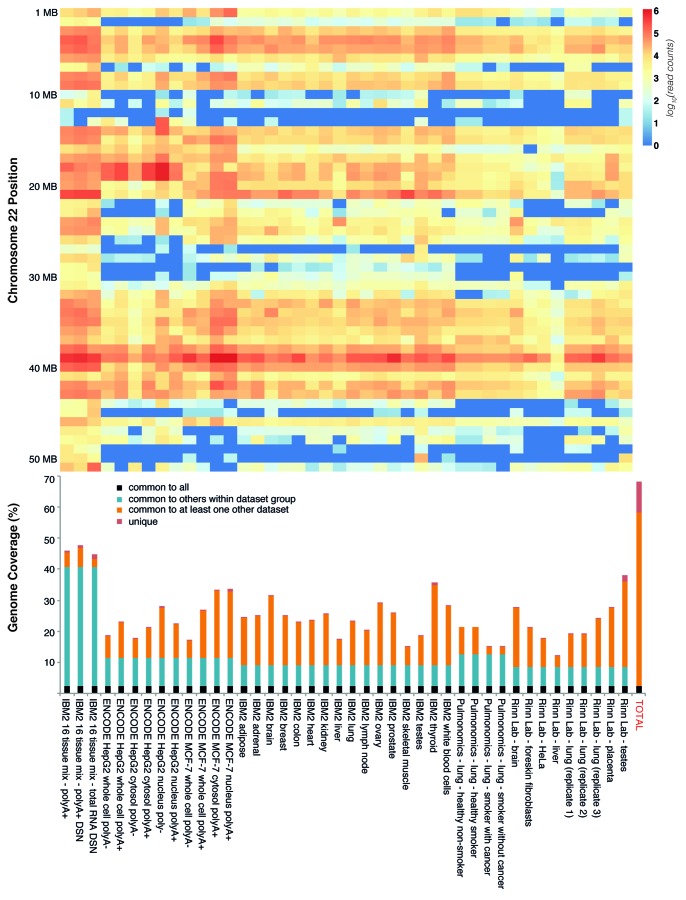
To gain additional insight into the extent of transcription in animals, we focused our subsequent analysis on H. sapiens due to the fact that it is one of the most ncDNA-dense genomes and has data available from 16 primary tissues and polyA-enriched expression normalized data sets (Table S2). The Tophat-Cufflinks pipeline has difficulty assembling transcripts that do not fit the standard statistical models consistent with traditional mRNAs. We therefore expanded our analysis and examined the overall coverage all of all mapped tags, and those falling into clusters of greater than 2, 4, 8 and 16 overlapping reads (Table S2). This revealed that if the uniquely mapped RNA-seq reads from all 42 data sets are considered (including those that are spliced), as much as 77% of the human genome is transcribed, which is only reduced to 70.24% if the analysis is restricted to clusters with at least 17 overlapping reads and Tophat-mapped spliced tags (Table 3 and Fig. 4). As little as 2.3% of this transcription is shared across all data sets, indicating that there is vast repertoire of tissue- and cell-specific transcription (Fig. 4B), and therefore that the extent of transcription we have presented here is likely a minimum value. Indeed, comparison of the MCF-7 and HepG2 ENCODE data sets revealed that ~147 Mb of transcribed bases (5.09% of the genome) were detected uniquely in polyA- prepared libraries.
Figure 4. Investigation of the extent of transcription in the human genome across 42 RNA-seq data sets. In the top (A) heatmap of RNA-seq expression is shown across chromosome 22 in 1 megabase bins, with the intensity displayed as a spectrum from log10(0) (blue) to log10(6) (red). The bottom panel shows total genomic coverage of each RNA-seq data set, which is derived from the tag clusters with at least 16 independent and overlapping reads plus Tophat mapped junctions with an anchor of at least 20 bases. Bar colors in the bottom panel are indicative of regions of the genome that are covered in all data sets (black, ~2.3%), those that are present in all members of a data set group (blue, i.e., the data sets were derived from the source), the proportion shared with another data set not in the data group (organge), and the proportion of genomic coverage that is unique to a particular data set (red). Note that in both the top and bottom panels the IBM2 16 tissue mixed data sets show the greatest extent and relative intensity of RNA-seq expression. Please see Supplemental Material for heatmaps of all 42 data sets across all human chromosomes.
We found that three of the 42 data sets, those generated by Illumina Research and Development (Table S2; available through the EBI’s ArrayExpress), were responsible for the majority of novel transcription in our meta-analysis. These data sets were generated as part of the Illumina Body Map 2 (IBM2) initiative and are derived from RNA from a mix of 16 tissues. They differ, however, in their preparation. While one was prepared using a traditional polyA selection (abbreviated for this work as 16-mRNA), two were prepared from mRNA and total RNA from 16 tissues that was then normalized using a duplex-specific nuclease (16-mDSN and 16-tDSN, respectively), a protocol that takes advantage of renaturation kinetics to preferentially degrade highly expressed transcripts and thereby facilitate sequencing of lowly expressed RNAs.60 When we examined expression across all chromosomes in 1 Mb bins, we found that these data sets consistently displayed the highest levels of transcriptional coverage, with the 16-tDSN library frequently showing expression in regions that were either not detected elsewhere or restricted to brain and testes (Fig. 4A and Supplemental Material). Indeed, excluding all spliced tags, the uniquely mapped and clustered 16-tDSN data alone covered more than 32% of the human genome.
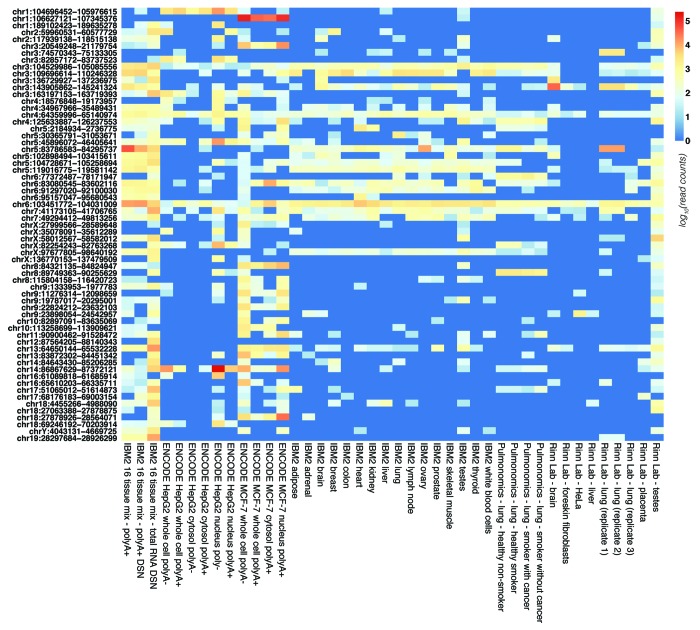
To investigate if the DSN libraries might shed further light on regions that have gone previously unannotated, we identified 178 gene deserts using published metrics (see “Materials and Methods”) and intersected them with clustered reads from all 42 RNA-seq libraries. We found that 63 of these regions were expressed, i.e., they had at least 1,000 RNA-seq tags in at least one library, with the 16-tDSN data set showing the most widespread and robust expression (Fig. 5). Intriguingly, and consistent with the data presented above indicating that a substantial portion of mammalian transcription is cell type- or condition-dependent, we also found cell line- and tissue-specific expression of particular gene deserts, including regions preferentially expressed in brain and testes (Fig. 5). To evaluate if this transcription might be biologically meaningful, we intersected all expressed gene deserts with the curated NHGRI GWAS SNP data61 and found that 42 overlapped with GWAS SNPs associated with phenotypes spanning from brain structure to prostate cancer (Table S4). For example, we found robust expression on a chromosome 17 gene desert in brain, liver and the 16-tDSN library showing two distinct and tissue specific clusters neighboring SNPs associated with thyrotoxic hypokalemic periodic paralysis, pediatric eosinophilic esophagitis, QT interval, sudden cardiac arrest and formal thought disorder in schizophrenia (Fig. S2A and Table S4). Likewise, we identified a region on chromosome 8 that is highly expressed in two independent brain data sets, shows continuous transcription of > 300 kb in the 16-DSN library and is associated with a GWAS SNP connected to schizophrenia (Fig. S2B and Table S4). These results indicate that previously enigmatic “intergenic” GWAS SNPs may be associated with RNA transcripts that have gone previously undetected and unannotated, many of which may act in trans.62
Figure 5. Heatmap of transcription across gene deserts. The relative expression of each of the 63 gene deserts with at least 1,000 RNA-seq read counts (from a single library) is shown for each of the 42 human RNA-seq data sets surveyed. Read intensity is scaled in log10 from 0 (blue) to greater than 5 (red). The IBM2 16 tissue mix total RNA DSN (16-tDSN) library reveals high levels of transcription across the vast majority of gene deserts.
Discussion
Here we have shown that: (1) the number of protein-coding genes and bases does not scale with biological complexity and is in fact relatively static across all multicellular animal lineages; (2) that there is a strong and statistically significant correlation between the proportion of the genome that is non-protein-coding and organismal complexity; and (3) that a meta-analysis of more than 170 RNA-seq data sets has revealed, consistent with other studies,4-8 that the vast majority of multicellular animal genomes are transcribed. Taken together, these findings suggest that non-protein-coding sequences house a set of information-rich instructions, many of which are likely to be regulatory in nature and transacted as RNA species, which have facilitated the emergence of biological complexity.19,57,58 Indeed, work on marine sponge and other basal metazoan species has shown, consistent with the universal genome hypothesis,51 that the protein-coding repertoire has changed little since the dawn of multicellularity,52,53 suggesting that there is a substantial problem of “missing information” that can be resolved if non-protein-coding sequences are as information rich as the work here suggests.
We note that the line of best fit for the data describing the correlation between biological complexity and the nc/tg ratio is a derivation of a Hill’s equation,56 i.e., a standard biochemical metric by which to assess the saturation of binding sites in a given protein, y = Kxn/(1+Kxn), which is itself a modified logistic function (Fig. 2B). These functions are used to describe a wide range of phenomena, including artificial neural networks, chemical reaction models and tumor growth and can also be employed to assess the entropic, and therefore information, content of a given system.63 If we assume that: (1) any given genome is a store of transactable information; (2) that genomes from multiple species that are dispersed across a spectrum (as in Fig. 2B) are representative of a semi-continuous distribution of complex information systems; and (3) that in animals, protein-coding genes are a storehouse of largely unchangeable effector molecules that are regulated by a suite of increasingly complex set of instructions embedded within non-protein-coding DNA sequences then, like many complex systems, the asymptote approached by our line of best fit may represent the boundary between maximum information content and complete disorder.64,65 Put more simply, an nc/tg ratio of 1.0 would represent a genome replete with information but with no effector agents, while a genome with a nc/tg ratio of 0.0 would be packed with effector molecules with little associated regulatory information. This model, in conjunction with the other results presented here, would suggest that the mammalian genome is nearly maximized with regulatory information bound within ncDNA.
Materials and Methods
Bioinformatics resources
All bioinformatic analyses were performed on either the Queensland Cyber Infrastructure Foundation’s High Performance Computing cluster (barrine, www.qcif.edu.au/about-us), or on the Genomics Virtual Laboratory system (www.nectar.org.au/genomics-virtual-laboratory-0). On each system we made use of the suite of backend tools available through a local mirror of the UCSC Genome Browswer,66 the Hanon Lab’s Fastx Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), Tophat and Cufflinks59,67,68 (see more below), Bam Tools (http://sourceforge.net/projects/bamtools/), BedTools,69 FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and a suite of in-house developed Perl, Python, Awk and Shell scripts. Additional details are available upon request.
Genome composition analysis
To ensure that the genomic composition analysis was robust, we restricted the analysis to well-annotated and assembled genomes. In total we interrogated 111 archaea, 1,516 bacterial, 60 fungi, 23 protists, 20 viridiplantae (i.e., green plants) and 50 metazoa genomes (for a complete catalog please see Table S1). The archaeal and bacterial genome annotation data sets were obtained from IMG (Version 3.5).70 Fungi, protist and viridiplantae data were obtained from Ensemble (Release 12)71 and/or directly from the Joint Genome Institute’s website (http://genome.jgi.doe.gov/). Metazoan genome annotations were obtained from three sources depending on the organism of interest: (1) directly from the from UCSC genome browser (v 243); (2) from the Ensemble genome browser (Release 12); or (3) directly from references detailing the genomic composition of a given species (see Table S1). For example, there is no comprehensive database for the marine sponge (i.e., it is not hosted by either Ensemble or UCSC), so the relevant genome information was taken from the literature (see “Results,” above).
In the vast majority of cases, protein-coding gene annotations were obtained from the same sources as the genomic data, although the details of the annotations sets sometimes differed (see Table S1). For example, the coding sequences (CDS) of all archaeal and bacterial species were obtained from IMG; however, the CDSs of Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, Danio rerio, Mus musculus and Homo sapiens were obtained from the Saccharomyces Genome Database (SGDgene), the Sanger Center (sangerGene), Fly Base (flyBaseGene), Ensemble gene (ensGene) and UCSC knownGene databases, respectively, through the UCSC Genome Browser portal (see Table S1). To ensure that coding bases in eukaroyote genomes were only counted once, regardless of the number of putative or known protein-coding isoforms they may be associated with, each annotation set was “collapsed down.” To accomplish this in fungi, protists, viridiplantae and the majority of Metazoa we merged the overlapping CDS regions as annotated in the appropriate gtf or gff annotation file. For those organisms with complete annotation sets in the UCSC Genome Browser (C. elegans = ce6, D. melanogaster = dm3, D. rerio = danRer7 and H. sapiens = hg19), their corresponding gene annotation track (sgdGene, sangerGene, flyBaseGene, Ensemble ensGene, UCSC knownGene) was collapsed using the UCSC backend tool featureBits. In one case the CDS annotations were manually curated to remove a high degree of putative false positives before the merge step, the gene annotations Branchiostoma floridae were parsed to remove all duplicates, and then only those with a gene ontology annotation were further analyzed.
RNA-seq analyses
We selected four well-studied model organisms for detailed transcriptomic analysis, roundworm (C. elegans), fruit fly (D. melanogaster), zebrafish (D. rerio) and man (H. sapiens). To ensure that we were able to catalog as completely as possible the full complexity of each species’ transcriptome, we collected RNA-seq from all available sources (e.g., cell lines, tissues, development stages, replicates data sets; see Table S2). In most cases, the raw RNA-seq data were all downloaded from the SRA database72 (Table S2). For each data set, we calculated the total gigabaes of raw sequencing data by multiplying the length of each sequence read by the total number of reads in the data set.
Using the transcriptome analysis pipeline recently published by Trapnell et al. in Nature Protocols59 as the foundation of our RNA-seq investigation, we utilized the Tophat (version 1.4.1) and Cufflinks (version 1.3.0) programs with default parameters to assemble de-novo transcriptomes.59,67,68 The Cufflinks output file gtf format was converted to BED format by in-house scripts, and a UCSC Genome Browser backend tool program, bedToExons, was used to fetch individual exons. The genomic coverage of RNA-seq transcripts was then calculated using the UCSC Genome Broswer backend program featureBits across the entire length of all transcripts (txStart-txStop) or across only “exons” (i.e., transcribed bases that result in a de-novo assembled transcript).
Human RNA-seq data sets were further interrogated to assess the extent of transcription in the human genome, under the assumption that the Tophat-Cufflinks pipeline would fail to assemble any transcripts that did not fit the standard/statistical models consistent with traditional mRNAs. This would be particularly true for data sets that specifically interrogated polyA-transcripts (e.g., the IBM2 16 tissue mix total RNA treated with duplex specific nuclease). Therefore, for each human RNA-Seq data set the Tophat-generated BAM file was converted to BED format using the BEDtools69 program bamToBed, which was then clustered using the BEDtools program, mergeBed. Clutering was done strand specifically for those data sets for which were generated strand-specifically (e.g., the IBM2 16 tissue mix data sets). Clusters were then filtered to isolate those that contained > 2, > 4, > 8 and > 16 overlapping mapped tags. To capture the full extent of transcription, a final data set was generated that contained the most conservative clustering, i.e., > 16 overlapping reads, plus the addition of Tophat mapped spiced tags with anchors of > 20 bp. This data set was used for the majority of subsequent analyses unless otherwise noted. The genomic coverage of all clustered data sets (and their genomic overlap) was calculated using the UCSC backend program featureBits (see Table S2).
Using the most conservative clustering metric (i.e., > 16 tags per cluster) plus mapped spliced reads, heatmaps were generated across 1 Mb bins of all human chromosomes using the R-package pheatmap (see Supplemental Material). Expression intensity is shown as counts are depicted as log10(raw mapped reads) for each data set in each 1 Mb bin.
Gene deserts were defined as gene-free regions of > 500 kb, as originally described by Norbrega et al.73 To obtain the genomic coordinates of gene deserts we employed BEDtools subtractBed to filter out human genome (hg19) assembly gaps, and to identify regions without annotations in the latest Refseq and Ensemble gene sets. These regions were then intersected with RNA-seq clusters of > 16 tags from all 40 human RNA-seq data sets. Only those that had > 1,000 mapped RNA-seq tags were considered “expressed.” GWAS regions were obtained from the UCSC NHGRI GWAS track,61 and intersections were performed using UCSC backend tool overlapSelect.
Processed RNA-seq data availability
Processed and assembled RNA-seq data are available through the Genomics Virtual Laboratory. Publicly available UCSC-viewable tracks for the C. elegans, D. melanogaster, D. rerio and H. sapiens Tophat-Cufflinks assembled transcripts of all mapped tags are available at the following URLs:
This data can be viewed by visiting the public UCSC Genome Browser instance (http://genome.ucsc.edu/), and navigating to Track Hubs > My Hubs > pasting in the URL and clicking “Add Hub.” Please note that the human hub (e.g., “humanSuper”) also includes wiggle density tracks derived from the Tophat-mapped BAM files.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
G.L. performed bioinformatics analyses and drafted the manuscript. J.S.M. assisted with study design and drafted the manuscript. R.J.T. conceived and designed the study, performed bioinformatics analyses and drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgments
R.J.T. is supported by an Australian Research Council Discovery Early Career Researcher Award DE1211916. This work was supported the NeCTAR Genomics Virtual Lab (GVL) and Australian Research Council Discovery Program Grant DP1213828.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/25134
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25134
References
- 1.McCLINTOCK B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol. 1951;16:13–47. doi: 10.1101/SQB.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 2.McCLINTOCK B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci USA. 1950;36:344–55. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–57. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 4.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nat Cell Biol. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- 10.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 11.Huda A, Bowen NJ, Conley AB, Jordan IK. Epigenetic regulation of transposable element derived human gene promoters. Gene. 2011;475:39–48. doi: 10.1016/j.gene.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Román AC, González-Rico FJ, Moltó E, Hernando H, Neto A, Vicente-Garcia C, et al. Dioxin receptor and SLUG transcription factors regulate the insulator activity of B1 SINE retrotransposons via an RNA polymerase switch. Genome Res. 2011;21:422–32. doi: 10.1101/gr.111203.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–69. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 15.Lynch VJ, Leclerc RD, May G, Wagner GP. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet. 2011;43:1154–9. doi: 10.1038/ng.917. [DOI] [PubMed] [Google Scholar]
- 16.Sela N, Kim E, Ast G. The role of transposable elements in the evolution of non-mammalian vertebrates and invertebrates. Genome Biol. 2010;11:R59. doi: 10.1186/gb-2010-11-6-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, et al. Affymetrix ENCODE Transcriptome Project. Cold Spring Harbor Laboratory ENCODE Transcriptome Project Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–32. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 19.Mattick JS, Taft RJ, Faulkner GJ. A global view of genomic information--moving beyond the gene and the master regulator. Trends Genet. 2010;26:21–8. doi: 10.1016/j.tig.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 24.Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30:99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol. 2010;17:1030–4. doi: 10.1038/nsmb.1841. [DOI] [PubMed] [Google Scholar]
- 28.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–8. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 29.Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–40. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamasaki S, Ivanov P, Hu G-F, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–95. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285:10959–68. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taft R, Mattick J. Increasing biological complexity is positively correlated with the relative genome-wide expansion of non-protein-coding DNA sequences. Genome Biology Pre-print Depository. 2003;5:1. doi: 10.1186/gb-2003-5-1-p1. [DOI] [Google Scholar]
- 34.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays. 2007;29:288–99. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 35.Schad E, Tompa P, Hegyi H. The relationship between proteome size, structural disorder and organism complexity. Genome Biol. 2011;12:R120. doi: 10.1186/gb-2011-12-12-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hedges SB, Blair JE, Venturi ML, Shoe JL. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 2004;4:2. doi: 10.1186/1471-2148-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel C, Chothia C. Protein family expansions and biological complexity. PLoS Comput Biol. 2006;2:e48. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haygood R, SMBE Tri-National Young Investigators Proceedings of the SMBE Tri-National Young Investigators’ Workshop 2005. Mutation rate and the cost of complexity. Mol Biol Evol. 2006;23:957–63. doi: 10.1093/molbev/msj104. [DOI] [PubMed] [Google Scholar]
- 39.Bell G, Mooers AO. Size and complexity among multicellular organisms. Biol J Linn Soc Lond. 1997;60:345–63. doi: 10.1111/j.1095-8312.1997.tb01500.x. [DOI] [Google Scholar]
- 40.Hahn MW, Wray GA. The g-value paradox. Evol Dev. 2002;4:73–5. doi: 10.1046/j.1525-142X.2002.01069.x. [DOI] [PubMed] [Google Scholar]
- 41.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, et al. modENCODE Consortium Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–97. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–8. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 43.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–15. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 44.Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–10. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 46.Hiller LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, et al. International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 47.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40(Database issue):D130–5. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scally A, Dutheil JY, Hillier LW, Jordan GE, Goodhead I, Herrero J, et al. Insights into hominid evolution from the gorilla genome sequence. Nature. 2012;483:169–75. doi: 10.1038/nature10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stein LD. Human genome: end of the beginning. Nature. 2004;431:915–6. doi: 10.1038/431915a. [DOI] [PubMed] [Google Scholar]
- 50.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40(Database issue):D13–25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sherman M. Universal genome in the origin of metazoa: thoughts about evolution. Cell Cycle. 2007;6:1873–7. doi: 10.4161/cc.6.15.4557. [DOI] [PubMed] [Google Scholar]
- 52.Richards GS, Simionato E, Perron M, Adamska M, Vervoort M, Degnan BM. Sponge genes provide new insight into the evolutionary origin of the neurogenic circuit. Curr Biol. 2008;18:1156–61. doi: 10.1016/j.cub.2008.06.074. [DOI] [PubMed] [Google Scholar]
- 53.Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466:720–6. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vizcaíno JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41(Database issue):D1063–9. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akama T, Suzuki K, Tanigawa K, Nakamura K, Kawashima A, Wu H, et al. Whole-genome expression analysis of Mycobacterium leprae and its clinical application. Jpn J Infect Dis. 2010;63:387–92. [PubMed] [Google Scholar]
- 56.Hill AV. The possible effects of the aggregation of molecules of hemoglobin on its dissociation curve. J Physiol. 1910;(Suppl):40. [ ] [Google Scholar]
- 57.Mattick JS. Deconstructing the dogma: a new view of the evolution and genetic programming of complex organisms. Ann N Y Acad Sci. 2009;1178:29–46. doi: 10.1111/j.1749-6632.2009.04991.x. [DOI] [PubMed] [Google Scholar]
- 58.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–92. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 59.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhulidov PA, Bogdanova EA, Shcheglov AS, Vagner LL, Khaspekov GL, Kozhemyako VB, et al. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 2004;32:e37. doi: 10.1093/nar/gnh031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glinskii AB, Ma J, Ma S, Grant D, Lim C-U, Sell S, et al. Identification of intergenic trans-regulatory RNAs containing a disease-linked SNP sequence and targeting cell cycle progression/differentiation pathways in multiple common human disorders. Cell Cycle. 2009;8:3925–42. doi: 10.4161/cc.8.23.10113. [DOI] [PubMed] [Google Scholar]
- 63.Balestrino A, Caiti A, Crisostomi E. Generalised Entropy of Curves for the Analysis and Classification of Dynamical Systems. Entropy. 2009;11:249–70. doi: 10.3390/e11020249. [DOI] [Google Scholar]
- 64.Langton CG. Computation at the edge of chaos: Phase transitions and emergent computation. Physica D. 1990;42:12–37. doi: 10.1016/0167-2789(90)90064-V. [DOI] [Google Scholar]
- 65.Mitchell M, Crutchfield JP, Hraber PT. Dynamics, computation, and the“ edge of chaos”: A re-examination. Santa Fe Institute Studies in the Sciences of Complexity. 1994;19:497–7. [Google Scholar]
- 66.Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40(Database issue):D918–23. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2010;38(Database issue):D382–90. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, et al. Ensembl’s 10th year. Nucleic Acids Res. 2010;38(Database issue):D557–62. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kodama Y, Shumway M, Leinonen R, International Nucleotide Sequence Database Collaboration The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40(Database issue):D54–6. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 74.Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–61. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grbić M, Van Leeuwen T, Clark RM, Rombauts S, Rouzé P, Grbić V, et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–92. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Künstner A, et al. The genome of a songbird. Nature. 2010;464:757–62. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.