Abstract
Cells disseminated from primary epithelial tumors into peripheral blood, called circulating tumor cells (CTCs), can be monitored to assess metastases and to provide a surrogate marker of treatment response. Here, we demonstrate how the flexible micro spring array (FMSA) device—a novel microfluidic device that enriches CTCs by two physical parameters: size and deformability—could be used in the rational development of treatment intervention and as a method to study the fundamental biology of CTCs. Cancer cells of different origins were spiked into healthy samples of donor blood to mimic blood samples of metastatic cancer patients. This spiked human blood was filtered using the FMSA device, and the recovered cells were successfully expanded in vitro and in a novel in vivo system. A series of experiments were performed to characterize these cells and to investigate the effect of chemotherapy on the resulting cultures. As few as 20 colon cancer cells in 7.5 mL blood could be isolated with the FMSA device, expanded both in vitro and in vivo and used at 25 cells per well to obtain significant and reliable chemosensitivity data. We also show that isolating a low number of viable patient CTCs and maintaining them in culture for a few weeks is possible. The isolation of viable cancer cells from human blood using the FMSA device provides a novel and realistic means for studying the biology of viable CTCs and for testing drug efficacy on these rare cells—a hypothesis that can be tested in future clinical trials.
Keywords: circulating tumor cells, drug sensitivity testing, personalized medicine, viable cell capture, microfluidic
Introduction
Most deaths resulting from tumors of epithelial origin (carcinomas) are caused by the hematogenous spread of cancer cells into distant organs and these cells’ subsequent growth into overt metastases.1 Although classically viewed as a late process in malignant progression, the dissemination of such cells—called circulating tumor cells (CTCs)—from primary carcinomas recently has been shown to be a relatively early event in cancer progression.2 It also has been shown that CTCs often have key biological differences, in regards to established prognostic markers, that make these cells radically different from the cancerous cells found at the primary tumor site.3 Being that an anticancer therapeutic regimen based on the molecular profile of the primary tumor may be ineffective in stemming the outgrowth of fundamentally different circulating tumor cells into metastases, the characterization of a patients’ CTCs holds potential as a novel, rapid and early method for the evaluation of cancer treatments.
In a series of prospective, multi-center clinical trials, the enumeration of blood-borne cells that are: (1) a round to oval shape (as determined by light scatter), (2) nucleus-possessing [as evidenced by 4’,6-diamidino-2-phenylindole (DAPI) staining], (3) positive for the expression of epithelial cell adhesion molecule (EpCAM) and (4) cytokeratins-8,-18,-19, but (5) negative for the expression of CD45 (by immunofluorescent detection) using the Veridex CellSearch system led to the establishment of CTCs as independent predictors of progression-free survival (PFS) and overall survival (OS) in metastatic breast, castration-resistant prostate and advanced colorectal cancers.4-6 Although technological advances in recent years have enabled isolation and enumeration of CTCs through a variety of other methods, including immunoaffinity separation, density-based enrichment and magneto-pheresis, only one CTC isolation and detection method currently has been approved by the US Food and Drug Administration (FDA): the Veridex CellSearch system. This system, in an automated manner, isolates cancer cells from blood using EpCAM and qualifies them as CTCs based on the above criteria. Although the enumeration of CTCs is an important clinical tool and can help with the monitoring of therapy (reduction in CTC counts correlate with response to therapy and better prognosis, whereas increase in CTC counts may predict tumor relapse or the emergence of drug resistance), their mere enumeration obscures their greater biological and clinical value.
Given the limitations of other CTC enrichment methods, we propose to use a novel approach that enriches CTCs by two physical parameters—size and deformability—in order to expand the functional use of these rare cells. Past studies have revealed that the shear modulus, stiffness, size and deformability of cancer cells is distinctively different from blood constituents.7 Aided by modern micro-fabrication tools, we are developing a new technology, called the flexible micro spring array (FMSA) device, which enables size-exclusion based viable CTC enrichment. By exploiting intrinsic differences between cancer cells and other blood constituents, the FMSA device overcomes limitations of other technologies: these include the EpCAM dependence of the Veridex CellSearch system and the antigen dependence of the CTC-chip, the need to lyse blood cells with the Epics Bioscience system and the ScreenCell system and the labor/cost of other available microfluidic approaches such as the ClearCell CTChip.8-10 The FMSA device is a unique approach that allows for the enrichment of viable CTCs with minimal cellular manipulation and without the use of antibodies. Because CTCs derived from carcinomas are generally larger and more rigid than hematopoietic blood cells, the FMSA device retains CTCs on the surface of the device—along with a minor fraction of large blood cells (mainly large granulocytes)—while the overwhelming majority of other blood cells flow through the device.11,12 The FMSA device is, essentially, a filtration device that allows for the exclusion of CTCs from whole blood. The device has microspring structures patterned by photolithography on a single layer parylene diaphragm. The flexible polymer structure mitigates the stresses experienced by cells during vacuum-driven filtration to encourage their health and survival afterwards. A low driving pressure is applied during enrichment to ensure minimal cell damage. As mentioned above, the device enriches CTCs by physical criteria, mainly size, and does not depend on CTC surface antigen expression. The incorporated system allows size-based separation of the cells at the micro-scale at non-lethal pressures while taking advantage of a high-throughput and rapid processing speed. The testing results of a model system of spiked samples from different cancers (similar to the system used in this manuscript) show this device can enrich CTCs from 7.5 mL of whole blood samples with 90% recovery, higher than 104 enrichment and better than 80% viability in approximately 10 min.13,14
In the present study, we use the FMSA device to isolate, culture, expand and functionally characterize viable cancer cells from blood. In accordance with a previously described method, various cancer cell lines of different origin were spiked into reconstituted healthy donor blood to mimic the blood of metastatic cancer patients.15 This spiked blood was filtered using the FMSA device, and the recovered cancer cells were either implanted into the flanks of immunodeficient mice for study or expanded in traditional cell culture systems. We show that the FMSA device can be used in this manner to establish both in vitro and in vivo cultures of spiked cancer cells isolated from blood samples, and that these cultures can subsequently be subjected to drug sensitivity testing. More importantly, we show that as few as 20 recovered (spiked) cancer cells in 7.5 mL of blood can be isolated with the FMSA device, expanded both in vitro and in vivo over the course of a few weeks, and used to generate large amounts of reliable drug sensitivity data. We follow this finding by showing that viable CTCs from patients with metastatic cancer can be isolated from blood using the FMSA device and subsequently kept in culture for several weeks, allowing for the type of chemosensitivity analysis we describe. Amounting to a significant step toward personalized cancer medicine, the opportunity to isolate viable CTCs using the FMSA device offers the potential to investigate the genomics, explore the heterogeneity and potentially test cancer treatments in real time and to rapidly feed information back to clinicians. This is a hypothesis that can be tested in the future in clinical trials. There remain some challenges for the field with patient-derived CTCs in terms of their ability to grow following recovery from blood.
Results
In vitro culture of spiked cancer cells isolated from blood using the FMSA device allows viable cell culture of recovered cancer cells
To facilitate the identification of cancer cells from blood, we spiked fluorescently-labeled cancer cells into reconstituted healthy donor blood and filtered the resulting spiked blood using the FMSA device. As shown in Figure 1A, physiologically relevant numbers of cancer cells spiked into 7.5 mL blood, here 100 GFP-labeled HCT-116 cells, can be isolated and detected on the FMSA device surface. Beyond permitting for the simple enumeration of CTCs, the FMSA device’s unique properties allow for the potential in vitro culture of CTCs: after gently shaking cells off of the FMSA, cancer cells can be cultured and expanded in vitro. As shown in Figure 1B, as few as 20 or 100 RFP-labeled MCF-10A-HRAS cells can be spiked into 7.5 mL of blood, isolated using the FMSA device, shaken off and used to form 1 or 6 actively proliferating colonies, respectively. Similar results were also obtained from the isolation of GFP-labeled HCT-116 cells from blood (Fig. 1C). From the filtration of 7.5 mL of blood spiked with 20, 100 and 1,000 GFP-labeled HCT-116 cells, 3, 28 and 102 colonies were formed, respectively.
Figure 1. Proliferation of cancer cells isolated from blood using the FMSA device. (A) Left: fluorescent image of GFP-labeled HCT-116 cells isolated from blood on FMSA surface. Right: composite brightfield-fluorescent image of those same cells. (B) Left: outgrowth of 2 RFP-labeled MCF-10A-HRAS colonies resulting from the isolation of RFP-labeled MCF-10A-HRAS cells from blood using the FMSA device (20 cells spiked in 7.5 mL blood). Right: quantification of the number of colonies resulting from the isolation and culture of different numbers of RFP-labeled MCF-10A-HRAS cells (20, 100 and 1,000) in 7.5 mL blood using the FMSA device (n = 3). (C) Left: outgrowth of 3 GFP-labeled HCT-116 colonies resulting from the isolation of GFP-labeled HCT-116 cells from blood using the FMSA device (20 cells spiked in 7.5 mL blood). Right: quantification of the number of colonies resulting from the isolation and culture of different numbers of GFP-labeled HCT-116 cells (20, 100 and 1,000) in 7.5 mL blood using the FMSA device (n = 3).
The isolation and ensuing culture of spiked cancer cells from blood using the FMSA device does not affect their chemosensitivity
One of the challenges facing the isolation of CTCs from blood is that current techniques cause much stress to the cells that their inherent viability and proliferability are altered. Such alterations mean that ensuing functional analysis of CTCs with other technologies is limited and otherwise problematic in terms of reflecting the behavior of the original tumor cells in the circulation. Here, we show that the FMSA device does not affect the viability, proliferation ability or chemosensitivity of spiked cancer cells isolated from blood. We spiked human HT-29 colon adenocarcinoma cells into healthy donor blood, isolated the cells with the FMSA device, cultured the cells in vitro for 5 d, seeded the cells in 96-well plates and treated the cells with different concentrations of approved anticancer agents. We detected no significant differences, with regards to either proliferability or chemosensitivity (Fig. 2A), between HT29 cells that had been spiked into blood and isolated with the FMSA device and the original HT29 cell line. As can be seen in Figure 2B, we observed similar results when comparing GFP-labeled HCT-116 cells recovered from blood using the FMSA device as opposed to the original GFP-labeled HCT-116 cell line.
Figure 2. The FMSA device does not affect the chemosensitivity of cancer cells isolated from blood. (A) 1 × 105 HT-29 human colon cancer cells were spiked into 7.5 mL blood and isolated with the FMSA device. Dose response curves comparing the effects of 5-FU (top), oxaliplatin (middle) and irinotecan (bottom) on cell viability against HT-29 cells isolated from blood and expanded in vitro (red) and the original HT-29 cell line. Cell viability measured with CellTiter Glo assay as described in “Materials and Methods.” (B) 1 × 105 HCT-116 human colon cancer cells were spiked into 7.5 mL blood and isolated with the FMSA device. Dose response curves comparing the effects of 5-FU (top), oxaliplatin (middle) and irinotecan (bottom) on cell viability against HCT-116 cells isolated from blood and expanded in vitro (red) and the original HCT-116 cell line. Cell viability measured with CellTiter Glo assay as described in “Materials and Methods.”
The FMSA device may be used for high-throughput CTC drug sensitivity testing
Given that the FMSA device allows for the isolation, culture, proliferation and drug sensitivity testing of spiked cancer cells from blood, and that the chemosensitivity data of the isolated cells is comparable to the original cells, we hypothesized that the FMSA device could be used to test patient CTCs against broad panels of anticancer agents. To test the feasibility of such high-throughput CTC chemosensitivity testing, 20 GFP-labeled HCT-116 cells were spiked into 7.5 mL blood, isolated from blood using the FMSA device, expanded in vitro for 15 d, plated in 384-well plates, subjected to chemotherapy testing and analyzed for viability using the CellTiter Glo assay (see “Materials and Methods”). Post-FMSA isolation, and after 15 d of culture, the GFP-labeled HCT-116 cells had colonized the original culture plates and grown to over 1.2 × 106 cells. We were able to seed this large number of cells, at different concentrations of cells per well, in 384-well plates and, using a trio of drugs (irinotecan, 5-fluorouracil and oxaliplatin) and the CellTiter Glo reagent, carry out high-throughput chemosensitivity testing (Fig. 3A). In doing so, we determined that only 25 cells per well are needed for statistically significant chemosensitivity testing, meaning that a minimal amount of cell culture is needed in between CTC isolation and chemosensitivity testing using this approach (Fig. 3B).
Figure 3. The FMSA device allows for high-throughput drug sensitivity testing on isolated and expanded CTCs. (A) A total of 20 GFP-labeled HCT-116 colon cancer cells were spiked into blood, isolated with the FMSA device, expanded in vitro for 15 d and seeded into 384-well plates. Left: sample Cell Titer Glo data showing feasibility of high-throughput drug sensitivity testing. Right: dose response curve of multi-agent drug sensitivity testing acquired with the Cell Titer Glo assay. (B) 20 GFP-labeled HCT-116 cancer cells were spiked into blood, isolated with the FMSA device, expanded in vitro for 15 d, seeded into 384-well plates at differing concentrations of cells per well and treated with various concentrations of oxaliplatin. Cell viability was measured with the Cell Titer Glo assay and statistics calculated as described in “Materials and Methods.”
CTCs isolated from patient blood can be kept in culture for several weeks
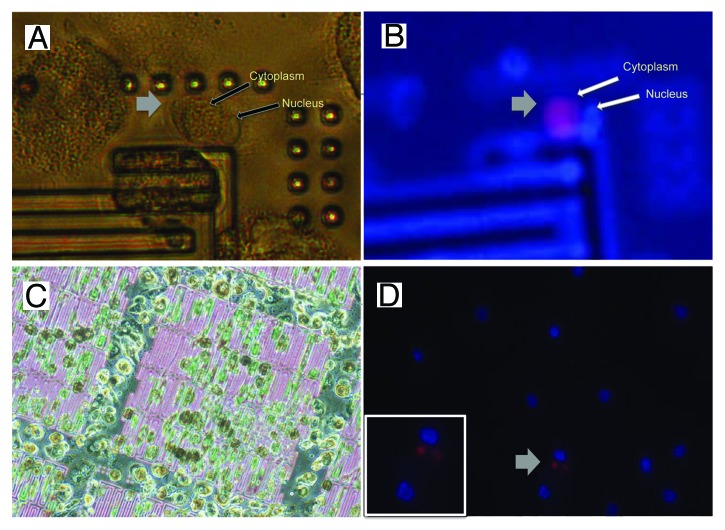
Much of the work outlined in this manuscript rests on the assumption that viable patient CTCs can be kept in culture and expanded similarly to cancer cells isolated from healthy donor blood. In order to show the feasibility of our approach, we filtered blood from several patients with metastatic cancers (from a variety of cancer types including small cell lung cancer, neuroendocrine carcinoma, colorectal cancer and carcinoma of unknown primary) that had proven CTCs (as confirmed by Veridex CellSearch analysis) using the FMSA device. Viable CTCs were successfully isolated and cultured with the FMSA device, or an alternate method (see “Materials and Methods”), from a patient with carcinoma of unknown primary (CUP) and a patient with metastatic colorectal cancer. Cells from a CUP patient isolated from blood using the FMSA device were kept in culture for 12 d and then stained for the presence of CTCs (Fig. 4). High-power phase microscopy showed that the putative CTC has both an intact nuclear membrane (Fig. 4A) enclosing a DAPI-stained nucleus (Fig. 4B) in addition to an intact plasma membrane. Although a specific viability assay was not performed, it was inferred that this cell survived in culture and was viable prior to fixation on day 12 based on the morphological criteria described above. A putative CTC from same patient demonstrated an unusual punctuate yet clearly cytoplasmic pattern of cytokeratin-8,-18 expression (Fig. 4D) as distinguished from the surrounding cells, which showed DAPI nuclear stain but demonstrated no cytoplasmic cytokeratin-8,-18. Blood from a normal donor that was depleted of some of the red blood cells (see “Materials and Methods”) demonstrated no expression of cytokeratin-8,-18 in peripheral blood mononuclear cells (Fig. S1, center). Similar results were obtained with the cells from a metastatic colorectal cancer patient after 23 d in culture (Fig. S1). Although unidentified peripheral blood mononuclear cells other than CTCs appeared to proliferate in vitro, even after 23 d of culture, CD45-negative and CK-8,18-positive cells (putative CTCs) were still viable in culture at reasonable numbers (Fig. S1). To our knowledge, CTCs have never been kept in culture in this manner before; the viable isolation and culture of CTCs with the FMSA device should allow for some of the in vitro chemosensitivity testing we outline above given the low number of required cells. However, it is clear that progress must be made to encourage the proliferation and growth of these cells.
Figure 4. Putative CTCs in culture from a patient with carcinoma of unknown primary (CUP) Cells in culture following isolation with the FMSA device from blood of a patient with carcinoma of unknown primary. (A) High-power phase image of putative CTC. (B) CK8, 18 expression (red) and nuclear stain (DAPI, blue) of the putative CTC in (A). (C) Phase microscopy of live cells on FMSA device on day 10 in culture, 2 d before fixing and staining. (D) CK-8, 18 expression (red) and nuclear stain (DAPI, blue) of cells from the same patient on the bottom of the culture dish (Inset: zoom) on day 12.
Spiked cancer cells isolated from blood using the FMSA device can be grown as tumors in vivo
In order to gain a more comprehensive understanding of the biology, behavior, heterogeneity and chemosensitivity of CTCs, and in order to explore other potential methods for CTC culture, we have also been developing models to culture and study these cells in vivo. Because the FMSA device is only 10 µm thick and extremely flexible, it is possible to use the device as a carrier for cancer cells isolated from blood and to implant the whole FMSA device including the isolated cancer cells in vivo. The FMSA device is made with a USP class VI polymer, parylene, suitable for long-term implantation. As depicted in Figure 5, after passing blood through the device, we have taken the entire biocompatible FMSA device, together with the trapped cells, adding a small amount of matrigel, and inserted the matrigel-covered FMSA subcutaneously, through a small incision, into the right flank of SCiD mice (more details in the Methods section). We have termed the resulting outgrowths “FMSA-derived tumors.” In all of our experiments, each mouse bearing an FMSA implant has also borne what we term a standard xenograft (the subcutaneous injection of a 1:1 mixture of matrigel:cancer cell suspension) on the left flank.
Figure 5. Cancer cell isolation from blood and in vivo implantation using the FMSA device. Current workflow for CTC isolation and in vivo implantation. Details of techniques describes in “Materials and Methods” and “Results” sections.
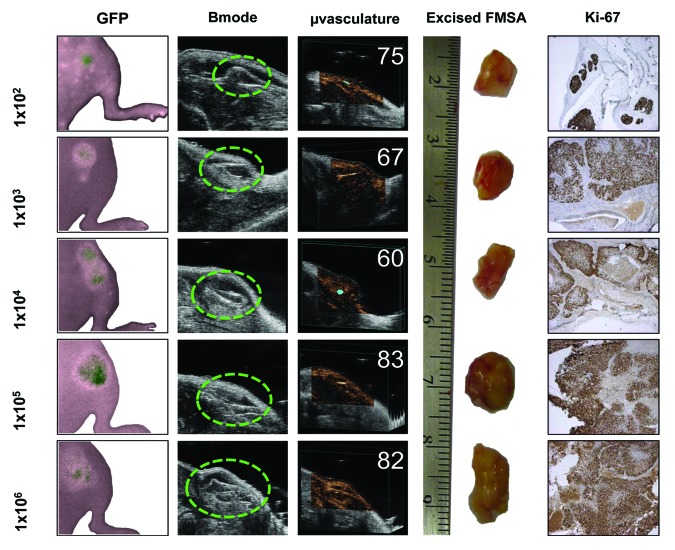
Using the method outlined above, we have successfully grow FMSA-derived tumors in SCiD mice resulting from the implantation of the FMSA device after filtration of 7.5 mL of blood containing as few as 100 spiked human colon cancer cells (Fig. 6A). The FMSA-derived tumors recapitulate the growth of standard xenografts (Fig. 6B). Furthermore, FMSA-derived tumors are measurable in as little as 2 wk, are well perfused with blood and can be excised for immunohistochemical analysis (Fig. 7). Clearly this approach is amenable to future testing with patient-derived viable CTCs.
Figure 6. Outgrowth of FMSA-derived tumors is comparable to standard xenografts Hairless SCiD mice were implanted subcutaneously with both a 1:1 v/v suspension of GFP-labeled HCT-116 cells:matrigel on their left flank and the FMSA on their right flank as per Figure 5. (A) Fluorescent signal of GFP-labeled HCT-116 tumors resulting from the injection of a cell suspension or FMSA implantation. The cell number injected (solid line) or spiked into 7.5 mL blood and filtered with the FMSA (dotted line) indicated on figure. (B) Normalized fluorescent signal of Figure 5A. (C) Calliper measurements of tumor size.
Figure 7. Visualization and study of the in vivo outgrowth of cancer cells isolated from blood using the FMSA device. Various amounts of GFP-labeled HCT-116 cells (indicated on left y-axis) were spiked into 7.5 mL blood and implanted into the right flank of hairless SCiD mice (A) Pseudo-color fluorescent imaging of FMSA-derived tumors at day 14 (B) axial/transverse sections of FMSA-derived tumors taken with ultrasound imaging at day 14 (C) calculation of tumor microvasculature perfusion percentage using ultrasound imaging and micro-bubbles at day 14 (D) excised FMSA-derived tumors (cm scale) at day 30 (E) Ki-67 immunohistochemical staining of FMSA-derived tumors.
FMSA-derived tumors can be used for in vivo drug sensitivity testing of CTCs
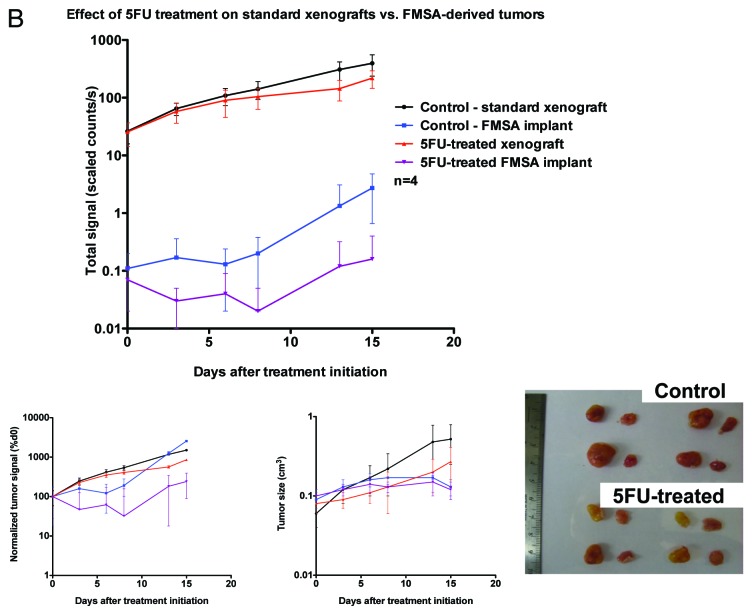
To determine whether it is possible to test the efficacy of chemotherapies on spiked cancer cells isolated from blood in vivo using the FMSA device, we subjected mice bearing both FMSA-derived tumors and standard xenografts to treatment by different chemotherapy regimens. We determined that it is indeed possible to measure a difference between the tumor outgrowth in mice bearing control FMSA-derived tumors vs. those bearing FMSA-derived tumors treated with either irinotecan or 5-fluorouracil (Figs. 8A and 8B). The difference in growth rates between control FMSA-derived tumors and chemotherapy-treated FMSA-derived tumors was comparable to that between control standard tumor xenografts and similarly chemotherapy-treated standard tumor xenografts. Although potentially less desirable due to cost, time and effort involved, this approach is certainly amenable for future testing using patient-derived CTCs.
Figure 8A. In vivo drug sensitivity of cancer cells isolated from human blood using the FMSA device. (A and B) Hairless SCiD mice were implanted subcutaneously with both a suspension of 1 × 106 HCT-116-GFP cells on their left flank and the FMSA on their right flank (1 × 106 cells spiked in 7.5 mL blood). n = 4 per treatment group. Treatment began at day 0, 7 d post implantation. 5-FU and CPT-11 were administered at 50 mg/kg IP once a week. (A and B) Top: fluorescent signal of HCT-116-GFP tumors resulting from the injection of a cell suspension or FMSA implantation, following CPT-11 (A) or 5-FU (B) treatment. (A and B) Bottom left: normalized fluorescent signal. (A and B) Bottom middle: calliper measurements of tumor size. (A) Bottom right: Representative mice following CPT-11 treatment bearing standard xenografts and FMSA implants (B) Bottom right: standard xenografts and FMSA implants excised from mice upon euthanasia following 5-FU treatment. (C) Ten hairless SCiD mice were implanted subcutaneously with both a suspension of 20 HCT-116-GFP cells on their left flank and the FMSA on their right flank (20 cells spiked in 7.5 mL blood). Two control mice were implanted with an FMSA device after filtration of 7.5 mL human blood containing no cancer cells. Treatment began at 30 d post implantation. CPT-11 was administered at 50 mg/kg IP once a week. Top: fluorescent signal of HCT-116-GFP tumors resulting from the injection of a cell suspension or FMSA implantation following CPT-11 treatment. Bottom left: normalized fluorescent signal. Bottom right: calliper measurements of tumor size.
Figure 8B. See Figure 8A legend.
To assess the limits of our system in terms of minimal numbers of circulating tumor cells and their potential to be recovered and to grow as tumors, we repeated the same experiments, but with a minimal number of cells: FMSA-derived tumors were grown from 20 GFP-labeled HCT-116 cells spiked into 7.5 mL of blood. After a month of uninhibited growth, enough mice had sufficiently sized FMSA-derived tumors and standard xenografts to allow for drug sensitivity testing. Of note, 5/10 mice developed significantly sized (> 3 mm) standard xenografts, and 2/10 mice developed comparably sized FMSA-derived tumors. Mice were split into two groups and treated with irinotecan for 3 wk. As before, we observed a clear difference between the growth rates of chemotherapy-treated and control FMSA-derived tumors, and this differential in growth rate was similar to that of the chemotherapy-treated and control standard xenografts in these same mice (Fig. 8C).
Figure 8C. See Figure 8A legend.
Discussion
Although patient-derived CTCs can be enumerated reliably using the FDA-approved Veridex CellSearch system, there exists, to our knowledge, no other technology that allows for the functional analyses of CTCs of different origins as described herein. Although other technologies allow for functional CTC analyses, their capture is biased toward certain cancer types or epitopes.16 The FMSA device solves the challenges of other microfabricated CTC filtration devices and is, in essence, a new technology optimized for unbiased viable CTC enrichment, culture, cell expansion and analysis.17-19 Testing with spiked blood samples (representative of patient samples) shows this device can enrich CTCs from 7.5 mL of whole-blood samples with 90% recovery, higher than 104 enrichment and better than 80% viability in approximately 10 min.13,14
The successful establishment of CTC cultures using the FMSA device, whose potential is demonstrated herein with spiked blood samples and patient samples, could in the future enable in vitro and in vivo drug efficacy testing and for the formation of personalized treatment plans for patients with metastatic cancer. Drug sensitivity testing on CTCs provides an attractive personalized cancer treatment strategy that tailors individual patient’s treatment by measuring the therapeutic response of CTCs to certain anticancer drugs. Using a high-throughput automated robotic system, most currently FDA-approved anticancer drugs could rapidly be tested against a single patient’s CTCs, feeding back large amounts of potentially invaluable information, in real time, to clinicians. As we have demonstrated, minimal culture would be needed before chemosensitivity data could be obtained, minimizing the chance for culture artifacts to influence data.
However, growing cells isolated from blood in the manner we have proposed is not trivial: cancer cells would sometimes not propagate when isolated from blood using the FMSA device and returned to their original ATCC-recommended culture conditions. The percent concentration of FBS occasionally had to be increased upwards of 20% to encourage initial cell growth. We met an even bigger challenge when attempting to establish cultures of patient CTCs, which were few in number and intractable to growth as compared with cancer cell lines. While we recovered apparently viable tumor cells, getting these cells to proliferate remains a problem for the field. Solving this problem is complex for stimulating the cells, or altering their environment may fundamentally change their phenotype. While the field moves toward a greater focus on genomics,20 we believe that there is still value in getting patient tumor cells to proliferate either in culture or in vivo, especially since this would allow for chemosensitivity testing. It is possible that as more experience is gained with our systems, that some tumor types may be more amenable to this strategy than others. Although each patient’s CTCs are likely to require their own set of optimized conditions, we speculate that a general set of culture conditions could be developed to optimize viable CTC proliferation without affecting these cells’ phenotypes.
Simultaneously demonstrating both CTC proliferation and tumor identity also remains a technical feat. While we utilized well-established carcinoma markers (cytokeratins-8,-18) to identify putative CTCs, the use of additional markers to type the CTCs remains problematic. There is for example, no single cancer type-specific marker that can be used for CUP.21,22 There currently is no approach for unequivocally identifying CTCs, not even the presence of CK-8,-18 and absence of CD45 in a single CTC. Furthermore, it is possible that culture conditions may change the marker expression pattern of rare surviving cells isolated from blood, including other phenomena such as epithelial-to-mesenchymal transition. As the study of CTCs continues, this is another challenge that the field is facing. This is particularly relevant for chemosensitivity testing that does require some expansion of the cells, as we have shown.
Due to the inherent challenges of in vitro CTC growth, proliferation and identification, we have proposed a novel means to functionally study CTCs: the in vivo implantation of the FMSA device. The implanted FMSA device should accurately mimic the natural tumor microenvironment due to the presence of a variety of cells (fibroblasts, leukocytes, etc.) on the filter and in the nearby mouse microenvironment. This microenvironment may provide soluble factors, cell-to-cell interactions and physical parameters that are not easy to be mimicked by in vitro culture, and thus may create a more favorable environment for CTC proliferation. The implantation of isolated cells in vivo also may encourage cancer stem cell survival, self-renewal, proliferation and development into tumors. We anticipated that orthotropic implantation of the FMSA device at typical sites of metastatic colorectal cancer would provide salient proof of this point, but we were dismayed by the lack of growth vs. subcutaneous implantation (data not shown). Lack of appropriate microsurgical technique to implant cells in the most suitable location and/or the inability of the FMSA to actually be implanted into the desired tissue are likely to blame. Nevertheless, as shown in our results, the growth of CTCs in vivo could in the future potentially enable determination of efficacy of treatments on a patient’s CTCs—tumors that develop can be treated with agents and their response evaluated in real time. Although it may take some time for tumors to grow in vivo, the ability to test therapies against spiked cancer cells isolated from blood in this setting is significant, because certain antitumor agents’ effects can best be recapitulated in vivo (for example, cetuximab). Growing patient-derived CTCs as tumors in vivo was not attempted in this study and is clearly a future direction that is anticipated based on the results we describe.
Beyond allowing for chemotherapy testing, our in vitro and in vivo models could allow for increased biological understanding of CTCs. Our in vivo model could allow us to study, among other things, the heterogeneity, tumorigenicity and homing behavior of CTCs. Our in vitro culture model also could allow for molecular and functional CTC analysis not currently possible with the small numbers of cells isolated with other technologies.
Overall, the potential of the FMSA device in allowing the in vitro and in vivo culture of spiked cancer cells isolated from blood and their rapid chemosensitivity testing has herein been demonstrated. This approach is novel and significant, because in conventional cancer treatment, patients typically receive broad-acting cytotoxic treatments based on the location of tumor origin. These therapies are currently based on simplistic algorithms largely empirically derived and applied to the tissue of origin and a few other standardized tumor measures. Differences within the underlying malignant cells can result in significantly different responses to the same therapy. The analysis and identification of these characteristics through the testing of enriched viable CTCs amounts to a paradigm shift toward personalized and targeted cancer therapy according to the specific genetic defects of each patient’s malignancy, determined inexpensively and in real time. The progress made here is to demonstrate feasibility of viable cancer cell recovery, as well as a potential for spiked cancer cells to be cultured, expanded and analyzed as well as grown in vivo. There are significant challenges that remain for the field that we encountered, including the inability of the patient-derived recovered apparently viable CTCs (from the tumor types we tested in preliminary attempts) to proliferate when cultured. This is not expected to limit genomic analysis that may predict sensitivity to therapeutics or prognosis but is certainly a major challenge for expansion and testing of recovered patient-derived cancer cells against approved or novel therapeutic agents.
Materials and Methods
Chemicals and reagents
Penicillin, streptomycin, DMEM, McCoy’s 5A, RPMI 1640, PBS and trypsin were purchased from Cellgro. Fetal bovine serum (FBS) was bought from Gemini Bio-Products. Irinotecan, 5-fluorouracil and oxaliplatin from differing commercial sources were obtained through the Penn State Hershey Cancer Institute Infusion Pharmacy.
Cell culture
Cell lines were obtained from ATCC (ATCC). Unless otherwise noted, cells were propagated in ATCC-recommended media supplemented with 10% (v/v) fetal bovine serum, 100 µg/mL of penicillin and 100 µg/mL of streptomycin (P/S) in a humidified incubator at 5% CO2 and 37°C. GFP-labeled HCT-116 cells and RFP-labeled MCF-10A-HRAS were derived from parental lines as previously described.23
Capture and culture of patient-derived circulating tumor cells
Circulating tumor cells were isolated using the FMSA device following filtration of 7.5 mL of patient blood collected in an EDTA-coated tube. Immediately after filtration, the FMSA device, along with captured cells, was transferred to a 35 mm dish and cultured in similar conditions outlined above: 37°C, 5% CO2 with RPMI or DMEM, 10–30% FBS and 1% P/S. Alternatively, 7.5 mL of patient blood in an EDTA tube was directly transferred into an 154 mM NH4Cl/10 mM KHCO3/2 mM EDTA solution, incubated with shaking, centrifuged, lysed red blood cells aspirated, the pellet washed with PBS and peripheral blood mononuclear cells and CTCs cultured together in adherent conditions as described.
Immunofluorescence of patient-derived circulating tumor cells
Patient-derived circulating tumor cells were fixed with Cytofix/Cytoperm (BD Biosciences) for 30 min at 4°C, washed with PBS, permeabilized with modified PBT (PBS with 0.1% BSA and 0.2–0.3% Triton-X-100), blocked with goat serum at room temperature and incubated with common leukocyte antigen (CD45) or Cytokeratin 8,18 mouse monoclonal antibodies (Cell Signaling Technology) in PBT overnight at 4°C. After washing (PBS, PBT), cells were incubated in secondary antibody, R-phycoerythrin or Alexa-Fluor 488 (Invitrogen), at room temperature for 2 h, washed with PBS and dH2O, stained with DAPI, washed in dH2O and mounted with fluorescence mounting medium.
Cell viability assays
Cells were seeded in black-walled 96-well plates at a density of 100 cells/well (unless otherwise indicated) in 100 μL of medium and incubated overnight to allow for proper attachment. Twenty-four hours later, the medium of each well was replaced with 100 μL of new medium containing treatment (5-FU, oxaliplatin or irinotecan). Drugs were added in PBS vehicle, which itself never exceeded 0.5% of each well. After 24 h of treatment, the CellTiter-Glo bioluminescent cell proliferation assay (Promega Corporation), a modification of the MTT assay, was used according to the manufacturer’s protocol to quantify cell viability.
In vivo studies
SCiD hairless outbred female mice (Charles River Laboratories) were used for all experiments. For FMSA implants, mice were anesthetized per veterinary recommendation using a ketamine-acepromazine-xylazine cocktail. The rear dorsal flank of the mouse was prepared by: (1) cleansing the site with betadiene scrub, (2) rinsing the site with alcohol and (3) painting the site with betadiene solution. A < 1 cm incision was made in the skin of the mouse, and the skin was lifted from the underlying tissues using blunt dissection to create a small subcutaneous pocket. The FMSA device was implanted into this pocket and positioned as to alleviate mouse discomfort. The incision was closed using two 7 mm stainless steel wound clips. Post-operative analgesia was given using bupivacaine and carprofen. For standard xenografts, 200 µL of a 1:1 v/v mixture of cancer cells in PBS:matrigel was injected subcutaneously in the rear flank of mice following ketamine-xylazine anesthesia.
For tumor monitoring: both a multispectral fluorescent small animal imaging machine (Maestro, Cri) and digital calipers were used to monitor tumors. Tumor volume was calculated according to the modified formula for ellipsoid volume (volume = π/6 × length × width2). Ultrasound imaging: the Vevo 770 system (VisualSonics) was used according to the manufacturer’s specifications. Microvasculature was measured using Vevo MicroMarker contrast microbubbles. For tissue analysis, tissue was harvested from euthanized mice and fixed in 4% paraformaldehyde for 48 h. Tissue was paraffin-embedded, sectioned and stained by the Morphologic and Molecular Pathology Core Research Lab at the Penn State Milton S. Hershey Medical Center.
Statistical analysis
All results shown represent the average ± SD from triplicate experiments, unless otherwise indicated. Statistical analyses were performed using GraphPad Prism and either an unpaired, two-tailed Student’s t-test or ANOVA. All comparisons were made relative to untreated controls, and significant differences are indicated as * p < 0.05.
Supplementary Material
Acknowledgments
This work was presented in part at the 102nd meeting of the American Association for Cancer Research meeting in Chicago, IL in April 2012. This work was supported by grants to W.S.E-D., as well as funds from the Penn State Hershey Cancer Institute. W.S.E-D. is an American Cancer Society Research Professor.
Blood for patient-derived circulating tumor cell study was collected under one of the following clinical protocols: IRB Protocol No. 34969EP; Principal Investigator: Joseph Drabick, MD; Title: General Protocol for Acquisition of Whole Blood for the Detection and Characterization of Circulating Tumor Cells and/or Tumor Specific Immunity/Immune Cell Activation for any Malignancy at any Stage of Disease (PSHCI 10-061). IRB Protocol No. 34902EP; Principal Investigator: Wafik S. El-Deiry, MD, PhD, FACP Title: Prevalence of Stem Cell and Prognostic Markers in Circulating Tumor Cells of Patients with Metastatic Colorectal Cancer Undergoing Chemotherapy (PSHCI 10-066).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25165
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Hüsemann Y, Geigl JB, Schubert F, Musiani P, Meyer M, Burghart E, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Baldus SE, Schaefer K-L, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16:790–9. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- 4.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 5.Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res. 2007;13:7053–8. doi: 10.1158/1078-0432.CCR-07-1506. [DOI] [PubMed] [Google Scholar]
- 6.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–21. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 7.Tan SJ, Yobas L, Lee GY, Ong CN, Lim CT. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed Microdevices. 2009;11:883–92. doi: 10.1007/s10544-009-9305-9. [DOI] [PubMed] [Google Scholar]
- 8.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–9. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci USA. 2010;107:18392–7. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn P, Bethel K. A fluid biopsy as investigating technology for the fluid phase of solid tumors. Phys Biol. 2012;9:010301. doi: 10.1088/1478-3975/9/1/010301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischer RL, Price PB, Symes EM. Novel Filter for Biological Materials. Science. 1964;143:249–50. doi: 10.1126/science.143.3603.249. [DOI] [PubMed] [Google Scholar]
- 12.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, et al. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16:5011–8. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harouaka R, Zhou M-D, Khan W, Yeh Y-T, Zheng S-Y. A flexible micro spring array (FMSA) device for the high throughput enrichment of viable CTCs. 103rd Annual Meeting of the American Association for Cancer Research; Chicago: AACR; 2012. [Google Scholar]
- 14.Harouaka R, Zhou M-D, Zhang J, Khan W, Zheng S-Y. Development of a microfiltration system for the improved detection and viable capture of metastatic circulating tumor cells. 102nd Annual Meeting of the American Association for Cancer Research; Orlando: AACR; 2011. [Google Scholar]
- 15.Patel AS, Allen JE, Dicker DT, Peters KL, Sheehan JM, Glantz MJ, et al. Identification and enumeration of circulating tumor cells in the cerebrospinal fluid of breast cancer patients with central nervous system metastases. Oncotarget. 2011;2:752–60. doi: 10.18632/oncotarget.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirby BJ, Jodari M, Loftus MS, Gakhar G, Pratt ED, Chanel-Vos C, et al. Functional characterization of circulating tumor cells with a prostate-cancer-specific microfluidic device. PLoS One. 2012;7:e35976. doi: 10.1371/journal.pone.0035976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng S, Lin HK, Liu J-Q, Balic M, Datar R, Cote RJ, et al. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A. 2007;1162:154–61. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 18.Lin HK, Zheng S, Williams AJ, Balic M, Groshen S, Scher HI, et al. Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin Cancer Res. 2010;16:5011–8. doi: 10.1158/1078-0432.CCR-10-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng S, Lin HK, Lu B, Williams A, Datar RH, Cote RJ, et al. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed Microdevices. 2011;13:203–13. doi: 10.1007/s10544-010-9485-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–3. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oien KA. Pathologic evaluation of unknown primary cancer. Semin Oncol. 2009;36:8–37. doi: 10.1053/j.seminoncol.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Matthew EM, Zhou L, Yang Z, Lamparella N, Dicker DT, Yiu LL, et al. A Multiplexed Marker-based Algorithm for Diagnosis of Carcinoma of Unknown Primary Using CTCs. 103rd Annual Meeting of the American Association for Cancer Research; Chicago: AACR; 2012. [Google Scholar]
- 23.Navaraj A, Finnberg N, Dicker DT, Yang W, Matthew EM, El-Deiry WS. Reduced cell death, invasive and angiogenic features conferred by BRCA1-deficiency in mammary epithelial cells transformed with H-Ras. Cancer Biol Ther. 2009;8:2417–44. doi: 10.4161/cbt.8.24.10850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.