Abstract
Members of the phylum Cyanobacteria inhabit ecologically diverse environments. However, the CRISPR-Cas (clustered regularly interspaced short palindromic repeats, CRISPR associated genes), an extremely adaptable defense system, has not been surveyed in this phylum. We analyzed 126 cyanobacterial genomes and, surprisingly, found CRISPR-Cas in the majority except the marine subclade (Synechococcus and Prochlorococcus), in which cyanophages are a known force shaping their evolution. Multiple observations of CRISPR loci in the absence of cas1/cas2 genes may represent an early stage of losing a CRISPR-Cas locus. Our findings reveal the widespread distribution of their role in the phylum Cyanobacteria and provide a first step to systematically understanding CRISPR-Cas systems in cyanobacteria.
Keywords: Cas, CRISPR, cyanobacteria, cyanophage, adaptive immunity
Introduction
CRISPR (clustered regularly interspaced short palindromic repeats) loci and cas (CRISPR associated) operons together form a heritable adaptive immunity system found in many bacteria and most archaea.1,2 The CRISPR-Cas system proceeds through three steps: acquisition, expression and interference. In the acquisition step, foreign nucleic acid fragments are incorporated as new direct repeat-spacer units into a CRISPR locus. The CRISPR locus can be transcribed constitutively or triggered by the invading virus or a foreign plasmid. The resulting mRNA is processed and then used as a guide for degradation of foreign nucleic acids. Genes cas1 and cas2 are widely used as diagnostic markers for the presence of CRISPR-Cas systems1,3,4 and are proposed to be involved only in the acquisition step but not in the interference process.5,6 Based on the phylogenetic analysis of the Cas1 protein, cas operon organization, signature genes other than cas1/2, and the interference mechanism, the CRISPR-Cas system has been classified into three major types (I, II and III), each having several subtypes.3 Regardless of the classification, this nucleic acid-based mechanism shares many functional similarities to RNA interference found in eukaryotic organisms. Recently, increasing attention from clinical microbiologists, ecologists and evolutionary biologists has been directed toward the CRISPR-Cas system because of its many potential uses such as the detection and genotyping of microbial pathogens,7-9 host identification in metagenomes, analysis of viral genomes10-14 and targeted genome engineering in both prokaryotic and eukaryotic cells.15-19 However, the CRISPR-Cas system in the Cyanobacteria, which is one of the most metabolically and morphologically diverse of the bacterial phyla, has not been systemically investigated. A recent sequencing initiative,20 aimed at improving the phylogenetic coverage and diversity of sequenced genomes of the Cyanobacteria, prompted us to survey CRISPR-Cas systems across this ecologically diverse phylum.
Results and Discussion
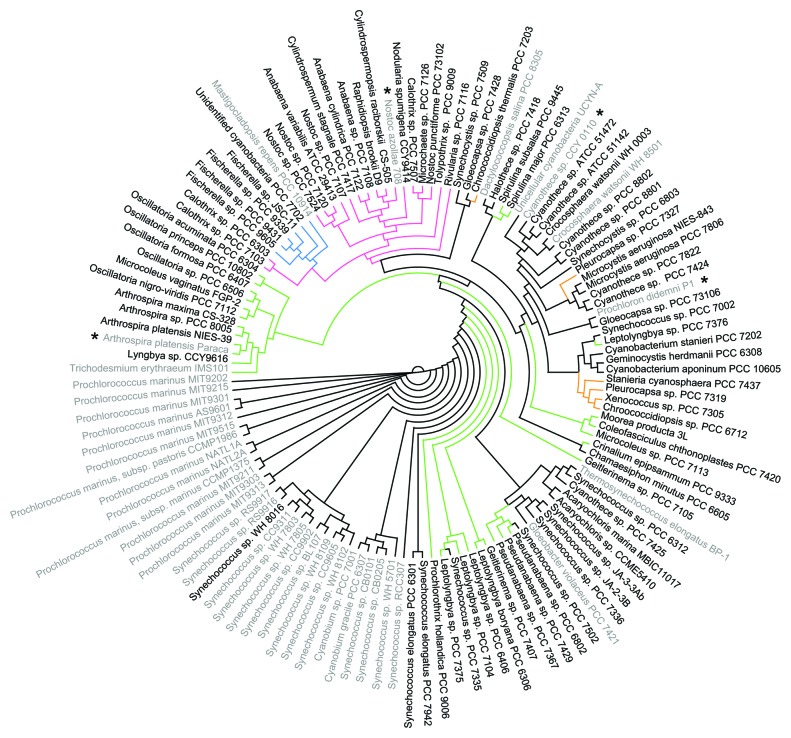
The phylum Cyanobacteria has been divided into five subsections based on cell morphology: I (Unicellular), unicellular strains that undergo binary fission; II (Baeocystous), unicellular strains that perform multiple fissions; III (Filamentous), filamentous strains that only contain vegetative cells; IV (Heterocystous), filamentous strains with differentiated cells (e.g., nitrogen fixing heterocysts) and V (Ramified), branching filamentous strains with differentiated cells.21 Subsection I can be further divided into two subclades based on the type of CO2 fixation enzyme, ribulose-1,5-bisphosphate carboxylase oxygenase (RuBisCO), that they harbor: the marine Synechococcus and Prochlorococcus (subsection I Pro/Syn) and subsection I non-Pro/Syn. Evidence of the CRISPR-Cas system was found in a majority of sequenced cyanobacterial genomes (86 out of 126) except the Pro/Syn subclade (Figs. 1 and 2; Table S1). This result revealed an apparent paradox about marine Synechococcus and Prochlorococcus: they live in an environment replete with cyanophages,22-24 but they almost exclusively lack CRISPRs (the only exception is Synechococcus sp WH8016, with one predicted CRISPR locus and one cas cluster but no cas1 or cas2 genes). Very recently, Weinberger et al. proposed a mathematical model suggesting a very high level of viral diversity will outrun the CRISPR-Cas immune system.25 Their model might be able to explain this paradox, but it has not been tested in this particular case. On the other hand, considering the relatively smaller genome size (p < 0.001 in comparison to all other five groups) of marine Synechococcus and Prochlorococcus, another possible explanation is that Pro/Syn might use other bacteriophage resistance mechanisms that involve less genetic load. For example, they could prevent phage adsorption or use restriction-modification (R-M) systems. However, both marine Synechococcus and Prochlorococcus have no or a limited number of R-M systems (the restriction enzyme data BASE, http://rebase.neb.com).26 Recent studies on the phage-resistant strains of marine Synechococcus and Prochlorococcus suggested this resistance is most likely due to the changes in genes involved in phage attachment to the cell surface.27,28 In addition, Stazic et al. observed that endogenous antisense RNAs protect a set of mRNAs from degradation during phage infection in Prochlorococcus MED4.29 These findings shed some light on how marine unicellular cyanobacteria (Pro/Syn) coexist with their phages long-term, but further investigation is needed in order to fully elucidate the underlying mechanism.
Figure 1. Species tree of all cyanobacterial genomes used in this analysis. The names of genomes containing at least one CRISPR locus with multiple spacer units and in which a cas operon could be found are colored black, while the names of all genomes in which at least one of these two conditions was not met are colored gray. Branches are colored according to morphological Subsection: Unicellular (subsection I; black), Baeocystous (subsection II; orange), Filamentous (subsection III; green), Heterocystous (subsection IV; magenta) and Ramified (subsection V; blue). The tree was generated using 31 concatenated conserved proteins (Shih et al., 2012). *Please see Table S1 for additional information.
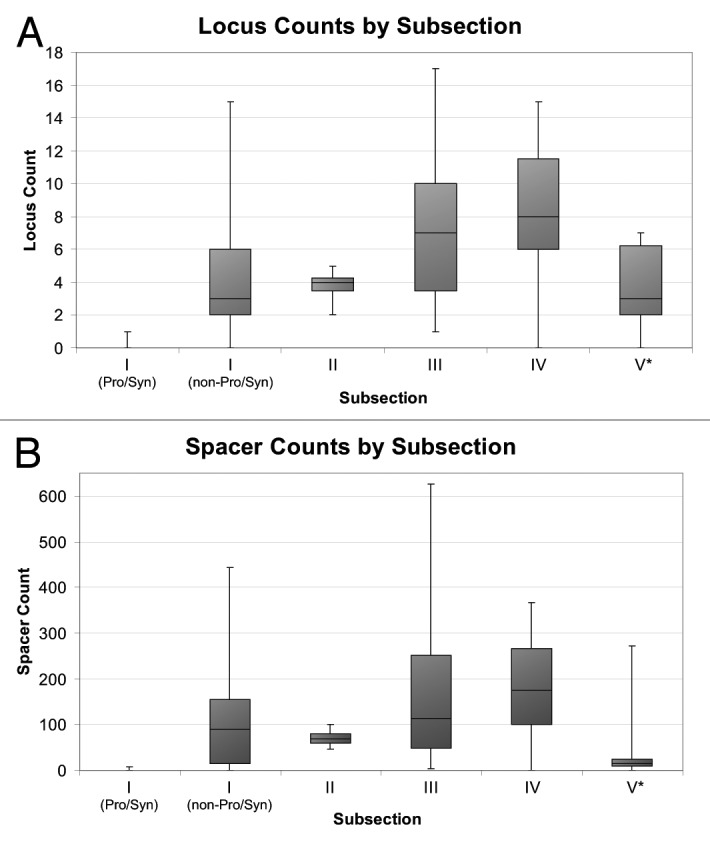
Figure 2. Box plot of CRISPR survey results by morphological subsection. Box plots depicting the range of numbers of (A) CRISPR loci and (B) Total number of spacers in the genome for all finished genomes in a given subsection. Draft genomes from each subsection were excluded. The whiskers contain the complete range of values, while the boxes contain the interquartile range, and the line within each box denotes the median count for the given subsection. Subsection I was divided into two categories: Pro/Syn (Prochlorococcus and Synechococcus) and non-Pro/Syn. *Due to the lack of finished subsection V genomes, draft genomes were used for subsection V counts.
After excluding the Pro/Syn clade, 85 (88.5%) of the remaining 96 (52 complete genomes and 44 draft genomes) of the Cyanobacteria with sequenced genomes are predicted to contain the CRISPR-Cas system (Fig. 1). These cyanobacteria inhabit a wide range of ecological niches. In general, cyanobacteria from subsection III and IV tend to have more CRISPR loci and a greater number of direct repeat-spacer units (Fig. 2). The numbers of CRISPR loci and direct repeat-spacer units did not appear to correlate with genome size; when counts are normalized for genome size, this trend continues for locus counts but is weaker for spacer counts (Fig. S1). However, when comparing the subsections for either the normalized or non-normalized data, the differences in these counts between the subsections are not statistically significant (p > 0.05), with the exception of subsection I Pro/Syn, where the counts are significantly less than those of the other subsections (p < 0.0001). Notably, subsection I strains (excluding marine Synechococcus and Prochlorococcus) contain similar numbers of spacers for their smaller genome size when compared with subsections III and IV (Fig. S1). The average lengths of a direct repeat sequence and a spacer found in members of the Cyanobacteria are approximately 34 and 40 nucleotides, respectively, typical values for other bacterial CRISPRs.4
In our survey, direct repeat sequences can be clustered into 409 distinct classes, among which only 140 (34.2%) are cataloged in the Rfam database (http://rfam.sanger.ac.uk/)30 (Table S2). These clusters hit to 12 of the 64 (18.8%) RNA families in the Rfam database. Excluding environmental sequences in RNA families, four (RF01371, RF01365, RF01347 and RF01329) of these 12 RNA families had previously contained only cyanobacterial direct repeats, while four (RF01318, RF01370, RF01343 and RF01331) had previously contained direct repeat sequences from cyanobacterial genomes as well as genomes of other phyla, and four (RF01322, RF01340, RF01342 and RF01359) had not previously contained any cyanobacterial direct repeats. Two hundred and sixty-nine (65.8%) of 409 cyanobacterial direct repeat clusters bore no significant similarity to an existing RNA family and represent novel direct repeats. Coleofasciculus chthonoplastes PCC 7420 contains the largest number of CRISPR loci observed in a cyanobacterial genome, with 23 predicted loci. This number is also higher than that of the reported current record holder, the thermophilic Archeaon Methanocaldococcus jannaschii, with 18 loci.1,2,31 The genome of Geitlerinema sp PCC 7105, a subsection III species that is a reference strain for marine species of Geitlerinema, contains 650 direct repeat-spacer units in its total of 15 CRISPR loci, and this is the highest number of units observed in any sequenced cyanobacterial genome. In contrast, the other sequenced Geitlerinema species, PCC 7407, contains only one CRISPR locus with 23 units (also discussed below). Unfortunately, the isolation sources of both Geitlerinema species are unknown,21 and it is not clear if such an extensive CRISPR system in Geitlerinema sp PCC 7105 is functional and why it needs to maintain so many direct repeat-spacer units. Two other species from subsection III also contain over 600 direct repeat-spacer units: Pseudanabaena sp PCC 7429 (with 610 units; isolated from sphagnum bog, near Kastanienbaum, Switzerland) and Spirulina sp PCC 9445 (with 625 units; isolated from hard sand of Lake Venere, Pantelleria island, Italy). It has been shown that for other CRISPR model organisms, such as Streptococcus thermophilus and Sulfolobus, the CRISPR loci are highly dynamic and can change rapidly.32,33 Constant challenge from largely diverse phages may result in the preservation of the corresponding CRISPR loci. Therefore, this may also explain the observation of many CRISPR loci in C. chthonoplastes PCC 7420, Pseudanabaena sp PCC 7429 and Spirulina sp PCC 9445. However, the majority of the cyanophages to which they may be exposed have not been characterized. According to the classification of the CRISPR-Cas system proposed by Makarova et al.,3 and using the signature cas gene of each subtype as a marker, 56 out of 86 CRISPR-Cas containing cyanobacterial genomes have subtype I-D system (Table S1), which is a rarely found subtype outside of the phylum Cyanobacteria. This can also be visualized on the phylogenetic tree of the Cas1 protein (Fig. S2). Subtypes I-A, III-A and III-B can be found in 22, 12 and 14 genomes, respectively. Subtypes I-B, I-F and II-A have not been found in the phylum Cyanobacteria. However, the accuracy of this subtype prediction is largely dependent on the quality of genome annotation. In many cases, subtype assignment is challenging, due to the diversity of CRISPR-Cas systems.
It has been previously reported that in many organisms, cas1 and cas2 genes are missing from the type III CRISPR-Cas operon, but Cas1 and Cas2 proteins could be provided in trans since these two genes can be found in an additional CRISPR-Cas operon of a different type (type I or type II) in the same genome.3 This scenario is also observed in cyanobacteria such as Oscillatoria sp PCC 7112. However, rather unexpectedly, the finished genomes of free-living cyanobacteria Geitlerinema sp PCC 7407 and Synechococcus sp WH8016 lack the cas1 and cas2 genes but have CRISPR loci and a putative operon containing other cas genes (Fig. 3A). This observation has not been previously reported, and it prompted us to survey all currently available complete bacterial and archaeal genomes in GenBank (2,045 non-cyanobacterial genomes as of September 5, 2012) for CRISPR-Cas systems. Our survey shows that 1,130 genomes (approximately 55%) were predicted to contain CRISPR loci, and 372 of these (approximately 33%) also lack cas1 and cas2 genes (Fig. 4). Of these, 73 (approximately 6.5% of genomes with CRISPRs) have other cas genes near the predicted CRISPR loci. This trend continues even when only genomes that contain multiple CRISPR loci, those with the least likelihood of false positives, are surveyed.
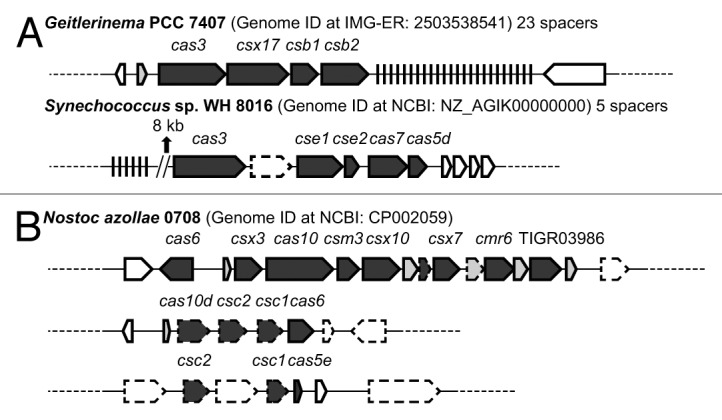
Figure 3. The cas gene and pseudogene organizations and CRISPR loci in Geitlerinema sp PCC 7407, Synechococcus sp WH8016 and Nostoc azollae 0708. (A) Although Geitlerinema sp PCC 7407 and Synechococcus sp WH8016 lack CRISPR signature genes cas1 and cas2, predicted CRISPR loci are present, and an operon containing cas genes, such as cas3 and cmr6, is co-present on the genomic scaffold. cas genes are shown in dark grey, genes encoding hypothetical proteins are shown in gray, and other genes are shown in white. Pseudogenes are shown with a dashed border. CRISPR loci are shown with short vertical black lines. Parallel diagonal lines represent a separation on the scaffold. Dashed lines indicate upstream or downstream sequences. Other cas genes found are csx17, csb1, csb2, which belong to subtype I-U, as well as cse1 and cse2, which belong to subtype I-E (see Table S4 in Makarova et al., 2011 for detail of classification and nomenclature of CRISPR-associated genes). (B) cas genes of Nostoc azollae 0708 are organized into three operons, each lacking cas1 and cas2 and containing at least two cas genes annotated as pseudogenes, but no CRISPR loci were predicted in this genome. Other cas genes found are csc1/2, csx10, csm3, cmr6 and csx3, which belong to Subtypes I-D, I-U, III-A, III-B and III-U, respectively (see Table S4 in Makarova et al. 2011 for detail of classification and nomenclature of CRISPR-associated genes). The gene csx7 (TIGR02581) belongs to RAMP superfamily,4 and the gene indicated as “TIGR03986” is a cas5-like RAMP.
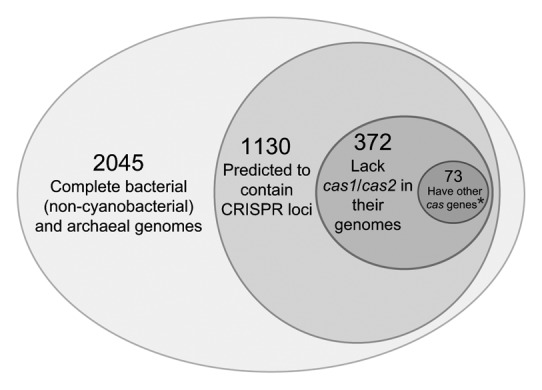
Figure 4. Venn diagram of the survey of GenBank for CRISR-Cas systems. All bacterial and archaeal genomes, excluding Cyanobacteria were examined for the presence of predicted CRISPR loci, for cas1 and cas2, and for presence of other cas genes. *These 73 genomes lack cas1 and cas2, but other cas genes are found in the vicinity of a CRISPR locus.
This result and our findings in cyanobacterial genomes suggest that using solely cas1 and cas2 genes as the diagnostic marker for identification may underestimate the presence of CRISPR-Cas defense systems. Although the underlying mechanism of the CRISPR-Cas system has not been fully elucidated, it has been shown that acquisition of new repeat-spacer units and loss of existing direct repeat-spacer units are highly dynamic in response to the environment.32-34 This leads to a possible explanation: the loss of cas1 and cas2 genes may be the first step in losing the CRISPR-Cas system. An alternative explanation is that these genomes have a different mechanism for acquisition of novel spacers that has not yet been discovered. We observed several interesting features in the genome of Nostoc azollae 0708 that could be explained by the first hypothesis. Nostoc azollae 0708 is an obligate symbiont; its genome is in an eroding state, containing many pseudogenes and fragmented operons.35,36 The cas genes of this genome are organized into three operons (Fig. 3B), each lacking cas1 and cas2 and containing at least two cas genes annotated as pseudogenes, but no CRISPR loci were predicted in this genome. Perhaps Nostoc azollae 0708 provides a snapshot of a step in the process of losing a CRISPR-Cas system: in the absence of selective pressure, the CRISPR locus, cas1 and cas2 are lost first, followed by the degradation of other cas genes. A similar example is that of Dactylococcopsis salina PCC 8305, a cyanobacterium originally isolated from a stratified heliothermal saline pool.37 Neither cas1 and cas2 genes nor any CRISPR loci are found in this finished genome, but three cas pseudogenes are present at two locations.
We attempted to identify sequences in publicly available databases homologous to the predicted spacers from the cyanobacteria. Of the 12,586 spacers queried, only 49 bore homology to sequences from refseq_genomic, env_nt or gss (Table S3). Of note, one spacer from Leptolyngbya sp PCC 6306 bore significant homology to a sequence in the refseq_genomic database from the genome of Phormidium phage Pf-WMP4, which is known to infect Leptolyngbya foveolarum.38 No significant homology was found to any other viral genomes in refseq_genomic (total of 3091 viral genomes, including 36 cyanophage genomes). When searched against env_nt, in several cases, duplications of CRISPR loci found in cyanobacteria appear in metagenomic sequences from similar environments. For example, large portions of two CRISPR loci from the CRISPR-replete genome of C. chthonoplastes PCC 7420, which was isolated from a salt marsh in Woods Hole, MA (see organism information at Genome Online Database, www.genomesonline.org, GOLD CARD ID: Gi01423), have strong homologs in three metagenomes isolated from saline microbial mats in Guerrero Negro, Baja California Sur, Mexico (Table S3).39 The conserved order of spacers in these homologous loci indicates that the CRISPR loci in these metagenomes share a common origin with those in C. chthonoplastes PCC 7420. However, because this organism has been observed in many microbial mats globally, it is also likely that this organism is present in this mat, thus explaining the presence of these CRISPR loci. Similarly, one locus from the genome of Synechococcus sp JA-3-3Ab is also homologous to a CRISPR locus in a contig of a metagenome isolated from the mushroom and octopus hotsprings in Yellowstone National Park. Strains closely related to this genome are known to be present in the corresponding metagenome, thus explaining this synteny.40 None of the spacers bore significant homology to non-cyanobacterial plasmids in the refseq_genomic database, though several spacers were homologous to plasmid sequences in other cyanobacterial genomes (Table S3). These results reveal that the phage communities challenging cyanobacteria remain largely uncharacterized.
The Cyanobacteria is arguably one of the most ecophysiologically diverse phyla, inhabiting a myriad of environments, such as freshwater, marine, hypersaline, desert and tundra. As one of the oldest lineages of life, the Cyanobacteria have diverged considerably in morphology, metabolism and lifestyle and play major roles in global biogeochemical cycles. The evidence that the CRISPR-Cas immunity system is found in the majority of cyanobacterial genomes sequenced to-date, with the only exception of the marine subclade, indicates that CRISPR-mediated phage-host interaction has been a previously underappreciated force in cyanobacterial evolution. Very recently, mechanisms of CRISPR-Cas processing in two cyanobacterial model strains were studied via RNaseq and northern hybridization;41,42 evolution of CRISPR-Cas systems in closely related cyanobacteria strains were also investigated via comparative genomic analysis.41,43,44 These studies are the commencement of our understanding of how CRISPR-Cas systems function in cyanobacteria.
Materials and Methods
CRISPR loci were predicted for 126 cyanobacterial genomes (54 draft genomes and 72 finished genomes) using an in-house implementation of CRISPRFinder45 run according to the default settings. CRISPR clusters were predicted by identifying and merging “maximal repeats,” units of one spacer flanked by two direct repeats. The consensus direct repeat for each CRISPR was determined, from which the sequence of each direct repeat in the CRISPR locus was determined. From this information, the spacer sequence was predicted, if the spacer length was 0.6–2.5 times the size of the direct repeat consensus. Finally, these possible CRISPR loci were predicted to be CRISPRs if the CRISPR locus did not appear to be a tandem repeat, the locus had last least three spacers, and at least two of the direct repeats were identical. The presence of CRISPR-Cas systems was confirmed by examining the co-existence of predicted CRISPR loci and the ubiquitous CRISPR-associated (cas) genes, namely cas1 and cas2, within a genome. Where only the former criterion was met, we manually inspected the genome to search for a putative cas operon. When a cas operon was observed, we considered the genome to have a CRISPR-Cas system. We did not observe any cases where cas1 and cas2 were present in a genome where there was no predicted CRISPR locus. A one-way ANOVA test with Tukey’s post-test was performed in comparison of non-normalized (Fig. 2) and normalized (Fig. S1) locus counts and spacer counts among different subsections. While counts in subsection I Pro/Syn are extremely different from those of all other subsections (p < 0.0001), differences between all other subsections are not statistically significant (p > 0.05).
All complete non-cyanobacterial bacterial and archaeal genomes (2,045 genomes) were downloaded from GenBank on September 5, 2012 and were also examined for the presence of CRISPRs using CRISPRFinder. Of these, 1,130 were predicted to contain CRISPRs. cas1 and cas2 genes were identified by means of searching the Escherichia coli K12 cas1 and cas2 genes against the nucleotide sequences of these genomes using tblastn46 at an e-value cutoff of 1. Additional cas1 and cas2 genes for each genome were also identified by retrieving the cas1 (PF01867) and cas2 (PF09827) Pfam domains47 and using the HMMER48 program hmmsearch on Mobyle49 at an e-value cutoff of one to search the NR protein database for matches to the domains.
To survey the GenBank genomes for presence of other cas genes in the vicinity of the predicted CRISPRs, tblastn with an e-value cut-off of 1e-02 was used to search a list of representative sequences for each cas gene listed by Makarova et al.3 Additional cas homologs were found using hmmsearch and the TIGRFAM50 models for each cas gene listed by Makarova et al., when available. This list was then filtered to only contain homologs found within 3,000 base pairs upstream or downstream of a predicted CRISPR.
Conserved CRISPR Direct Repeat (DR) sequences for each cyanobacterial genome (586 sequences) were extracted from the CRISPRFinder results and stringently clustered at 95% sequence identity and 95% sequence coverage (with mean DR size of 34 nucleotides, this on average permits one to two nucleotide difference in length and sequence) using BLASTCLUST with a word size of 7. These sequences were sorted into 409 clusters. All sequences for each cluster were aligned using the default settings of R-Coffee,51 and a consensus sequence was generated using ViennaRNA 2.1.1.52 The consensus sequence for each cluster was selected and searched against Rfam 11.053 using Rfam Scan at an e-value cutoff of 1. One hundred and forty of these clusters had significant hits to CRISPR direct repeats deposited in Rfam. These clusters hit to a total of 12 out of the 64 direct repeats RNA families currently in Rfam. Secondary structure predictions of cluster consensus sequences were generated using RNAalifold in the ViennaRNA package with no lonely pairs. These were used to predict if the consensus sequence forms a stem-loop structure (Table S2).
To survey CRISPR spacers for sequences that had homologs in publicly available sequence databases, copies of the NCBI blast databases NCBI Reference Sequence Project genomic sequences (refseq_genomic), environmental sample sequences (env_nt) and the Genome Survey Sequence (gss) were downloaded from NCBI. Spacers were searched against these sequence databases and all other cyanobacterial genomes examined in this study using blastall at an e-value of 1e-6.
Phylogenetic analysis on Cas1 proteins was performed by using 184 Cas1 protein sequences from the phylum Cyanobacteria and 215 non-cyanobacterial representatives of Cas proteins used in Makarova et al. review.3 All sequences were downloaded from IMG-ER (http://img.jgi.doe.gov/er), and the maximum likelihood tree was constructed using the PHYML program.54
Supplementary Material
Acknowledgments
We thank Patrick Shih for providing the cyanobacterial species tree used in Figure 1 and Jan Zarzycki for critical reading of the manuscript. We also acknowledge Christine Pourcel and Christine Drevet for providing the in-house version of CRISPRFinder used in this analysis for the identification of cyanobacterial CRISPRs. C.A.K. and F.C. were supported by the NSF (MCB0851094).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24571
References
- 1.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–6. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 3.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 7.Fabre L, Zhang J, Guigon G, Le Hello S, Guibert V, Accou-Demartin M, et al. CRISPR typing and subtyping for improved laboratory surveillance of Salmonella infections. PLoS One. 2012;7:e36995. doi: 10.1371/journal.pone.0036995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hauck Y, Soler C, Jault P, Mérens A, Gérome P, Nab CM, et al. Diversity of Acinetobacter baumannii in four French military hospitals, as assessed by multiple locus variable number of tandem repeats analysis. PLoS One. 2012;7:e44597. doi: 10.1371/journal.pone.0044597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Abadia E, Refregier G, Tafaj S, Boschiroli ML, Guillard B, et al. Mycobacterium tuberculosis complex CRISPR genotyping: improving efficiency, throughput and discriminative power of ‘spoligotyping’ with new spacers and a microbead-based hybridization assay. J Med Microbiol. 2010;59:285–94. doi: 10.1099/jmm.0.016949-0. [DOI] [PubMed] [Google Scholar]
- 10.Rho M, Wu YW, Tang H, Doak TG, Ye Y. Diverse CRISPRs evolving in human microbiomes. PLoS Genet. 2012;8:e1002441. doi: 10.1371/journal.pgen.1002441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberger AD, Sun CL, Pluciński MM, Denef VJ, Thomas BC, Horvath P, et al. Persisting viral sequences shape microbial CRISPR-based immunity. PLoS Comput Biol. 2012;8:e1002475. doi: 10.1371/journal.pcbi.1002475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berg Miller ME, Yeoman CJ, Chia N, Tringe SG, Angly FE, Edwards RA, et al. Phage-bacteria relationships and CRISPR elements revealed by a metagenomic survey of the rumen microbiome. Environ Microbiol. 2012;14:207–27. doi: 10.1111/j.1462-2920.2011.02593.x. [DOI] [PubMed] [Google Scholar]
- 13.Anderson RE, Brazelton WJ, Baross JA. Using CRISPRs as a metagenomic tool to identify microbial hosts of a diffuse flow hydrothermal vent viral assemblage. FEMS Microbiol Ecol. 2011;77:120–33. doi: 10.1111/j.1574-6941.2011.01090.x. [DOI] [PubMed] [Google Scholar]
- 14.Snyder JC, Bateson MM, Lavin M, Young MJ. Use of cellular CRISPR (clusters of regularly interspaced short palindromic repeats) spacer-based microarrays for detection of viruses in environmental samples. Appl Environ Microbiol. 2010;76:7251–8. doi: 10.1128/AEM.01109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–2. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 16.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–9. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–9. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc Natl Acad Sci USA. 2012 doi: 10.1073/pnas.1217107110. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. J Gen Microbiol. 1979;111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- 22.Proctor LM, Fuhrman JA. Viral Mortality of Marine-Bacteria and Cyanobacteria. Nature. 1990;343:60–2. doi: 10.1038/343060a0. [DOI] [Google Scholar]
- 23.Suttle CA, Chan AM. Marine Cyanophages Infecting Oceanic and Coastal Strains of Synechococcus - Abundance, Morphology, Cross-Infectivity and Growth-Characteristics. Mar Ecol Prog Ser. 1993;92:99–109. doi: 10.3354/meps092099. [DOI] [Google Scholar]
- 24.Suttle CA, Chan AM. Dynamics and Distribution of Cyanophages and Their Effect on Marine Synechococcus spp. Appl Environ Microbiol. 1994;60:3167–74. doi: 10.1128/aem.60.9.3167-3174.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinberger AD, Wolf YI, Lobkovsky AE, Gilmore MS, Koonin EV. Viral diversity threshold for adaptive immunity in prokaryotes. MBio. 2012;3:e00456–12. doi: 10.1128/mBio.00456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE--a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38(Database issue):D234–6. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoddard LI, Martiny JB, Marston MF. Selection and characterization of cyanophage resistance in marine Synechococcus strains. Appl Environ Microbiol. 2007;73:5516–22. doi: 10.1128/AEM.00356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avrani S, Wurtzel O, Sharon I, Sorek R, Lindell D. Genomic island variability facilitates Prochlorococcus-virus coexistence. Nature. 2011;474:604–8. doi: 10.1038/nature10172. [DOI] [PubMed] [Google Scholar]
- 29.Stazic D, Lindell D, Steglich C. Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection. Nucleic Acids Res. 2011;39:4890–9. doi: 10.1093/nar/gkr037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burge SW, Daub J, Eberhardt R, Tate J, Barquist L, Nawrocki EP, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41(Database issue):D226–32. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–73. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 32.Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, et al. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Sanchez MJ, Sauvage E, Da Cunha V, Clermont D, Ratsima Hariniaina E, Gonzalez-Zorn B, et al. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol Microbiol. 2012;85:1057–71. doi: 10.1111/j.1365-2958.2012.08172.x. [DOI] [PubMed] [Google Scholar]
- 34.Kuno S, Yoshida T, Kaneko T, Sako Y. Intricate interactions between the bloom-forming cyanobacterium Microcystis aeruginosa and foreign genetic elements, revealed by diversified clustered regularly interspaced short palindromic repeat (CRISPR) signatures. Appl Environ Microbiol. 2012;78:5353–60. doi: 10.1128/AEM.00626-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ran L, Larsson J, Vigil-Stenman T, Nylander JA, Ininbergs K, Zheng WW, et al. Genome erosion in a nitrogen-fixing vertically transmitted endosymbiotic multicellular cyanobacterium. PLoS One. 2010;5:e11486. doi: 10.1371/journal.pone.0011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsson J, Nylander JA, Bergman B. Genome fluctuations in cyanobacteria reflect evolutionary, developmental and adaptive traits. BMC Evol Biol. 2011;11:187. doi: 10.1186/1471-2148-11-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walsby A, van Rijn J, Cohen Y. The Biology of a New Gas-Vacuolate Cyanobacterium, Dactylococcopsis salina sp. nov., in Solar Lake. Proc R Soc Lond B Biol Sci. 1983;217:417–47. doi: 10.1098/rspb.1983.0019. [DOI] [Google Scholar]
- 38.Liu X, Shi M, Kong S, Gao Y, An C. Cyanophage Pf-WMP4, a T7-like phage infecting the freshwater cyanobacterium Phormidium foveolarum: complete genome sequence and DNA translocation. Virology. 2007;366:28–39. doi: 10.1016/j.virol.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 39.Kunin V, Raes J, Harris JK, Spear JR, Walker JJ, Ivanova N, et al. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol Syst Biol. 2008;4:198. doi: 10.1038/msb.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klatt CG, Wood JM, Rusch DB, Bateson MM, Hamamura N, Heidelberg JF, et al. Community ecology of hot spring cyanobacterial mats: predominant populations and their functional potential. ISME J. 2011;5:1262–78. doi: 10.1038/ismej.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hein S, Scholz I, Voß B, Hess WR. Adaptation and modification of three CRISPR loci in two closely related cyanobacteria. RNA Biol. 2013;10 doi: 10.4161/rna.24160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scholz I, Lange SJ, Hein S, Hess WR, Backofen R. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PLoS One. 2013;8:e56470. doi: 10.1371/journal.pone.0056470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romano C, D’Imperio S, Woyke T, Mavromatis K, Lasken R, Shock EL, et al. Comparative Genomic Analysis of Phylogenetically Closely-Related Hydrogenobaculum sp. from Yellowstone National Park. Appl Environ Microbiol. 2013 doi: 10.1128/AEM.03591-12. [epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trautmann D, Voss B, Wilde A, Al-Babili S, Hess WR. Microevolution in cyanobacteria: re-sequencing a motile substrain of Synechocystis sp. PCC 6803. DNA Res. 2012;19:435–48. doi: 10.1093/dnares/dss024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35(Web Server issue):W52-7. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39(Web Server issue):W29-37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Néron B, Ménager H, Maufrais C, Joly N, Maupetit J, Letort S, et al. Mobyle: a new full web bioinformatics framework. Bioinformatics. 2009;25:3005–11. doi: 10.1093/bioinformatics/btp493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, et al. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35(Database issue):D260–4. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilm A, Higgins DG, Notredame C. R-Coffee: a method for multiple alignment of non-coding RNA. Nucleic Acids Res. 2008;36:e52. doi: 10.1093/nar/gkn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39(Database issue):D141–5. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.