Abstract
CRISPR-Cas is a rapidly evolving RNA-mediated adaptive immune system that protects bacteria and archaea against mobile genetic elements. The system relies on the activity of short mature CRISPR RNAs (crRNAs) that guide Cas protein(s) to silence invading nucleic acids. A set of CRISPR-Cas, type II, requires a trans-activating small RNA, tracrRNA, for maturation of precursor crRNA (pre-crRNA) and interference with invading sequences. Following co-processing of tracrRNA and pre-crRNA by RNase III, dual-tracrRNA:crRNA guides the CRISPR-associated endonuclease Cas9 (Csn1) to cleave site-specifically cognate target DNA. Here, we screened available genomes for type II CRISPR-Cas loci by searching for Cas9 orthologs. We analyzed 75 representative loci, and for 56 of them we predicted novel tracrRNA orthologs. Our analysis demonstrates a high diversity in cas operon architecture and position of the tracrRNA gene within CRISPR-Cas loci. We observed a correlation between locus heterogeneity and Cas9 sequence diversity, resulting in the identification of various type II CRISPR-Cas subgroups. We validated the expression and co-processing of predicted tracrRNAs and pre-crRNAs by RNA sequencing in five bacterial species. This study reveals tracrRNA family as an atypical, small RNA family with no obvious conservation of structure, sequence or localization within type II CRISPR-Cas loci. The tracrRNA family is however characterized by the conserved feature to base-pair to cognate pre-crRNA repeats, an essential function for crRNA maturation and DNA silencing by dual-RNA:Cas9. The large panel of tracrRNA and Cas9 ortholog sequences should constitute a useful database to improve the design of RNA-programmable Cas9 as genome editing tool.
Keywords: tracrRNA, CRISPR-Cas, type II system, Cas9 (Csn1), RNA processing, RNA maturation, small non-coding RNA, bacteria, adaptive immunity, mobile genetic elements
Introduction
CRISPR-Cas (clustered regularly interspaced short palindromic repeats, CRISPR-associated) is a widespread RNA-mediated heritable and adaptive immune system against mobile genetic elements (phages, plasmids) in bacteria and archaea.1-15 A CRISPR-Cas locus is typically composed of an operon encoding the Cas proteins and a repeat-spacer array consisting of interspersed identical repeat sequences and unique invader-targeting spacer sequences. CRISPR-Cas immunity operates in three steps with the principle that an intruder once memorized by the system will be remembered and silenced upon a repeated infection.16 During the adaptation phase, a part of an invading nucleic acid sequence is incorporated as a new spacer within the repeat-spacer array and the infection is thus memorized.9,16-25 During the expression phase, the repeat-spacer array is transcribed as a precursor CRISPR RNA (pre-crRNA) molecule that undergoes processing to generate short mature crRNAs, each complementary to a unique invader sequence.26-35 During the interference phase, the individual crRNAs guide Cas protein(s) to cleave the cognate invading nucleic acids in a sequence-specific manner for their ultimate destruction.28,36-54
The CRISPR-Cas systems have recently been classified into three distinct types (I‒III), with further division into several subtypes according to specific combinations of cas genes.55-58 The classification reflects an evolution of the defense system into subtype-specific molecular mechanisms for expression and maturation of crRNAs and interference with invaders.55,56 Types I and III share some common features, with crRNAs and Cas proteins being the only known components required for the steps of expression and interference. Processing of pre-crRNAs involves a first cleavage within the repeats by an endoribonuclease of the Cas6 family and, in type III, the intermediate crRNAs are further maturated to produce shorter repeat-spacer crRNAs. In both types I and III, the mature crRNAs guide a complex of several Cas proteins to the cognate-invading nucleic acids and a Cas endonuclease of the ribonucleoprotein complex cleaves the target nucleic acids.27-37,41-53
Type II CRISPR-Cas has evolved distinct pre-crRNA processing and interference mechanisms. Our recent analysis of the type II-A system in the human pathogen Streptococcus pyogenes demonstrated that processing of pre-crRNA requires base-pairing of every pre-crRNA repeat with a small, non-coding, RNA, tracrRNA (trans-activating crRNA), encoded in the vicinity of the cas genes and repeat-spacer array.26 The tracrRNA:pre-crRNA repeat duplexes once formed are cleaved by the double-stranded RNA-specific endoribonuclease RNase III in the presence of Cas9 (formerly Csn1), hallmark protein of type II systems.26 The resulting intermediate crRNAs composed of repeat-spacer-repeat sequences are further trimmed into short mature crRNAs consisting of unique spacer-repeat sequences in a second maturation event that is yet to be described.26 Each mature crRNA remains duplexed to the processed tracrRNA, forming a dual-RNA structure that is associated with Cas9 in a ternary silencing complex.38 Cas9 is an unusual endonuclease13,16,17,38-40,54-56,58 that can be programmed by the dual-tracrRNA:crRNA structure to cleave site-specifically cognate target DNA using two distinct endonuclease domains (HNH and RuvC/RNase H-like domains).38
Our previous analysis of tracrRNA occurrence across genomes led to the identification of tracrRNA orthologs associated to type II CRISPR-Cas systems in several bacterial species.26 Expression of primary and mature forms of tracrRNA was validated experimentally by northern blot analysis in Streptococcus thermophilus, Streptococcus mutans, Listeria innocua and Neisseria meningitidis.26 Although diverse in length and sequence, tracrRNA orthologs share the common feature to contain an anti-pre-crRNA repeat sequence (anti-repeat).26 In this study, we first searched for all putative type II CRISPR-Cas loci existing in publicly available genomes by screening for sequences homologous to Cas9, the hallmark protein of the type II system. We constructed a phylogenetic tree from a multiple sequence alignment of the identified Cas9 orthologs. The CRISPR repeat length and gene organization of cas operons of the associated type II systems were analyzed in the different Cas9 subclusters. A subclassification of type II loci was proposed and further divided into subgroups based on the selection of 75 representative Cas9 orthologs. We then predicted tracrRNA sequences mainly by retrieving CRISPR repeat sequences and screening for anti-repeats within or in the vicinity of the cas genes and CRISPR arrays of selected type II loci. Comparative analysis of sequences and predicted structures of chosen tracrRNA orthologs was performed. Finally, we determined the expression and processing profiles of tracrRNAs and crRNAs from five bacterial species.
Results
Type II CRISPR-Cas systems are widespread in bacteria
In addition to the tracrRNA-encoding DNA and the repeat-spacer array, type II CRISPR-Cas loci are typically composed of three to four cas genes organized in an operon (Fig. 1). Cas9 is the signature protein characteristic for type II and is involved in the steps of expression and interference as mentioned above.13,16,17,26,38-40,54-56,58 Cas1 and Cas2 are core proteins that are shared by all CRISPR-Cas systems and are implicated in spacer acquisition.18,21,58 Csn2 and Cas4 are present in only a subset of type II systems and were suggested to play a role in adaptation.16 To retrieve a maximum number of type II CRISPR-Cas loci, possibly containing tracrRNA, we first screened publicly available genomes for sequences homologous to already described Cas9 proteins.55-58 Two hundred and thirty five putative Cas9 orthologs were identified in 203 bacterial species (Table S1). We discarded the incomplete, possibly truncated sequences, and selected a set of 75 diverse sequences representative of all retrieved Cas9 orthologs for further analysis (Table S1 and Materials and Methods). Consistent with previous observations,55-58 Cas9 orthologs were not found in archaeal genomes.
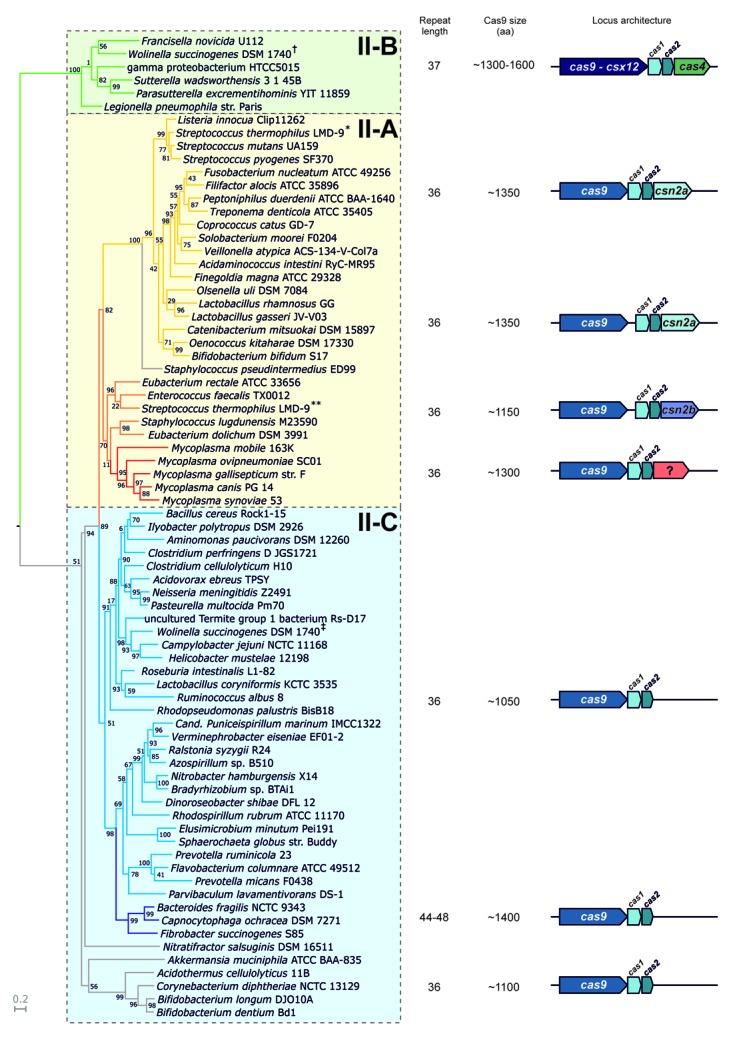
Figure 1. Phylogenetic tree of representative Cas9 sequences. See also Fig. S1 and Table S1. Bootstrap values calculated for each node are indicated. Same color branches represent selected subclusters of similar Cas9 orthologs. CRISPR repeat length in nucleotides, average Cas9 protein size in amino acids (aa) and consensus locus architecture are shown for every subcluster. *- gi|116628213, **- gi|116627542, †- gi|34557790, ‡- gi|34557932. Type II-A is characterized by cas9-csx12, cas1, cas2, cas4. Type II-B is characterized by cas9, cas1, cas2 followed by a csn2 variant. Type II-C is characterized by a conserved cas9, cas1, cas2 operon.
Next, we performed a multiple sequence alignment of the selected Cas9 orthologs (Fig. S1). The comparative analysis revealed high diversities in amino acid composition and protein size. The Cas9 orthologs share only a few identical amino acids and all retrieved sequences have the same domain architecture with a central HNH endonuclease domain and splitted RuvC/RNase H domain.13,16,17,38-40,54-56,58 The lengths of Cas9 proteins range from 984 (Campylobacter jejuni) to 1,629 (Francisella novicida) amino acids, with typical sizes of ~1,100 or ~1,400 amino acids. Due to the high diversity of Cas9 sequences, especially in the length of the inter-domain regions, we selected only well-aligned, informative positions of the prepared alignment to reconstruct a phylogenetic tree of the analyzed sequences (Fig. 1; Fig. S1 and Materials and Methods). Cas9 orthologs grouped into three major, monophyletic clusters with some outlier sequences. The observed topology of the Cas9 tree is well in agreement with the current classification of type II loci,56 with previously defined type II-A and type II-B forming separate, monophyletic clusters. To further characterize the clusters, we examined in detail the cas operon compositions and CRISPR repeat sequences of all listed strains.
Cas9 subclustering reflects the diversity in type II CRISPR-Cas loci architecture
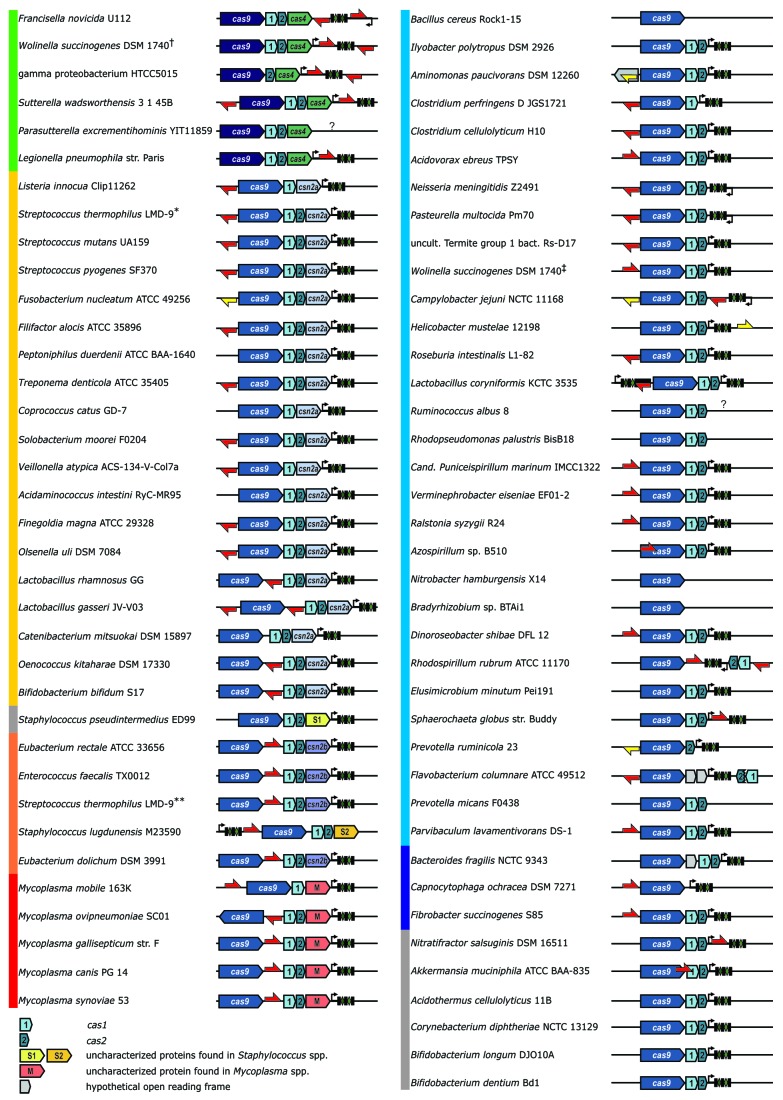
A deeper analysis of selected type II loci revealed that the clustering of Cas9 ortholog sequences correlates with the diversity in CRISPR repeat length. For most of the type II CRISPR-Cas systems, the repeat length is 36 nucleotides (nt) with some variations for two of the Cas9 tree subclusters. In the type II-B cluster (Fig. 1, green branches) that comprises loci encoding the long Cas9 ortholog, previously named Csx12, the CRISPR repeats are 37 nt long. The small subcluster composed of sequences from bacteria belonging to the Bacteroidetes phylum (Fig. 1, dark blue branches) is characterized by unusually long CRISPR repeats, up to 48 nt in size. Furthermore, we noticed that the subclustering of Cas9 sequences correlates with distinct cas operon architectures, as depicted in Figure 1. The third major cluster (Fig. 1, blue branches) and the outlier loci (Fig. 1, gray branches) consist mainly of the minimum operon composed of the cas9, cas1 and cas2 genes, with an exception of some incomplete loci that are discussed later. All other loci of the two first major clusters are associated with a fourth gene, mainly csn2-like, specific to type II-A and cas4, specific to type II-B (Fig. 1, clusters in yellow-orange-red and green, respectively). We identified genes encoding shorter variants of the Csn2 protein, Csn2a, within loci similar to type II-A S. pyogenes CRISPR01 (strain SF370, M1GAS) and S. thermophilus CRISPR3 (Fig. 1, yellow).13,26,55,59-61 The longer variant of Csn2, Csn2b, was found associated with loci similar to type II-A S. thermophilus CRISPR1 (Fig. 1, orange branches).55,62,63 Interestingly, we identified additional putative cas genes encoding proteins with no obvious sequence similarity to previously described Csn2 variants. One of those uncharacterized proteins is exclusively associated with type II-A loci of Mycoplasma species (Figs. 1 and 2, last gene of the cas operon in red). Two others were found encoded in type II-A loci of Staphylococcus species (Fig. 2, last gene of the cas operon in yellow and orange). Thus, in all cases, the cas operon architecture diversity is consistent with the subclustering of Cas9 sequences.
Figure 2. Architecture of type II CRISPR-Cas from selected bacterial species. The vertical color bars group the loci that code for Cas9 orthologs belonging to the same tree subcluster (compare with Figure 1). Thick black bar, leader sequence; black rectangles and green diamonds, repeat-spacer array. Note, that for simplicity the depicted repeat-spacer arrays do not represent the actual amount of spacers. Predicted anti-repeats are represented by arrows indicating the direction of putative tracrRNA ortholog transcription; red and yellow, high and low complementarity to the pre-crRNA repeat, respectively. Note that for the loci that were not verified experimentally, the CRISPR repeat-spacer array is considered here to be transcribed from the same strand as the cas operon. The transcription direction of the putative tracrRNA ortholog is indicated accordingly. *- gi|116628213, **- gi|116627542, †- gi|34557790, ‡- gi|34557932.
These characteristics together with the general topology of the Cas9 tree divided into three major, distinct, monophyletic clusters, led us to propose a new, further division of the type II CRISPR-Cas system into three subtypes. Type II-A is associated with Csn2-like protein, type II-B is associated with Csx12-like Cas9 and Cas4 and type II-C only contains the minimal set of the cas9, cas1 and cas2 genes, as depicted in Figure 1.
In silico predictions of novel tracrRNA orthologs
Type II loci selected earlier based on the 75 representative Cas9 orthologs were screened for the presence of putative tracrRNA orthologs. Our previous analysis performed on a restricted number of tracrRNA sequences26 revealed that neither the sequences of tracrRNAs nor their localization within the CRISPR-Cas loci seemed to be conserved. However, as mentioned above, tracrRNAs are characterized by an anti-repeat sequence capable of base-pairing with each of the pre-crRNA repeats to form tracrRNA:pre-crRNA repeat duplexes that are cleaved by RNase III in the presence of Cas9.26 To predict novel tracrRNAs, we took advantage of this characteristic and used the following workflow: (1) screen for potential anti-repeats (sequence base-pairing with CRISPR repeats) within the CRISPR-Cas loci, (2) select anti-repeats located in the intergenic regions, (3) validate CRISPR anti-repeat:repeat base-pairing, and (4) predict promoters and Rho-independent transcriptional terminators associated with the identified tracrRNAs.
To screen for putative anti-repeats, we retrieved repeat sequences from the CRISPRdb database,64 or, when the information was not available, we predicted the repeat sequences using the CRISPRfinder software.65 The CRISPR prediction tools are based on the assumption that the repeat spacer array and the cas operon are transcribed from the same strand. However, in our previous study, we showed experimentally that the transcription direction of the repeat-spacer array compared with that of the cas operon varied among loci.26 Here, RNA sequencing analysis confirmed this observation. In some of the analyzed loci, namely in F. novicida, N. meningitidis and C. jejuni, the repeat-spacer array is transcribed in the opposite direction of the cas operon (see paragraph “Deep RNA sequencing validates expression of novel tracrRNA orthologs” and Figs. 2 and 3) while in S. pyogenes, S. mutans, S. thermophilus and L. innocua, the array and the cas operon are transcribed in the same direction. These are the only type II repeat-spacer array expression data available to date. Because tracrRNA and crRNA base pair at the level of anti-repeat:repeat, the directionality of pre-crRNA repeats allows to determine the orientation of cognate tracrRNAs. To predict the transcription direction of other repeat-spacer arrays, we considered the previous observation according to which the last repeats of the arrays are usually mutated.66 This remark is in agreement with the current spacer acquisition model, in which typically the first repeat of the array is duplicated upon insertion of a spacer sequence during the adaptation phase.18,67 We observed, however, that the predicted orientation of transcription for the N. meningitidis and C. jejuni repeat-spacer array would be opposite to the orientation determined experimentally (RNA sequencing and northern blot analysis). Moreover, in most of the cases we could detect potential promoters on both ends of the arrays. Based on these facts, we considered transcription of the repeat-spacer arrays to occur in the same direction as transcription of the cas operon, if not validated otherwise.
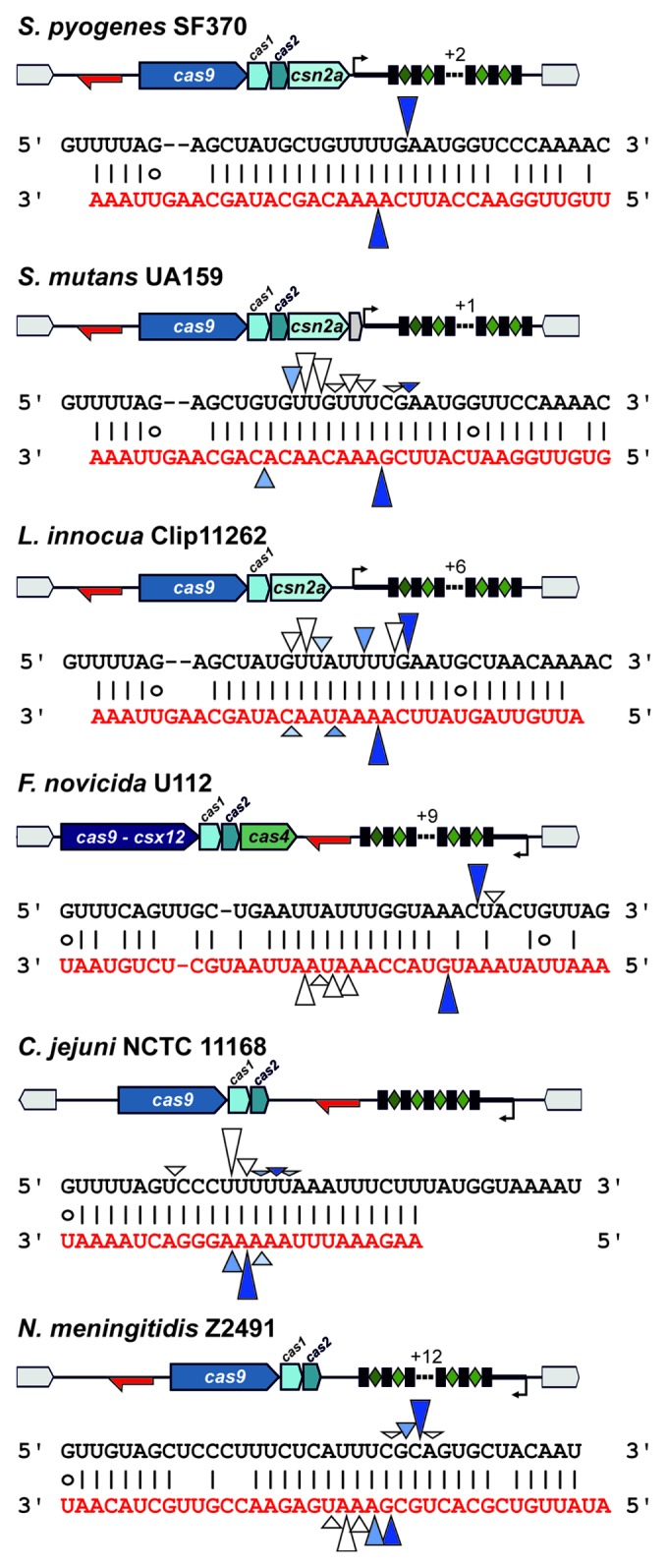
Figure 3. tracrRNA and pre-crRNA co-processing in selected type II CRISPR-Cas systems. CRISPR loci architectures with verified positions and directions of tracrRNA and pre-crRNA transcription are shown. Black sequences, pre-crRNA repeats; red sequences, tracrRNA sequences base-pairing with crRNA repeats. Putative RNA processing sites as revealed by RNA sequencing are indicated with arrowheads. For each locus, arrowhead sizes represent relative amounts of the retrieved 5′ and 3′-ends (see Table S3). Blue arrowhead pairs represent putative RNase III co-processing sites with dark blue indicating the putative primary processing sites.
We then screened the selected CRISPR-Cas loci including sequences located 1 kb upstream and downstream on both strands for possible repeat sequences that did not belong to the repeat-spacer array, allowing up to 15 mismatches. On average, we found one to three degenerated repeat sequences per locus that would correspond to anti-repeats of tracrRNA orthologs and selected the sequences located within the intergenic regions. The putative anti-repeats were found in four typical localizations: upstream of the cas9 gene, in the region between cas9 and cas1 and upstream or downstream of the repeat-spacer array (Fig. 2). For every retrieved sequence, we validated the extent of base-pairing formed between the repeat and anti-repeat (Table S2) by predicting the possible RNA:RNA interaction and focusing especially on candidates with longer and perfect complementarity region forming an optimal double-stranded structure for RNase III processing. To predict promoters and transcriptional terminators flanking the anti-repeat, we set the putative transcription start and termination sites to be included within a region located maximally 200 nt upstream and 100 nt downstream of the anti-repeat sequence, respectively, based on our previous observations.26 Due to the limitations of the in silico prediction algorithms, we could not determine the putative promoters and terminators for some of the analyzed anti-repeats, even though the tracrRNA ortholog expression could be validated experimentally, as exemplified by the C. jejuni locus (see paragraph “Deep RNA sequencing validates expression of novel tracrRNA orthologs”). We suggest considering promoter and transcriptional terminator predictions only as a supportive, but not essential, step of the guideline described above.
A plethora of tracrRNA orthologs
We predicted putative tracrRNA orthologs for 56 of the 75 loci selected earlier. The results of predictions are depicted in Figure 2. As already mentioned, the direction of tracrRNA transcription shown in this figure is hypothetical and based on the indicated direction of repeat-spacer array transcription. As previously stated, sequences encoding putative tracrRNA orthologs were identified either upstream, within or downstream of the cas operon, or downstream of the repeat spacer arrays including the putative leader sequences, the latter position found commonly in type II-B loci (Fig. 2). However, we observed that anti-repeats of similar localization within CRISPR-Cas loci can be transcribed in different directions (as observed when comparing e.g., Lactobacillus rhamnosus and Eubacterium rectale or Mycoplasma mobile and S. pyogenes or N. meningitidis) (Fig. 2). Notably, loci grouped within a same subcluster of the Cas9 guide tree share a common architecture with respect to the position of the tracrRNA-encoding gene. We identified anti-repeats around the repeat-spacer array in type II-B loci, and mostly upstream of the cas9 gene in types II-A and II-C with several notable exceptions for the putative tracrRNA located between cas9 and cas1 in three distinct subclusters of type II-A.
Some type II CRISPR-Cas loci have defective repeat-spacer arrays and/or tracrRNA orthologs
For six type II loci (Fusobacterium nucleatum, Aminomonas paucivorans, Helicobacter mustelae, Azospirillum sp., Prevotella ruminicola and Akkermansia muciniphila), we identified potential anti-repeats with weak base-pairing to the repeat sequence or located within the cas open reading frames (Fig. 2). Notably, in these loci, a weak anti-repeat within the open reading frame of the gene encoding a putative ATPase in A. paucivorans, a strong anti-repeat within the first 100 nt of the cas9 gene in Azospirillum sp. B510 and a strong anti-repeat overlapping with both cas9 and cas1 in A. muciniphila were identified (Fig. 2). For 12 additional loci (Peptoniphilus duerdenii, Coprococcus catus, Acidaminococcus intestini, Catenibacterium mitsuokai, Staphylococcus pseudintermedius, Ilyobacter polytropus, Elusimicrobium minutum, Bacteroides fragilis, Acidothermus cellulolyticus, Corynebacterium diphteriae, Bifidobacterium longum and Bifidobacterium dentium), we could not detect any putative anti-repeat. There is no available information on pre-crRNA expression and processing in these CRISPR-Cas loci. Thus, the functionality of type II systems in the absence of a clearly defined tracrRNA ortholog remains to be addressed. For seven analyzed loci we could not identify any repeat spacer array (Parasutterella excrementihominis, Bacillus cereus, Ruminococcus albus, Rhodopseudomonas palustris, Nitrobacter hamburgensis, Bradyrhizobium sp. BTAi1 and Prevotella micans) (Fig. 2) and in three of those (Bradyrhizobium sp. BTAi1, N. hamburgensis and B. cereus) we detected cas9 as a single gene with no other cas genes in the vicinity. For these three loci, we failed to predict any small RNA sequence upstream or downstream of the cas9 gene. In the case of R. albus and P. excrementihominis, the genomic contig containing cas9 is too short to allow prediction of the repeat spacer array.
Deep RNA sequencing validates expression of novel tracrRNA orthologs
To verify the in silico tracrRNA predictions and determine tracrRNA:pre-crRNA co-processing patterns, RNAs from selected Gram-positive (S. mutans and L. innocua) and Gram-negative (N. meningitidis, C. jejuni and F. novicida) bacteria were analyzed by deep sequencing. Sequences of tracrRNA orthologs and processed crRNAs were retrieved (Fig. S2,Table S3). Consistent with previously published differential tracrRNA sequencing data in S. pyogenes,26 tracrRNA orthologs were highly represented in the libraries, ranging from 0.08‒6.2% of total mapped reads. Processed tracrRNAs were also more abundant than primary transcripts, ranging from 66% to more than 95% of the total amount of tracrRNA reads (Fig. S2, Table S3).
To assess the 5′-ends of tracrRNA primary transcripts, we analyzed the abundance of all 5′-end reads of tracrRNA and retrieved the most prominent reads upstream or in the vicinity of the 5′-end of the predicted anti-repeat sequence. The 5′-ends of tracrRNA orthologs were further confirmed using promoter prediction algorithm. The identified 5′-ends of tracrRNAs from S. mutans, L. innocua and N. meningitidis correlated with both in silico predictions and northern blot analysis of tracrRNA expression.26 The most prominent 5′-end of C. jejuni tracrRNA was identified in the middle of the anti-repeat sequence. Five nucleotides upstream, an additional putative 5′-end correlating with the in silico prediction and providing longer sequence of interaction with the CRISPR repeat sequence was detected. We retrieved relatively low amount of reads from the F. novicida library that corresponded almost exclusively to processed transcripts. Analysis of the very small amount of reads of primary transcripts provided a 5′-end that corresponded to one of the strongest in silico promoter predictions. Northern blot probing of F. novicida tracrRNA further confirmed the validity of the predictions showing the low abundance of transcripts of around 90 nt in length (data not shown). The results are listed in Table 1. For all examined species, except N. meningitidis, primary tracrRNA transcripts were identified as single small RNA species of 75‒100 nt in length. In the case of N. meningitidis, we found a predominant primary tracrRNA form of ~110 nt and a putative longer transcript of ~170 nt represented by a very low amount of reads and detected previously as a weak band by northern blot analysis.26
Table 1. Selected tracrRNA orthologs.
| Strainsa | Transcript | 5′-endb | 3′-end | Length (nt) | ||
|---|---|---|---|---|---|---|
| RNA-seq |
Predicted |
|||||
| First read |
Most prominent |
|||||
|
S. pyogenes SF370 |
primary |
- |
854 546 |
- |
854 376 |
171 |
| primary |
- |
854 464 |
- |
89 |
||
| processed |
- |
854 450 |
- |
~75 |
||
|
C. jejuni NCTC 11168 |
primary |
1 455 497 |
1 455 502 |
1 455 497 |
1 455 570 |
~75 |
| processed |
- |
1 455 509 |
- |
~60 |
||
|
L. innocua Clip11262 |
primary |
2 774 774 |
2 774 774 |
2 774 773 |
2 774 863 |
~90 |
| processed |
- |
2 774 788 |
- |
~75 |
||
|
S. mutans UA159 |
primary |
1 335 040 |
1 335 040 |
1 355 039 |
1 335 141 |
~100 |
| processed |
- |
1 335 054 1 335 062 |
- |
~85 ~80 |
||
|
N. meningitidis Z2491 |
primary |
614 158 |
614 162 |
614 154 |
614 333 |
~175 |
| primary |
614 223 |
614 225 |
614 223 |
~110 |
||
| processed |
- |
614 240 |
- |
~90 |
||
|
F. novicida U112 |
primary |
817 144 |
- |
817 145 817 154 |
817 065 |
~80 |
| processed |
- |
817 138 817 128 |
- |
~75 ~65 |
||
|
S. thermophilus LMD-9 |
primary |
- |
- |
1 384 330 |
1 384 425 |
~95 |
| primary |
- |
- |
646 654 |
646 7662 |
~110 |
|
|
P. multocida Pm70 |
primary |
- |
- |
1 327 287 |
1 327 396 |
~110 |
| M. mobile 163K | primary | - | - | 49 470 | 49 361 | ~110 |
a tracrRNA orthologs of S. thermophilus, P. multocida and M. mobile were predicted in silico. bRNA-seq, revealed by RNA sequencing (Table S3); first read, first 5′-end position retrieved by sequencing; most prominent, abundant 5′-end according to RNA-seq data; predicted, in silico prediction of transcription start site; underlined, 5′-end chosen for the primary tracrRNA to be aligned. 5′-ends of processed tracrRNAs were validated by primer extension analysis (data not shown). cEstimated 3′-end according to RNA-seq data and transcriptional terminator prediction.
tracrRNA and pre-crRNA co-processing sites lie in the anti-repeat:repeat region
We examined the processed tracrRNA transcripts by analyzing abundant tracrRNA 5′ ends within the predicted anti-repeat sequence and abundant mature crRNA 3′-ends (Fig. 3; Table S3). In all species, we identified the prominent 5′-ends of tracrRNA orthologs that could result from co-processing of the tracrRNA:pre-crRNA repeat duplexes by RNase III. We also identified the processed 5′-ends of crRNAs that most probably result from a second maturation event by putative trimming, consistently with previous observations.26 Noteworthy, in the closely related RNA pairs of S. pyogenes, S. mutans and L. innocua, we observed the same processing site around the G:C basepair in the middle of the anti-repeat sequence. In both S. mutans and L. innocua, we detected additional prominent tracrRNA 5′-ends and crRNA 3′-ends that could suggest further trimming of the tracrRNA:crRNA duplex, with 3′-end of crRNA being shortened additionally to the already mentioned 5′-end trimming, following the RNase III-catalyzed first processing event. Similarly, in C. jejuni, we found only a small amount of crRNA 3′-ends that would fit to the RNase III processing patterns and retrieved the corresponding 5′-ends of processed tracrRNA. Thus, the putative trimming of tracrRNA:crRNA duplexes after initial cleavage by RNase III would result in a shorter repeat-derived part in mature crRNAs, producing shorter tracrRNA:crRNA duplexes stabilized by a triple G:C base-pairing for interaction with the endonuclease Cas9 and subsequent cleavage of target DNAs. The N. meningitidis RNA duplex seems to be processed at two primary sites further to the 3′ end of the CRISPR repeat, resulting in a long repeat-derived part in mature crRNA and stable RNA:RNA interaction despite the central bulge within the duplex. Interestingly, the tracrRNA:pre-crRNA duplex of F. novicida seems to be cleaved within the region of low complementarity and some of the retrieved abundant 5′ ends of tracrRNA suggest its further trimming without concomitant trimming of crRNA. Differences in primary transcript sizes and in the location of processing sites result in various lengths of processed tracrRNAs ranging from ~65‒85 nt. The coordinates and sizes of the prominent processed tracrRNA transcripts are shown in Table 1 and Table S3. The observed processing patterns of tracrRNA and crRNA are well in agreement with the previously proposed model of two maturation events. The putative further trimming of some of the tracrRNA 5′-ends and crRNA 3′-ends could stem from the second maturation event or alternatively, be an artifact of the cDNA library preparation or RNA sequencing. The nature of these processings remains to be investigated further.
Sequences of tracrRNA orthologs are highly diverse
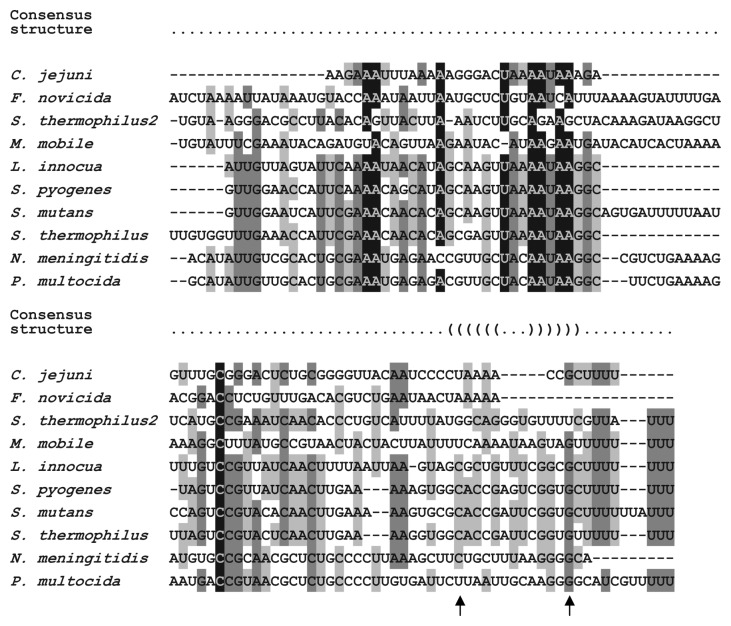
Sequence similarities of selected tracrRNA orthologs were also determined. We performed multiple sequence alignments of primary tracrRNA transcripts of S. pyogenes (89 nt form only), S. mutans, L. innocua and N. meningitidis (110 nt form only), S. thermophilus, P. multocida and M. mobile (Table 1 and Fig. 4). We observed high diversity in tracrRNA sequences but significant conservation of sequences from closely related CRISPR-Cas loci. tracrRNAs from L. innocua, S. pyogenes, S. mutans and S. thermophilus share on average 77% identity and tracrRNAs from N. meningitidis and P. multocida share 82% identity according to pairwise alignments (data not shown). The average identity of the analyzed tracrRNA sequences is 56%, comparable to the identity of random RNA sequences. This observation further confirms that the prediction of tracrRNA orthologs based on sequence similarity can be performed only in the case of closely related loci. We also sought for possible tracrRNA structure conservation but could not find any significant similarity except one co-variation and conserved transcriptional terminator structure (Fig. 4).
Figure 4. Sequence diversity of tracrRNA orthologs. tracrRNA sequence multiple alignment. S. thermophilus and S. thermophilus2, tracrRNA associated with gi|116628213 and gi|116627542 Cas9 orthologs, accordingly. Black, highly conserved; dark gray, conserved; light gray, weakly conserved. Predicted consensus structure is depicted on the top of the alignment. Arrows indicate the nucleotide co-variations. S. pyogenes SF370, S. mutans UA159, L. innocua Clip11262, C. jejuni NCTC 11168, F. novicida U112 and N. meningitidis Z2491 tracrRNAs were validated by RNA sequencing and northern blot analysis.26S. thermophilus LMD-9 tracrRNA was validated by northern blot analysis. P. multocida Pm70 tracrRNA was predicted from high similarity of the CRISPR-Cas locus with that of N. meningitidis Z2491. M. mobile 163K tracrRNA was predicted in silico from strong predictions of transcriptional promoter and terminator.
Discussion
This work describes tracrRNA as a unique family of small non-coding RNAs with conserved function but no obvious structure or sequence conservation. tracrRNAs are inherent to the type II CRISPR-Cas systems and critical for the steps of expression and interference. We demonstrated previously that tracrRNAs act in trans and share the common characteristic to contain an anti-repeat sequence allowing base-pairing with each repeat of related pre-crRNAs. According to previous in silico predictions,68 in contrast to types I and III, type II CRISPR repeats are only weakly palindromic. Hence, they lack per se the distinct characteristic of types I and III repeats to form stem-loop structures required for Cas6-like cleavage of pre-crRNA within the repeats. Base-pairing of tracrRNA with pre-crRNA repeats would compensate this deficiency by providing an intermolecular structure that activates a first processing of pre-crRNA by RNase III-mediated cleavage, leading to concomitant processing of tracrRNA itself.26 Following maturation, the dual-tracrRNA:crRNA structure associated with Cas9 endonuclease constitutes a ternary silencing complex that targets cognate invading DNA. These functional characteristics make the tracrRNA family distinct from other families of non-coding RNAs that ultimately affect the maintenance or function of either mRNAs or proteins.69-71
Diversity and rapid evolution of type II CRISPR-Cas loci within a same species or even a same strain67,72,73 and in different bacteria62 were already reported, however only on a limited number of selected type II systems. Here, we analyzed 75 representative type II CRISPR-Cas loci selected from 235 retrieved putative Cas9-containing type II loci from 203 bacterial species. Based on our analysis of the distinct monophyletic clusters of the Cas9 tree, we updated the subclassification of type II CRISPR-Cas loci from two to three subtypes; II-A, II-B and II-C. The subtyping correlates also with the different loci architectures, especially for the presence or absence of the fourth gene within the cas operon. The newly proposed type II-C is characterized by the minimal cas operon consisting only of the cas9, cas1 and cas2 genes. Remarkably, within type II-A, we identified three novel putative Cas proteins. Two of them are found exclusively in the staphylococcal type II systems (Figs. 1 and 2, yellow and orange), the third one is detected only in the mycoplasmal CRISPR-Cas loci (Figs. 1 and 2, red). These Cas proteins do not share any obvious similarities with Cas4 or Csn2, the only two described proteins other than the core Cas9, Cas1 and Cas2 of the type II systems. Further studies should reveal their potential functions in type II CRISPR-Cas adaptive immunity.
We observed a general correlation between tracrRNA localization and subclustering of Cas9 sequences. Sequence similarities of tracrRNA orthologs belonging to loci with closely related Cas9 proteins raise the question whether Cas9 proteins and tracrRNAs have functionally co-evolved. In a previous study, we showed on a small amount of samples that tracrRNA:crRNA pairs and Cas9 orthologs of closely related species (e.g., S. pyogenes, S. thermophilus and L. innocua) are partially exchangeable in an in vitro target DNA cleavage assay, but are not functional when coupled with the RNA or Cas9 ortholog of distant species, i.e., N. meningitidis.38 Moreover, the predicted base-pairing patterns, with respect to size and positions of bulges within the tracrRNA:crRNA duplexes are similar for closely related species of S. pyogenes, S. thermophilus and L. innocua, but distinct from N. meningitidis.38 This observation suggests functional evolution of tracrRNA, crRNA and Cas9, which future studies should validate.
Despite the similarity of some of the tracrRNA orthologs, we generally observed high diversity in both their sequence and exact localization within the CRISPR-Cas loci. This diversity taken together with the conserved anti-repeat feature raises questions about the origin of tracrRNA and the mechanisms that led to divergence in tracrRNA localization within type II loci. We hypothesize that tracrRNA could be derived from some degenerated repeat sequence. Some of the analyzed loci show significant architectural rearrangements compared with the consensus architecture observed for their subclusters. Possible scenarios leading to the observed diversity could be proposed. In Lactobacillus gasseri, we detected two potential anti-repeats, one upstream of cas9, characteristic for most of the type II-A loci and one between cas9 and cas1 as found in the small subset of the systems clustering with L. gasseri (Fig. 2). Thus, we hypothesize that L. gasseri CRISPR-Cas locus could be an intermediate locus, where the tracrRNA gene would have relocated in two positions typical to the type II-A in a scenario involving duplication or second acquisition of the anti-repeat (note that the detected anti-repeats are not identical; see Table S2). In the cluster comprising the loci of Mycoplasma spp., we observed potential series of rearrangements including an orientation of the cas9 gene that could have led to a change in tracrRNA position (Fig. 2). Finally, the type II locus of Lactobacillus coryniformis contains a second repeat-spacer array located upstream of cas9 and containing a last repeat, highly degenerated (11 mismatches to the repeat sequence). We suggest that this repeat located at the distal position of the CRISPR array could be the source of the anti-repeat of the associated tracrRNA. We could not predict any promoter that would drive its transcription. The representation of this rearranged locus is unfortunately too low to draw any definite conclusions.
tracrRNA is an essential component of the dual-RNA:Cas9 ternary silencer complex, which was proposed as an attractive programmable tool for site-specific genome modification.38,74-77 The large panel of various tracrRNA and Cas9 proteins should represent a valuable source of sequences to improve the design of dual-RNA:Cas9 and derived single-guide RNA:Cas9. Where does tracrRNA originate from, how did tracrRNAs and Cas9 proteins evolve and how has the type II system spread are significant questions for future studies and discussions.
Materials and Methods
Bacterial strains and culture conditions
The following media were used to grow bacteria on plates: TSA [trypticase soy agar, Trypticase™ Soy Agar (TSA II) BD BBL, Becton Dickinson] supplemented with 3% sheep blood for S. mutans (UA159) and BHI (brain heart infusion, BD Bacto™ Brain Heart Infusion, Becton Dickinson) agar for L. innocua (Clip11262). When cultivated in liquid cultures, THY medium [Todd Hewitt Broth (THB, Bacto, Becton Dickinson)] supplemented with 0.2% yeast extract (Servabacter®) was used for S. mutans, BHI broth for L. innocua, BHI liquid medium containing 1% vitamin-mix VX (Difco, Becton Dickinson) for N. meningitidis (Z2491), MH (Mueller Hinton Broth, Oxoid) Broth including 1% vitamin-mix VX for C. jejuni (NCTC 11168; ATCC 700819) and TSB (Tryptic Soy Broth, BD BBL™ Trypticase™ Soy Broth) for F. novicida (U112). S. mutans was incubated at 37∞C, 5% CO2 without shaking. Strains of L. innocua, N. meningitidis and F. novicida were grown aerobically at 37°C with shaking. C. jejuni was grown at 37∞C in microaerophilic conditions using campygen (Oxoid) atmosphere. Bacterial cell growth was followed by measuring the optical density of cultures at 620 nm (OD620 nm) at regular time intervals using a microplate reader (BioTek PowerWave™).
Sequencing of bacterial small RNA libraries
C. jejuni NCTC 11168 (ATCC 700819), F. novicida U112, L. innocua Clip11262, N. meningitidis Z2491 and S. mutans UA159 were cultivated until mid-logarithmic growth phase and total RNA was extracted with TRIzol (Sigma-Aldrich). Ten µg of total RNA from each strain were treated with TURBO™ DNase (Ambion) to remove any residual genomic DNA. rRNAs were removed by using the Ribo-Zero™ rRNA Removal Kits® for Gram-positive or Gram-negative bacteria (Epicentre) according to the manufacturer's instructions. Following purification with the RNA Clean and Concentrator™-5 kit (Zymo Research), the libraries were prepared using ScriptMiner™ Small RNA-Seq Library Preparation Kit (Multiplex, Illumina® compatible) following the manufacturer's instructions. RNAs were treated with the Tobacco Acid Pyrophosphatase (TAP) (Epicentre). Columns from RNA Clean Concentrator™-5 (Zymo Research) were used for subsequent RNA purification and the Phusion® High-Fidelity DNA Polymerase (New England Biolabs) was used for PCR amplification. Specific user-defined barcodes were added to each library [RNA-Seq Barcode Primers (Illumina®-compatible) Epicentre] and the samples were sequenced at the Next Generation Sequencing (CSF NGS Unit; csf.ac.at/) facility of the Vienna Biocenter (Illumina single end sequencing).
Analysis of tracrRNA and crRNA sequencing data
The RNA sequencing reads were split up using the illumina2bam tool and trimmed by (1) removal of Illumina adaptor sequences (cutadapt 1.0)78 and (2) removal of 15 nt at the 3′-end to improve the quality of reads. After removal of reads shorter than 15 nt, the cDNA reads were aligned to their respective genome using Bowtie by allowing two mismatches: C. jejuni (GenBank: NC_002163), F. novicida (GenBank: NC_008601), N. meningitidis (GenBank: NC_003116), L. innocua (GenBank: NC_003212) and S. mutans (GenBank: NC_004350). Coverage of the reads was calculated at each nucleotide position separately for both DNA strands using BEDTools-Version-2.15.0.79 A normalized wiggle file containing coverage in read per million (rpm) was created and visualized using the Integrative Genomics Viewer (IGV) tool (www.broadinstitute.org/igv/) (Fig. S2). Using SAMTools flagstat,80 the proportion of mapped reads was calculated on a total of 9914184 reads for C. jejuni, 48205 reads for F. novicida, 13110087 reads for N. meningitidis, 161865 reads for L. innocua and 1542239 reads for S. mutans. A file containing the number of reads starting (5′) and ending (3′) at each single nucleotide position was created and visualized in IGV. For each tracrRNA ortholog and crRNA, the total number of reads retrieved was calculated using SAMtools.80
Cas9 sequence analysis, multiple sequence alignment and guide tree construction
Position-specific iterated (PSI)-BLAST program81 was used to retrieve homologs of the Cas9 family in the NCBI non redundant database. Sequences shorter than 800 amino acids were discarded. The BLASTClust program82 set up with a length coverage cutoff of 0.8 and a score coverage threshold (bit score divided by alignment length) of 0.8 was used to cluster the remaining sequences (Table S1). This procedure produced 78 clusters (48 of those were represented by one sequence only). One (or rarely a few representatives) were selected from each cluster and multiple alignment for these sequences was constructed using the MUSCLE program83 with default parameters, followed by a manual correction on the basis of local alignments obtained using PSI-BLAST81 and HHpred programs.84 A few more sequences were unalignable and thus excluded from the final alignments. The confidently aligned blocks (Fig. S1) with 272 informative positions were used for maximum likelihood tree reconstruction using the FastTree program85 with the default parameters: JTT evolutionary model, discrete gamma model with 20 rate categories. The same program was used to calculate the bootstrap values.
Analysis of CRISPR-Cas loci
The CRISPR repeat sequences were retrieved from the CRISPRdb database or predicted using the CRISPRfinder tool.64,65 The cas genes were identified using the BLASTp algorithm and/or verified with the KEGG database (www.kegg.jp/).
In silico prediction and analysis of tracrRNA orthologs
The putative anti-repeats were identified using the Vector NTI® software (Invitrogen) by screening for additional, degenerated repeat sequences that did not belong to the repeat-spacer array on both strands of the respective genomes allowing up to 15 mismatches. The transcriptional promoters and Rho-independent terminators were predicted using the BDGP Neural Network Promoter Prediction program (www.fruitfly.org/seq_tools/promoter.html) and the TransTermHP software,86 respectively. The multiple sequence alignments were performed using the MUSCLE program83 with default parameters. The alignments were analyzed for the presence of conserved structure motifs using the RNAalifold algorithm of the Vienna RNA package 2.0.87,88
Supplementary Material
Acknowledgments
We are grateful to Kira Makarova for her help with Cas9 sequence analyses and phylogenetic tree reconstruction, for stimulating discussions and critical reading of the manuscript. We thank Anders Sjöstedt for his gift of F. novicida and help with the cultivation of bacterial strains. We are grateful to Aman Zare and Ido Tamir for their support with RNA sequencing data processing. We thank Ines Fonfara from the Charpentier laboratory for her critical comments on the manuscript. This work was funded by the Swedish Research Council [#K2010-57X-21436-01-3, #K2013-57X-21436-04-3 and #621-2011-5752-LiMS (E.C.)], the Kempe Foundation (E.C.), the University of Vienna (K.C.), Umeå University (#Dnr: 223-2728-10, #Dnr: 223-2836-10, #Dnr: 223-2989-10 (E.C.)] and the MIMS (E.C.). K.C. is a fellow of the Austrian Doctoral Program in RNA Biology.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24321
References
- 1.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–97. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 2.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–7. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–93. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 4.Bikard D, Marraffini LA. Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages. Curr Opin Immunol. 2012;24:15–20. doi: 10.1016/j.coi.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jore MM, Brouns SJ, van der Oost J. RNA in defense: CRISPRs protect prokaryotes against mobile genetic elements. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 8.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–39. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 9.Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202–9. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Garrett RA, Vestergaard G, Shah SA. Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol. 2011;19:549–56. doi: 10.1016/j.tim.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Richter C, Chang JT, Fineran PC. Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR) / CRISPR associated (Cas) systems. Viruses. 2012;4:2291–311. doi: 10.3390/v4102291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 13.Barrangou R, Horvath P. CRISPR: new horizons in phage resistance and strain identification. Annu Rev Food Sci Technol. 2012;3:143–62. doi: 10.1146/annurev-food-022811-101134. [DOI] [PubMed] [Google Scholar]
- 14.Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–6. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 15.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–63. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 16.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 17.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 18.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–76. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 22.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O’Toole GA. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol. 2012;194:5728–38. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez-Sanchez MJ, Sauvage E, Da Cunha V, Clermont D, Ratsima Hariniaina E, Gonzalez-Zorn B, et al. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol Microbiol. 2012;85:1057–71. doi: 10.1111/j.1365-2958.2012.08172.x. [DOI] [PubMed] [Google Scholar]
- 24.Erdmann S, Garrett RA. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol. 2012;85:1044–56. doi: 10.1111/j.1365-2958.2012.08171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westra ER, Brouns SJ. The rise and fall of CRISPRs--dynamics of spacer acquisition and loss. Mol Microbiol. 2012;85:1021–5. doi: 10.1111/j.1365-2958.2012.08170.x. [DOI] [PubMed] [Google Scholar]
- 26.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–7. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–96. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–8. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc Natl Acad Sci USA. 2011;108:21218–22. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–64. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garside EL, Schellenberg MJ, Gesner EM, Bonanno JB, Sauder JM, Burley SK, et al. Cas5d processes pre-crRNA and is a member of a larger family of CRISPR RNA endonucleases. RNA. 2012;18:2020–8. doi: 10.1261/rna.033100.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–92. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 34.Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–7. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 35.Nam KH, Haitjema C, Liu X, Ding F, Wang H, DeLisa MP, et al. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure. 2012;20:1574–84. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108:10098–103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westra ER, van Erp PB, Künne T, Wong SP, Staals RH, Seegers CL, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–82. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–13. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beloglazova N, Petit P, Flick R, Brown G, Savchenko A, Yakunin AF. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30:4616–27. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howard JA, Delmas S, Ivančić-Baće I, Bolt EL. Helicase dissociation and annealing of RNA-DNA hybrids by Escherichia coli Cas3 protein. Biochem J. 2011;439:85–95. doi: 10.1042/BJ20110901. [DOI] [PubMed] [Google Scholar]
- 45.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–36. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 46.Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–56. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mulepati S, Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) J Biol Chem. 2011;286:31896–903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–42. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011;108:10092–7. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–9. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–8. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–12. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 54.Magadán AH, Dupuis ME, Villion M, Moineau S. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS One. 2012;7:e40913. doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koo Y, Jung DK, Bae E. Crystal structure of Streptococcus pyogenes Csn2 reveals calcium-dependent conformational changes in its tertiary and quaternary structure. PLoS One. 2012;7:e33401. doi: 10.1371/journal.pone.0033401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ellinger P, Arslan Z, Wurm R, Tschapek B, MacKenzie C, Pfeffer K, et al. The crystal structure of the CRISPR-associated protein Csn2 from Streptococcus agalactiae. J Struct Biol. 2012;178:350–62. doi: 10.1016/j.jsb.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 61.Nam KH, Kurinov I, Ke A. Crystal structure of clustered regularly interspaced short palindromic repeats (CRISPR)-associated Csn2 protein revealed Ca2+-dependent double-stranded DNA binding activity. J Biol Chem. 2011;286:30759–68. doi: 10.1074/jbc.M111.256263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horvath P, Coûté-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, Barrangou R. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int J Food Microbiol. 2009;131:62–70. doi: 10.1016/j.ijfoodmicro.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 63.Lee KH, Lee SG, Eun Lee K, Jeon H, Robinson H, Oh BH. Identification, structural, and biochemical characterization of a group of large Csn2 proteins involved in CRISPR-mediated bacterial immunity. Proteins. 2012;80:2573–82. doi: 10.1002/prot.24138. [DOI] [PubMed] [Google Scholar]
- 64.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grissa I, Vergnaud G, Pourcel C. CRISPRfinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35(Web Server issue):W52-7. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–75. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 67.Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–12. doi: 10.1128/JB.01415-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Le Rhun A, Charpentier E. Small RNAs in streptococci. RNA Biol. 2012;9:414–26. doi: 10.4161/rna.20104. [DOI] [PubMed] [Google Scholar]
- 70.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–28. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delaney NF, Balenger S, Bonneaud C, Marx CJ, Hill GE, Ferguson-Noel N, et al. Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet. 2012;8:e1002511. doi: 10.1371/journal.pgen.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, et al. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008;9:R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Villion M, Moineau S. The double-edged sword of CRISPR-Cas systems. Cell Res. 2013;23:15–7. doi: 10.1038/cr.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carroll D. A CRISPR approach to gene targeting. Mol Ther. 2012;20:1658–60. doi: 10.1038/mt.2012.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brouns SJJ. Molecular biology. A Swiss army knife of immunity. Science. 2012;337:808–9. doi: 10.1126/science.1227253. [DOI] [PubMed] [Google Scholar]
- 77.Barrangou R. RNA-mediated programmable DNA cleavage. Nat Biotechnol. 2012;30:836–8. doi: 10.1038/nbt.2357. [DOI] [PubMed] [Google Scholar]
- 78.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 79.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wheeler D, Bhagwat M. BLAST QuickStart: example-driven web-based BLAST tutorial. Methods Mol Biol. 2007;395:149–76. doi: 10.1007/978-1-59745-514-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244-8. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kingsford CL, Ayanbule K, Salzberg SL. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 2007;8:R22. doi: 10.1186/gb-2007-8-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hofacker IL, Fekete M, Stadler PF. Secondary structure prediction for aligned RNA sequences. J Mol Biol. 2002;319:1059–66. doi: 10.1016/S0022-2836(02)00308-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.