Abstract
The Cascade complex for CRISPR-mediated antiviral immunity uses CRISPR RNA (crRNA) to target invading DNA species from mobile elements such as viruses, leading to their destruction. The core of the Cascade effector complex consists of the Cas5 and Cas7 subunits, which are widely conserved in prokaryotes. Cas7 binds crRNA and forms the helical backbone of Cascade. Many archaea encode a version of the Cascade complex (denoted Type I-A) that includes a Csa5 (or small) subunit, which interacts weakly with the core proteins. Here, we report the crystal structure of the Csa5 protein from Sulfolobus solfataricus. Csa5 comprises a conserved α-helical domain with a small insertion consisting of a weakly conserved β-strand domain. In the crystal, the Csa5 monomers have multimerized into infinite helical threads. At each interface is a strictly conserved intersubunit salt bridge, deletion of which disrupts multimerization. Structural analysis indicates a shared evolutionary history among the small subunits of the CRISPR effector complexes. The same α-helical domain is found in the C-terminal domain of Cse2 (from Type I-E Cascade), while the N-terminal domain of Cse2 is found in Cmr5 of the CMR (Type III-B) effector complex. As Cmr5 shares no match with Csa5, two possibilities present themselves: selective domain loss from an ancestral Cse2 to create two new subfamilies or domain fusion of two separate families to create a new Cse2 family. A definitive answer awaits structural studies of further small subunits from other CRISPR effector complexes.
Keywords: CRISPR, Csa5, structure, CRISPR interference, Cascade
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPRs) are a prokaryotic mechanism for preventing horizontal gene transfer (HGT) and infection by mobile genetic elements.1-3 The CRISPR array consists of identical repeats separated by short “spacers” derived from previously encountered foreign genetic elements.4-7 Linked to the CRISPR array are CRISPR-associated (cas) genes, which encode the proteins that mediate the CRISPR response to the insertion of foreign double-stranded DNA (dsDNA) or single-stranded RNA (ssRNA) into the cell.8-10 The response can be divided into three stages: adaptation, the integration of new spacers into the CRISPR array; expression, the transcription of the array and processing of the transcript to short, mature CRISPR RNAs (crRNA); interference, the recognition and degradation of foreign dsDNA or ssRNA.
cas genes are arranged in loci, which can be classified into one of three main systems (types I, II and III) based on the identity and order of genes in the locus.10 The CRISPR/Cas systems can be further subdivided into a total of 10 subtypes (Fig. S1), all of which, except type III-B, target dsDNA.11-13 Uniquely type III-B targets ssRNA.14,15 The type I interference complex is known as the CRISPR-associated complex for antiviral defense (Cascade). The exemplar is the type I-E complex (eCascade) from E. coli,13 which consists of Cas5, Cas6e, Cas7, Cse1 and Cse2 in a 1:1:6:1:2 stoichiometry with the six Cas7 subunits forming a central helical spine to which crRNA and all the other subunits bind.16 This topology appears to be conserved in the type I-C and I-F systems.17,18
Significantly less is known about the effector complex of type I-A systems (aCascade, also known as the archaeal Cascade), which consists of Cas5, Cas7, Csa5 and in some systems Cas8a1 or Cas8a2. It has not yet been possible to purify a full aCascade complex; the Sulfolobus solfataricus (Sso) aCascade partly dissociates during purification,19 while the Thermoproteus tenax complex is insoluble after overexpression, with minimal soluble material after refolding.20 Only a stable core complex of Cas5 and Cas7 remains after the partial dissociation of Sso aCascade. The Cas5/Cas7 complex forms helical oligomers reminiscent of the Cas7 backbone of eCascade. Interestingly this core complex is capable of binding crRNA and a heteroduplex formed from crRNA and protospacer, a mimic of the R-loop structure formed during interference.19 Csa5 has been shown to interact with this complex, though its role within aCascade is unknown.19
The genome of S. solfataricus encodes three possible aCascade complexes as well as one CSM complex and three CMR complexes.21 Here, we report the crystal structure and initial functional characterization of the archaeal-specific Cascade subunit Csa5. Structural homology shows that Csa5 and Cse2, a subunit of eCascade, have one domain in common. In addition, the second domain of Cse2 is shown to have a fold related to the small subunit (Cmr5) of the archaeal type III-B system. Altogether, these observations highlight the conservation of structural elements in diverse CRISPR effector complexes.
Results
Csa5 interacts with the Cas5/Cas7 complex
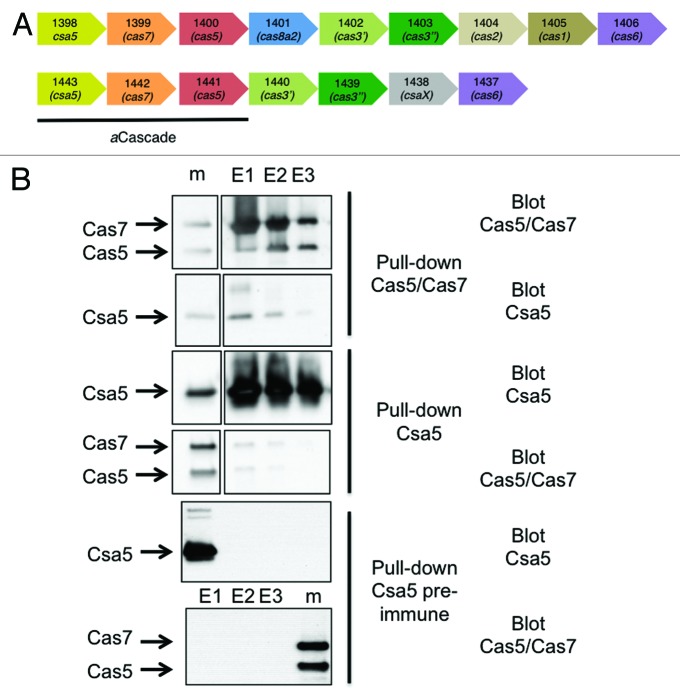
The two copies of S. solfataricus aCascade studied in this paper are encoded by the genes sso1398-1400 and sso1441-1443 (Fig. 1A). The two Csa5 paralogs share 11% sequence identity. Experiments described here utilize the Sso1441-1443 proteins unless otherwise indicated. Immunoprecipitation of the Cas5/Cas7 core complex from S. solfataricus cell lysates using affinity-purified antibodies raised against the recombinant complex co-precipitated small amounts of Csa5 (Fig. 1B). The reciprocal experiment using antibodies against Csa5 pulled down both Cas5 and Cas7. The small amounts that are co-immunoprecipitated are indicative of a weak interaction between Csa5 and Cas5/Cas7, consistent with the dissociation of Csa5 from the complex during purification.19
Figure 1. Gene organization and protein interactions of archaeal Cascade subunits. (A) Two of the type I-A cas loci from S. solfataricus showing the gene numbers and families. (B) Co-immunopreciptiation of Csa5 and Cas5/Cas7 from S. solfataricus cell extracts. Antibodies raised against Cas5/Cas7 pull down Csa5 from cell extracts (top two panels). Antibodies raised against Csa5 pull down small amounts of Cas5/Cas7 from cell extracts (middle two panels). Csa5 pre-immune complex did not pull down any of the proteins (bottom two panels). The marker lanes (m) contain recombinant proteins as size markers.
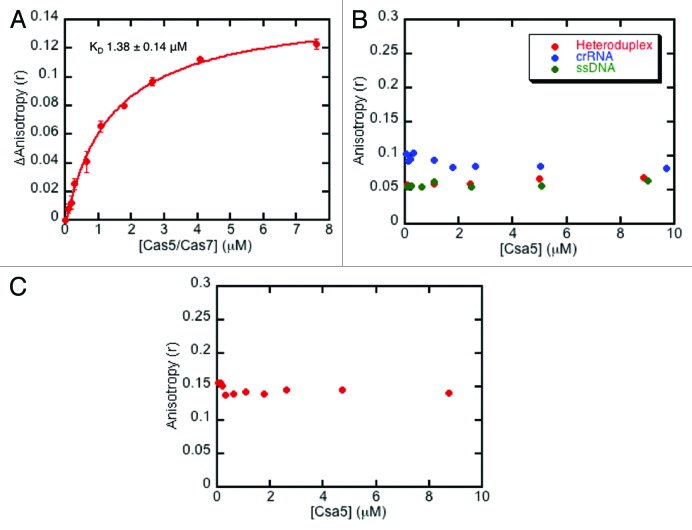
Lintner et al.19 showed that cellular Cas5/Cas7 (Sso1441-1442) bound to crRNA, raising the possibility that Csa5 only binds to Cas5/Cas7 in the presence of crRNA. Fluorescently labeled oligonucleotides were used for fluorescence anisotropy experiments to determine interactions between Cas5/Cas7 and Csa5. Consistent with previous studies,19 the Cas5/Cas7 complex bound to fluorescently labeled crRNA with a dissociation constant (KD) of 1.38 ± 0.14 μM (Fig. 2A). However, Csa5 itself did not form a stable interaction with crRNA, ssDNA or a heteroduplex formed from crRNA and ssDNA containing a protospacer in vitro (Fig. 2B), nor did we observe convincing evidence for binding to a pre-formed complex of Cas5/Cas7/crRNA (Fig. 2C).
Figure 2. Analysis of the interactions of aCascade with nucleic acids using fluorescence anisotropy. (A) The average change in anisotropy upon binding of the Cas5/Cas7 complex to fluorescent crRNA. Experiments were performed in triplicate and error bars represent the standard error of the data. The data were fitted to a binding curve with a KD of 1.38 ± 0.14 μM (error estimated from curve fitting). (B) Csa5 does not interact with crRNA, ssDNA or a heteroduplex of crRNA and protospacer. (C) Csa5 does not interact with a pre-formed complex of Cas5/Cas7/crRNA.
The crystal structure of Csa5
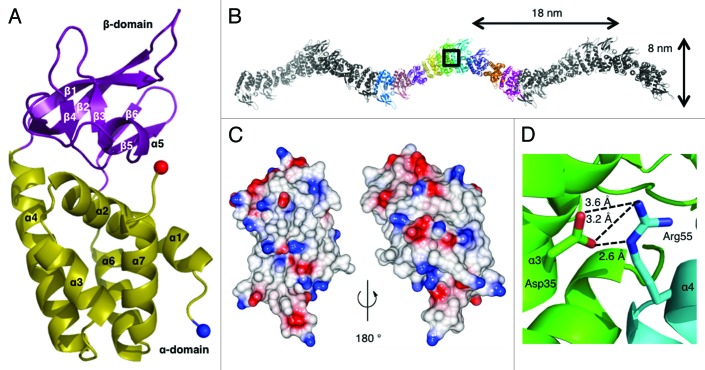
The limited solubility of Csa5 encoded by Sso1443 hindered its crystallization and so its paralog, Sso1398, was crystallized. The structure was solved using a selenomethionyl variant with diffraction recorded to a resolution of 2.7 Å (Fig. 3A and Table 1). The asymmetric unit (ASU) containes nine monomers and each protein monomer is constructed of two domains, denoted α and β (Fig. 3B). The α-domain is an all α-helical arrangement consisting of α1-α4, formed by residues 5–64 and α6 and α7 formed by residues 130–162. The β-domain is formed by residues 65–129 and is an insertion between α4 and α6. The β-domain contains a single helix (α5) and six β-strands (β1-β6) arranged as three anti-parallel two-stranded β-sheets (Fig. 3A). Residues 1–4 were disordered in all monomers; the loops connecting β1-β2 and β5-β6 were disordered in some monomers. The residues that are conserved across the Csa5 family are located in the α-domain, although the short β6 strand may show some conservation (Fig. S2). The β-domain insertions of Csa5 homologs vary in length from 20‒67 residues and show minimal sequence conservation, even between closely related strains (Fig. S2).
Figure 3. The crystal structure of Sso1398. (A) The Sso1398 monomer consists of the α-domain (yellow) and the β-domain (purple). Secondary structure elements are labeled and the N- and C-termini are shown as blue and red spheres, respectively. (B) The crystal contains chains of helical oligomers of Sso1398, shown here across three ASUs with the molecules of one ASU shown in colors. The helical pitch and width are indicated. (C) Electrostatic surface potential of a monomer of Sso1398 calculated using CCP4MG.41 (D) Close up of the box in (B). A conserved salt bridge forms at the dimer interface between Asp35 (green) and Arg55 (cyan) and contributes significantly to the stability of the oligomer.
Table 1. Data collection and refinement statistics.
| Data collection | Sso1398SeMet | Refinement | Sso1398SeMet |
|---|---|---|---|
| Wavelength (Å) |
0.98 |
Rwork/Rfree |
0.20/0.22 |
| Space group |
C 1 2 1 |
Mean B-value (Å2) |
|
|
a, b, c (Å) |
285.0, 53.7, 216.7 |
All atoms |
43 |
| α, β, γ (°) |
90.0, 124.7, 90.0 |
Protein |
43 |
| Resolution (Å) |
2.72 |
Water |
26 |
| I/σI |
12.3 (2.3) |
PEG |
55 |
| Rmerge |
0.109 (0.814) |
Rmsd |
|
| Completeness |
100.0 (100.0) |
Bond lengths (Å) |
0.01 |
| Multiplicity |
6.7 (7.0) |
Angles (°) |
1.13 |
| Anomalous completeness |
99.5 (99.8) |
|
|
| Anomalous multiplicity | 3.5 (3.6) |
Data are presented as averages with statistics for the highest resolution shell in parentheses.
Strikingly the nine monomers of the ASU are assembled in a continuous helical oligomer that, due to crystal symmetry, creates an infinite helical thread (Fig. 3B). The continuous helix has a pitch of 18 nm and a width of 8 nm. The crystal is composed of bundles of these infinite threads. Within the ASU, the nine monomers are essentially identical, superimposing with an average Cα rmsd of 0.21 Å. The nine interfaces within the ASU show subtle differences, suggesting a level of plasticity at the interface (Fig. S3). Analysis of the electrostatic potential of the solvent-exposed surface does not reveal any regions of significant positive charge (Fig. 3C), consistent with the observed lack of binding to nucleic acids.
Oligomerization of Csa5 proteins
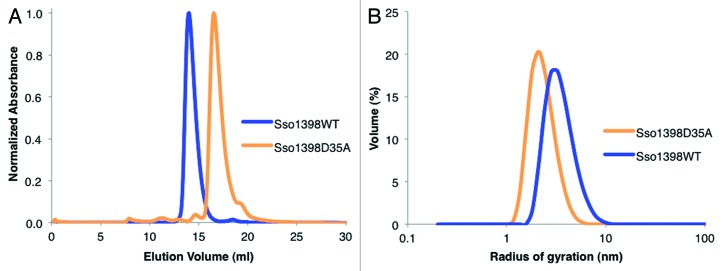
The interfacing residues are predominantly located in the α-domain with a few contacts from β1, β2 and β6. Analysis of the interfaces within the helices using the PDBe PISA server22 showed that 1100 Å2 of surface area from each monomer was buried within a “dimer.” The interfaces are hydrophilic and are scored in PISA as unstable in solution. In our analysis, we noted an intersubunit salt bridge between two conserved residues, Asp35 and Arg55 (Figs. 3D; Fig. S2). Disrupting this salt bridge (Sso1398D35A) led to the protein eluting from a calibrated gel filtration column with a larger retention volume than the wild-type protein (16.4 ml and 13.9 ml, respectively) (Fig. 4A), indicating that the mutant is smaller. The estimated weights of both native and mutant are physically unrealistic (26 and 6 kDa, respectively, where a monomer is 19 kDa) and we attribute this to the unusual shape of the protein. Dynamic light scattering experiments showed that the mutant had a smaller radius of gyration than the wild-type, supporting the gel filtration data (Fig. 4B).
Figure 4. The oligomerization of Csa5. (A) Gel filtration elution profiles for Sso1398WT and Sso1398D35A using 2 mg ml−1 protein loaded onto a Superose 12 10/300 column. The proteins ran aberrantly on gel filtration and so the masses of the oligomers could not be accurately estimated. (B) Representative radius distributions of Sso1398WT and Sso1398D35A determined by dynamic light scattering.
As both residues are conserved, we reasoned the salt bridge may be a conserved feature, so we examined the effect of the corresponding D32A mutation in Sso1443 (Csa5 itself). Recombinant wild-type Csa5 had a solubility limit of 1 mg ml−1, whereas Csa5D32A exhibited a > 10-fold increase in solubility. Csa5D32A eluted from a gel filtration column consistent with a monomeric species, an observation independently supported by isothermal calorimetry (Fig. S4).
Structural homology to other Cas proteins
The structure of Sso1398 was compared with known structures and while the PDBeFold server23 did not find any significant matches, the DALI server24 detected homology for the α-domain while the β-domain does appear to be unique. The α-domain (excluding α1) matched the C-terminal domain of Cse2 from Thermus thermophilus25 (PDB 2ZCA, Z score 4.5, 2.7 Å over 73 residues) and to a lesser extent Cse2 from Thermobifida fusca26 (PDB 4H79, Z-score 2.9, 2.9 Å over 73 residues). It is also similar to the N-terminal domain of StySJI M, a Type I HsdM subunit from a restriction enzyme of Bacteroides thetaiotaomicron (PDB 2OKC, Z score 3.3, 2.6 Å over 68 residues); the domain itself has no assigned function.
Discussion
Our data show that the interaction of the Csa5 subunit with the core Cascade structure is below the detection limit of the biophysical techniques we employed. Pull-down experiments also point to the interaction being weak. Of course, we cannot exclude the possibility that additional factors such as Cas6 and/or the presence of target DNA are required for assembly. However, in general our data fit with an emerging picture that Csa5 easily dissociates from the aCascade complex.
The structure of the paralog Sso1398 shows a remarkable arrangement in the crystal. The continuous helical oligomers are reminiscent of the Cas7 helical backbone that appears to be a conserved feature of Cascade complexes.16-18 The S. solfataricus Cas7 oligomer exhibits a helical pitch of 14 nm,19 which is broadly compatible with the Csa5 pitch of 18 nm. The helical threads are stabilized by a conserved pair of residues that form an intersubunit salt bridge. The absolute conservation of these residues is striking, as only one other residue (Ala158, located in the hydrophobic core) is completely conserved across the Csa5 family (Fig. S2). Mutation of the salt bridge both in the paralog and in Csa5 itself results in a profound change in the biophysical properties of the proteins. In both proteins, the mutant is smaller and less prone to aggregation. Since the change is seen in both proteins and the salt bridge is conserved, we hypothesize that the interface (but not the infinite threads) has a functional significance that will be revealed when the structure of the aCascade complex is determined.
Cse2 forms a dimer in eCascade, which creates a positively charged surface for interacting with nucleic acids.16 This face is formed by both domains of the protein and, therefore, is only partially conserved in Csa5. This surface is not significantly positively charged in Sso1398 or in a homology model of Csa5. Although Csa5 possesses a weakly positively charged face at the base of the α-domain (Fig. S5), the residues that form it are not well-conserved across the family. Cse2 itself is capable of binding both dsDNA and ssRNA non-specifically26 and an eCascade-crRNA complex lacking Cse2 shows reduced binding affinity for target plasmid DNA.27 It has been hypothesized that Cse2 stabilizes the R-loop nucleotide structure by either binding the RNA/DNA heteroduplex directly or indirectly by binding to non-complementary DNA strand. In contrast, Csa5 did not bind to any nucleic acid tested, including a DNA/RNA heteroduplex and ssDNA, both of which are present in an R-loop. Therefore, despite the structural homology between Cse2 and Csa5, these two proteins may play different roles within their respective Cascade complexes.
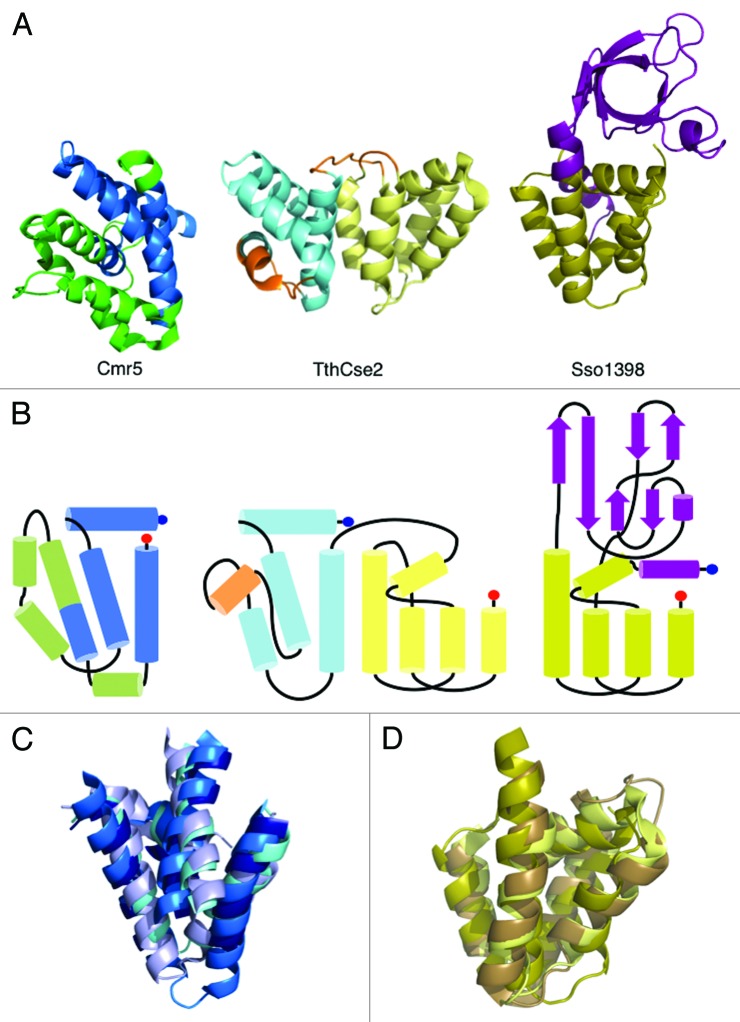
In a recent bioinformatics study, Makarova et al. suggested that several small Cas proteins, which were predicted to be largely helical, could be combined into a new family (Cas11).28 The putative Cas11 family has been proposed to comprise Csa5 (type I-A), Cse2 (type I-E), Csm2 (type III-A) and Cmr5 (type III-B) as well as the C-terminal helical domain of Cas8 (various subtypes). Crystal structures are now available for three of these protein families: Csa5 (Sso1398), Cse2 (TthCse2 and TfuCse2) and Cmr5 “T. thermophilus (PDB 2ZOP)25 and Archaeoglobus fulgidus (PDB 2OEB),” allowing an assessment of these predictions. The α-domain of Sso1398 is indeed homologous to the C-terminal domain of Cse2 (Fig. 5) but neither it nor the β-domain bears any resemblance to either Cmr5 structure. However, the N-terminal domain of Cse2 is homologous to four helices of Cmr5 (Cα rmsds of 3.2 Å over 50 residues and 3.0 Å over 60 residues to AfuCmr5 and TthCmr5, respectively) (Fig. 5). Sequence conservation between the structures is limited to residues of the hydrophobic core; the lack of conservation in exposed residues would argue against a common function. This is supported by the different properties of Csa5 and Cse2 combined with the observation that Cmr5 is not essential for the function of the CMR complex,15 whereas loss of Cse2 significantly impacts the activity of eCascade.27
Figure 5. Structural homology between Csa5, Cse2 and Cmr5. (A) The structures of AfuCmr5 (PDB 2OEB, right), TthCse2 (PDB 2ZCA, middle) and Csa5 (right). Structural features conserved between Cmr5 and Cse2 are shown in shades of blue and between Cse2 and Csa5 in shades of yellow. Non-conserved features are shown in green (Cmr5), orange (Cse2) and purple (Csa5). (B) Connector diagrams of the three proteins highlighting the conserved connectivity of the structural elements. (C) Superposition of the conserved helices of AfuCmr5, TthCse2, TthCmr5 (navy) and TfuCse2 (pale blue). (D) Superposition of the conserved helices of TthCse2, TfuCse2 (brown) and Csa5.
Analysis of the structures reveals a nuanced set of relationships among the small subunits. Of the two domains of Cse2, one is found in the Cmr5 family and the other in the Csa5 family; there is no structural relationship between the Cmr5 and Csa5 families. This is consistent with either selective loss of one or other domain from the ancestral Cse2 (Cas11) family during the diversification of the CRISPR/Cas systems to form the Cmr5 and Csa5 lineages, or alternatively a fusion of the Cmr5 and Csa5 families to create the Cse2 lineage as the Csa5 and Cmr5 proteins have nothing in common beyond a potential shared ancestor in Cse2. To further elucidate the evolutionary relationships between the so-called small subunits of the interference complexes, the structure of Csm2 would be particularly helpful, as it is a key remaining missing piece. The full-length structure of Cas8 is also likely to be useful in firmly establishing such relationships.
Materials and Methods
Cloning
sso1398 was cloned into the pDEST14 plasmid29 and sso1443 into pET151/D-TOPO (Invitrogen) with cleavable N-terminal His6-tags. Mutations were introduced using published30 or standard QuikChange (Stratagene) protocols. The oligonucleotide sequences used for cloning and mutagenesis are available on request.
Expression and purification
Sso1398, Sso1443 and their variants were expressed in C43 (DE3) E. coli in LB medium using 1.0 or 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), respectively, to induce protein expression. The cultures were incubated at 37°C throughout (Sso1398) or 25°C (Sso1443) after induction and the cells harvested after overnight incubation. The selenomethionyl derivative of Sso1398 was expressed in B834 (DE3) E. coli grown in M9 minimal medium supplemented with Selenomethionine Nutrient Mix (Molecular Dimensions) and 50 mg L−1 (L)-selenomethionine. The expression conditions were as with the native protein except the cells were harvested after 30 h. Sso1441 and Sso1442 were co-expressed in E. coli as described previously.19
All proteins were purified using previously published protocols.19,29 Briefly, cell pellets were resuspended and then lysed by sonication (MSE Soniprep) or with a cell disruptor (Constant Systems Ltd.). The lysate was cleared by centrifugation and passed over a Ni-NTA column (Qiagen). The eluted protein was incubated with TEV protease and then re-applied to the column before application to a Superdex S75 gel filtration column (GE Healthcare) equilibrated in 20 mM Tris pH 7.5, 150 mM NaCl (Sso1398) or 20 mM Tris pH 7.5, 500 mM NaCl, 10% glycerol (Sso1443). One mM β-mercaptoethanol was added to all buffers during purification of the selenomethionyl derivative. All proteins were concentrated to ≥ 10 mg ml−1 except for wild-type Sso1443, which was to 1 mg ml−1 due to low solubility.
Pull-down interactions
For the production of antibodies, 1 mg of purified recombinant Csa5 or Cas5/Cas7 protein was used to raise polyclonal antibodies in sheep (Scottish National Blood Transfusion Service). The antibodies were affinity purified using cyanogen bromide-activated-Sepharose 4B resin (Sigma). The target protein (Cas5/Cas7 or Csa5) and resin were incubated overnight in 0.1 M NaHCO3 pH 8.3, 0.5 M NaCl at 4°C. The resin was washed with buffer, resuspended in 1 M ethanolamine pH 8.0 and incubated at 4°C overnight. The resin was washed with 0.1 M NaHCO3 pH 8.3, 0.5 M NaCl and then 10 mM Tris pH 4.0 for four cycles followed by incubation with the immune serum in 10 mM Tris pH 7.5 at 4°C overnight. The resin was washed with 10 mM Tris pH 7.5 and then 10 mM Tris pH 7.5, 0.5 M NaCl. The antibodies were eluted in two stages, the first of which was 100 mM glycine pH 2.5 directly into 1 M Tris pH 8.0. The resin was further washed with 10 mM Tris pH 8.8 and the antibodies then eluted with 100 mM triethylamine pH 11.5 into 1 M Tris pH 8.0. The purified antibodies were dialyzed into PBS and then cross-linked to Protein G Sepharose (Sigma) by incubating together in TBS buffer followed by cross-linking with 20 mM dimethyl pimelimidate in 0.2 M triethanolamine. The beads were washed with trithenaolamine and ethanolamine before incubation with S. solfataricus P1 cell lysate prepared as described previously.19 After overnight incubation at 4°C, the beads were washed with TBS and the bound protein eluted with sodium citrate pH 2.5. Samples were run on NuPAGE SDS-PAGE gels (Invitrogen) and blotted onto iBlot stacks (Invitrogen). The samples were blocked with 5% milk powder in PBS, 0.05% TWEEN for 1 h and then incubated with the primary antibody of choice. The blots were incubated with 1:10,000 Immunopure rabbit anti-goat HRP (Thermo Scientific) in PBS, 0.1% TWEEN for 1 h and then visualized with SuperSignal West Pico kit (Pierce) according to the manufacturer’s instructions.
Fluorescence anisotropy
Synthetic oligonucleotides were purchased from Integrated DNA Technologies of fluorescently labeled crRNA from spacer 26 of CRISPR locus A of S. solfataricus P1, non-labeled complementary target ssDNA with PAM sequence and fluorescently labeled non-target ssDNA (see Table S1 for sequences). crRNA and protospacer were mixed 1:1, heated at 80°C for 5 min and cooled slowly to form a heteroduplex. Fluorescence anisotropy measurements (λex 490 nm, λem 535 nm) were recorded using a Varian Cary Eclipse fluorescence spectrophotometer at room temperature. Protein solutions were titrated into 20 nM crRNA, 20 nM non-target ssDNA or 20 nM heteroduplex in 20 mM Tris pH 7.5, 150 mM NaCl, 5% glycerol, 1 mM EDTA. Measurements were recorded over a range of protein concentrations (0.04 ‒11 μM). The data were analyzed using KaleidaGraph (Synergy Software) and, where appropriate, fitted to binding curves.
Crystallization of Sso1398
The selenomethionyl derivative of Sso1398 was crystallized at 2 mg ml−1 in vapor diffusion sitting drop experiments at 20°C. The crystals grew in a condition of 1:1 protein-to-precipitant with a reservoir of 0.1 M sodium acetate pH 4.0, 0.23 M potassium chloride and 39% PEG500. The crystals were cryo-cooled in liquid nitrogen directly from the drop. A single-wavelength anomalous diffraction data set was collected at the Se-K absorption edge at 100 K on the microfocus beamline I24, Diamond Light Source. The data were processed with xia231 and the phases solved using Phenix AutoSolve.32 The electron density map was further modified using Parrot33 before chain tracing with Buccaneer,34 both as part of the CCP4 suite.35 The chains were manually assigned followed by automated model building with Phenix AutoBuild.32 The model was refined by cycles of manual correction in COOT36 and refinement with REFMACv5.637 using TLS constraints generated using the TLSMD web server.38,39 The structure was validated with MolProbity.40
Dynamic light scattering
DLS experiments were performed using a Zetasizer μV system (Malvern Instruments) with a quartz SUPRASIL® cuvette. The experiments were performed at room temperature and in triplicate. The data were analyzed using the Zetasizer software.
Please cite references 43‒46 in the text or delete the reference. Also, please add a callout for Table 1 in the text.
Supplementary Material
Acknowledgments
This work was funded by a grant from the Biotechnology and Biological Sciences Research Council (BBSRC) (REF: BB/G011400/1) to M.F.W. and J.H.N. and a BBSRC-funded studentship to J.R. The authors would like to thank Drs Christophe Rouillon and Jose Peregrina for assistance with anisotropy experiments.
Glossary
Abbreviations:
- aCascade
Type I-A Cascade
- ASU
asymmetric unit
- Cas
CRISPR-associated
- Cascade
CRISPR-associated complex for antiviral defence
- CRISPR
clusters of regularly interspaced short palindromic repeats
- crRNA
CRISPR RNA
- Cmr
CRISPR module RAMP
- Csa
CRISPR subtype Apern
- Cse
CRISPR subtype Ecoli
- CSM
CRISPR subtype Mtube
- ds
double-stranded
- eCascade
Type I-E Cascade
- HGT
horizontal gene transfer
- ITC
isothermal calorimetry
- ss
single-stranded
- Sso
Sulfolobus solfataricus
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Accession Numbers
The data and coordinates of the structure of Sso1398 were deposited in the Protein Data Bank under the accession code 3ZC4.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23854
References
- 1.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 3.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 4.Mojica FJM, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36:244–6. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 5.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–82. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 6.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–61. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 7.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–63. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 8.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–75. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 9.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 12.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–13. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiedenheft B, Lander GC, Zhou KH, Jore MM, Brouns SJJ, van der Oost J, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–9. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nam KH, Haitjema C, Liu X, Ding F, Wang H, DeLisa MP, et al. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure. 2012;20:1574–84. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou KH, Barendregt A, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011;108:10092–7. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lintner NG, Kerou M, Brumfield SK, Graham S, Liu HT, Naismith JH, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–56. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plagens A, Tjaden B, Hagemann A, Randau L, Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J Bacteriol. 2012;194:2491–500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci USA. 2001;98:7835–40. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–68. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 24.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545-9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agari Y, Yokoyama S, Kuramitsu S, Shinkai A. X-ray crystal structure of a CRISPR-associated protein, Cse2, from Thermus thermaphilus HB8. Proteins-Structure Function and Bioinformatics. 2008;73:1063–7. doi: 10.1002/prot.22224. [DOI] [PubMed] [Google Scholar]
- 26.Nam KH, Huang Q, Ke A. Nucleic acid binding surface and dimer interface revealed by CRISPR-associated CasB protein structures. FEBS Lett. 2012;586:3956–61. doi: 10.1016/j.febslet.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westra ER, Nilges B, van Erp PBG, van der Oost J, Dame RT, Brouns SJJ. Cascade-mediated binding and bending of negatively supercoiled DNA. RNA Biol. 2012;9:1134–8. doi: 10.4161/rna.21410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oke M, Carter LG, Johnson KA, Liu H, McMahon SA, Yan X, et al. The Scottish Structural Proteomics Facility: targets, methods and outputs. J Struct Funct Genomics. 2010;11:167–80. doi: 10.1007/s10969-010-9090-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu HT, Naismith JH. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J Appl Cryst. 2010;43:186–90. doi: 10.1107/S0021889809045701. [DOI] [Google Scholar]
- 32.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang KYJ, Cowtan K, Main P. Combining constraints for electron-density modification. Macromolecular Crystallography. Pt B. 1997;277:53–64. doi: 10.1016/s0076-6879(97)77006-x. [DOI] [PubMed] [Google Scholar]
- 34.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr. 2006;62:1002–11. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 35.Bailey S, Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–50. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 39.Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Cryst. 2006;39:109–11. doi: 10.1107/S0021889805038987. [DOI] [Google Scholar]
- 40.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNicholas S, Potterton E, Wilson KS, Noble MEM. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67:386–94. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.