Abstract
Cas3 nuclease-helicase is part of CRISPR immunity systems in many bacteria and archaea. In type I CRISPR, Cas3 nuclease degrades invader DNA that has been base-paired to crRNA as an R-loop within a “Cascade” complex. An R-loop is a DNA-RNA hybrid that includes a displaced single-strand DNA loop. Purified Cas3 from E. coli and the archaeon M. thermautrophicus can process R-loops without DNA/RNA sequence specificity and without Cascade. This has potential to affect other aspects of microbial biology that involve R-loops. Regulatory RNAs and host cell proteins modulate replication of ColE1 plasmids (e.g., pUC) from R-loop primers. We observed that Cas3 could override endogenous control of a ColE1 replicon, stimulating uncontrolled (“runaway”) replication and resulting in much higher plasmid yields. This effect was absent when using helicase-defective Cas3 (Cas3K320L) or a non-ColE1 plasmid, and was dependent on RNaseHI. Cas3 also promoted formation of plasmid multimers or concatemers, a phenotype consistent with deregulated ColE1 replication and typical of cells lacking RNaseHI. These effects of Cas3 on ColE1 plasmids are inconsistent with it unwinding R-loops in vivo, at least in this assay. We discuss a model of how Cas3 might be able to regulate RNA molecules in vivo, unless it is targeted to CRISPR defense by Cascade, or kept in check by RecG and RNaseHI.
Keywords: CRISPR, ColE1, Cas3, RNaseHI, RecG, R-loop
Introduction
CRISPR (clustered regularly interspersed short palindromic repeat) DNA loci and Cas (CRISPR-associated) proteins can provide a system of adaptive immunity against phage and plasmids in bacteria1-5 and archaea.6,7 CRISPR/Cas systems are diverse,8 but in each case, transcription and nucleolytic processing of a CRISPR locus generates short (50–70 nucleotides) RNA (crRNA) that is the bedrock of CRISPR immunity. One major CRISPR/Cas sub-group (type I) targets crRNA to complementary sequences in invader DNA, reviewed most recently in references 9 and 10. Complementarity of host crRNA-to invader DNA is made possible from prior exposure to invader leading to incorporation of 25–45 base pairs of invader DNA as a “spacer” into a CRISPR locus.
Synonymous with type I CRISPR systems are ribonucleoprotein complexes that catalyze interference reactions in which crRNA is targeted to invader DNA. In E. coli, “Cascade” ribonucleoprotein complex is essential for interference, as is the nuclease-helicase Cas3.2 E. coli Cascade comprises five proteins: Cse1, Cse2, Cas7, Cas5 and Cas6e (also known as CasA-E, respectively)2,11 that form R-loops independently of ATP, harnessing energy within supercoiled DNA.12 DNA targeting by E. coli Cascade is most efficient if DNA has a protospacer adjacent motif (PAM) immediately next to the fully complementary spacer-protospacer sequence.12,13 Cascade catalyzed crRNA interference reactions generate R-loop intermediates, RNA-DNA hybrids that contain a displaced ssDNA loop.14,15 Nucleolytic degradation of ssDNA by Cas3 completes the reaction, destroying invader DNA and recycling Cascade. In addition to nuclease activity, purified Cas3 from E. coli and the archaeon M. thermautotrophicus can form and process R-loop substrates in vitro, acting as an ATP-dependent helicase and ATP-independent annealase.16 There is also evidence from archaea that Cas3 helicase activity is required for most efficient Cas3 nuclease activity.17 Like most superfamily 2 helicases and annealases, Cas3 lacks DNA/RNA sequence specificity, and does not require Cascade for helicase or nuclease activities.16,18-20 In E. coli, it is thought that Cas3 is targeted to crRNA R-loops through physical interaction with Cascade.12
Several biological processes require formation and processing of R-loops.21 In bacteria, replication of ColE1 plasmids is initiated from plasmid-encoded RNA transcript (RNAII) that forms a leading strand RNA primer (RNAII) as an R-loop with ori, the DNA origin of replication.22 RNaseHI activates this precise initiation of replication by nucleolytic processing of RNAII.22,23 Control is exerted by several host factors, including plasmid-encoded RNAI, which is antisense to the 5′ end of RNAII, blocking R-loop formation and reducing plasmid copy number.24-26 This RNAI-RNAII duplex is stabilized by Rop protein, although in commercial ColE1 plasmids (e.g., pUC) the gene encoding Rop (rom) has been removed to enhance copy number.
In E. coli, RecG helicase reduces ColE1 plasmid replication by dissociating R-loops.27,28 Recently described R-loop unwinding activity of E. coli Cas3 helicase-nuclease, and R-loop formation by Cascade complex, led us to consider if these proteins influence replication of ColE1 plasmids. We observed that Cas3 promoted runaway ColE1 plasmid replication, requiring Cas3 helicase activity. Cas3 expression antagonized RNaseHI, observed as a concatamer phenotype, but also required RNaseHI for its ability to stimulate plasmid copy number. This activity of Cas3 was contrary to that expected for a helicase that, like E. coli RecG, unwinds R-loops but is discussed in the light of alternative RNA processing activities.
Results
Cas3 stimulated ColE1 plasmid copy number
We measured pUC19 ColE1 plasmid yields as readout of replication proficiency after extraction from E. coli MG1655 grown as overnight cultures in the presence of ampicillin. Transcription of E. coli cse1-cas6e, encoding Cascade, is repressed by HNS5 and regulation of ygcB, encoding Cas3, has not been characterized. Therefore, Cas3 and Cascade were expressed ectopically under control of the lac promoter from within pUC19 as constructs listed in Table 1.
Table 1.E. coli strains and plasmids used in this study.
| E. coli strain | Relevant details | Reference |
|---|---|---|
| EB304 |
MG1655 Δcas3::apra |
This work: detailed in Materials and Methods |
| IIB796 |
MG1655 ΔrnhA733::kan |
This work: P1 into MG1655 from original E. coli stock center ΔrnhA strain |
| RCe442 |
MG1655 ΔproB:: ParaBAD rnhA+-frt > kan > frt |
39
|
| BW25113 |
Δ(araBAD)567 Δ(rhaBAD)568 rph-1 λ- ΔlacZ4748(::rrnB-3)hsdR514 Bacte |
For recombineering.38 |
| IIB947 |
BW25ΔΔ3 lacUV5-cas3 cat:: araBp8-casA |
This work: P1 into BW25113 from BW4011440 for recombineering |
| N4256 |
MG1655 ΔrecG::kan |
41
|
| IIB955 |
MG1655 pcnBΔ1::kan |
This work: derived from strain JVS-2058,42 by P1 into MG1655 |
| IIB967 |
MG1655 araB::T7 RNAP-tetA |
This work: P1 into MG1655 from BL21-AI |
| IIB968 | MG1655 ΔrecG::kan araB::T7 RNAP-tetA | This work: P1 into IIB967 from N4256 |
| Plasmids | ||
|---|---|---|
| pUC19 |
Empty plasmid vector |
43
|
| pEB526/pCas3 |
cas3 in pUC19 |
This work |
| pJLH11/pCas3K320L |
cas3K320L in pUC19 |
This work |
| pJLH12/pCas3K78L-K320L |
cas3K78L-K320L in pUC19 |
This work |
| pJLH18/pCas3K78L |
cas3K78L in pUC19 |
This work |
| pIIB14/pCascade |
cse1-cas6e in pUC19 |
This work |
| pIIB24/pCasC |
casC in pUC19 |
This work |
| pEB547 |
cas3 in pRSF-1b |
16
|
| pEB574 | cas3 in pACYCDuet-1 | This work |
Yields of pUC19 encoding Cas3 or Cascade (pCas3/pCascade) were compared with controls: empty plasmid (pUC19), plasmid-encoding stable catalytically inactive Cas3 (pCas3K320L/pCas3K78L) and pUC19 encoding incomplete Cascade (pCasC). Results are presented in Figure 1 and Table 2. Cas3 (pCas3) stimulated plasmid copy number that was 4-fold higher than empty pUC19 (respectively, 261.3 ng/µl ± 16.7 ng; 68.4 ± 22.5 ng/µl). Cas3 ATPase/helicase activity was required for this increase in yield; when identical tests were made on cells expressing Cas3 K320L (pCas3K320L), which lacks ATPase and helicase activity,16 plasmid copy number was similar to pUC19 (72.9 ± 3.4 ng/µl). Previous biochemical analysis16 showed that E. coli Cas3K320L protein overexpressed and purified in the same way as wild-type Cas3. We therefore think it unlikely that lack of Cas3K320L protein or its instability is an explanation for differences in plasmid yield between pCas3 and pCasK320L in the assay reported here. Cells expressing nuclease defective Cas3 (pCas3K78L) showed plasmid copy number that was similar to pCas3 (221 ± 18.8 ng/μl), indicating that Cas3 nuclease activity is not required for the observed effect on plasmid yield. pCas3 or pCas3K320L had little effect on plasmid stability, as cells generally retained the plasmid when measured from colony viabilities after plating on ampicillin or non-selective agar (Table 2). Expression of pCas3K78L corresponded to much-reduced plasmid stability for reasons unknown. Ethidium bromide staining of uncut pCas3 after agarose gel electrophoresis showed additional slowly migrating DNA compared with pUC19 or pCas3K320L (Fig. 1A). This is consistent with formation of multimeric plasmids. There was also an intriguing and reproducible lack of supercoiled plasmid observable from only pCas3K320L (Fig. 1A). Both of these observations on plasmid topology are addressed later in the results.
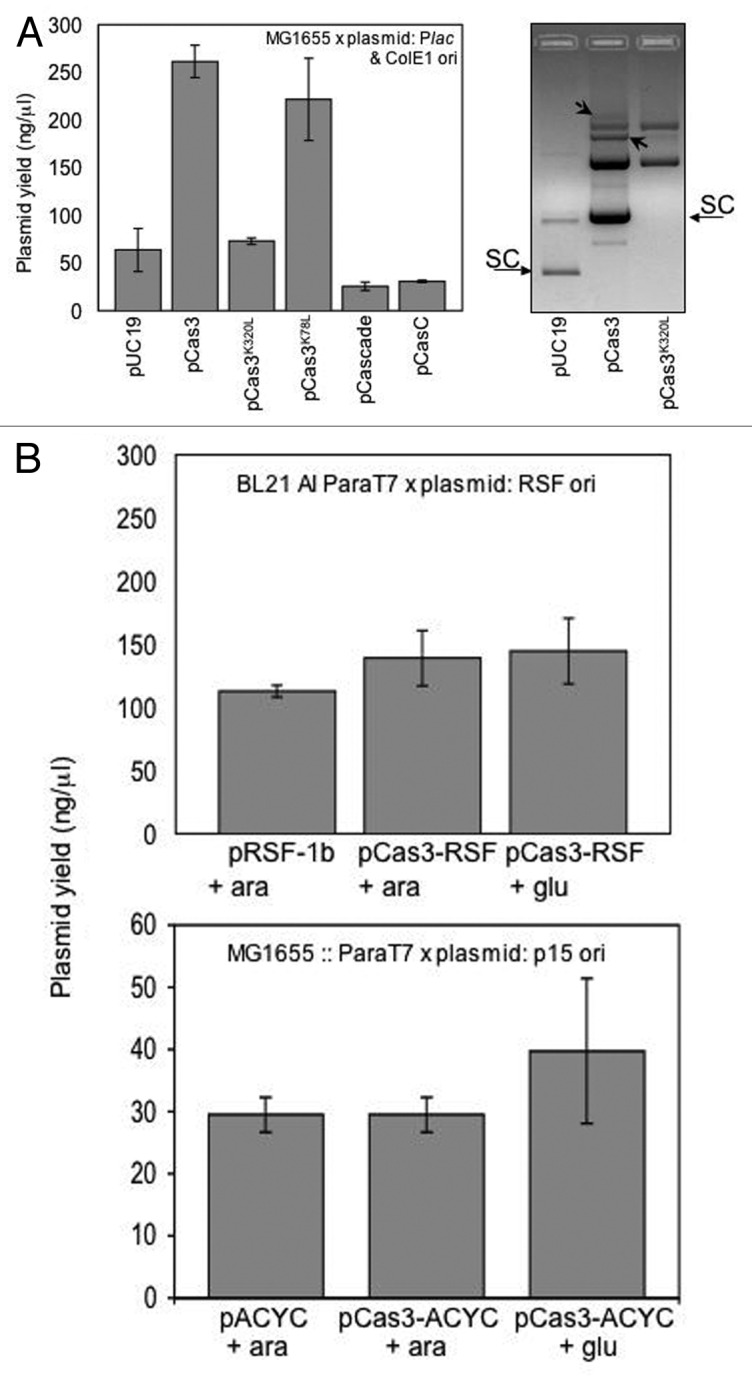
Figure 1. See also Table 2. Cas3 promotes ColE1 plasmid copy number. (A) Yields of the ColE1-based plasmid pUC19 were measured after extraction from E. coli MG1655 cells. Cells contained either pUC19 as empty plasmid vector, or pUC19 expressing Cas3, Cas3K320LL (helicase defective), Cas3K78L (nuclease defective), Cascade or Cas7 as indicated. Results are means of three independent tests with standard deviation error bars. The panel shows a typical outcome of gel electrophoresis loading 4 μl of uncut plasmid extracts (pUC, pCas3 or pCas3K320LL) in an ethidium bromide-stained agarose gel (0.8%). Arrows within the pCas3 lane point to slow-migrating plasmid multimers (see later Results section). Supercoiled plasmid is arrowed (SC); note that cas3 adds 2.9 kb to the size of pUC19, accounting for slower migration of pCas3 SC species compared with SC pUC19. No DNA size ladder is present because plasmids are uncut. (B) Yields of plasmids based on pRSF-1b or pACYC-Duet were measured after extraction from, respectively, BL21 AI or strain IIB967. pRSF-1b or PACYC-Duet were either empty plasmid, or expressed Cas3, as indicated. Arabinose (+ ara) was used to induce T7 polymerase that is needed to transcribe cas3 in each plasmid. Glucose (+ glu) gives tight repression of T7 polymerase. Results are means of two independent tests with error bars for standard deviation from the mean.
Table 2. Plasmid yields and stability (see also Figs. 1 and 2). Cells were grown as overnight cultures for extraction of pUC19-based ColE1 plasmids, or pRSF or p15A non-ColE1 plasmids, expressing Cas3 or Cascade as indicated.
| E. coli strain | Plasmid | Plasmid concentration (ng/µl)a) | OD600 | Viability cells per ml (×107)c) | AmpR, KanR or ChlmR cells (% total)b) |
|---|---|---|---|---|---|
| MG1655 |
pUC19 |
68.4 (± 22.5) |
1.3 (± 0.02) |
154 (± 22) |
100.0 |
| pCas3+ |
261.3 (± 17.0) |
2.0 (± 0.07) |
123 (± 32) |
94.0 |
|
| pCas3K320L |
72.9 (± 3.4) |
2.3 (± 0.01) |
109 (± 3) |
95.5 |
|
| |
pCas3K78L |
221.8 (± 43.0) |
1.4 (± 0.17) |
78.5 (± 23) |
5.3 |
| |
pCascade |
25.6 (± 4.4) |
1.9 (± 0.13) |
173(± 7) |
0.1 |
| |
pCasC |
30.8 (± 1.3) |
1.5 (± 0.12) |
72.5(± 33) |
5.0 |
| BL21 AI |
pRSF1b |
112.7 (± 4.6) |
1.4 (± 0.05) |
70 (± 10) |
89.7 |
| |
pCas3-RSF (arabinose) |
139.0 (± 21.8) |
1.1 (± 0.12) |
32.5 (± 5) |
85.7 |
| |
pCas3-RSF (glucose) |
144.5 (± 26.0) |
1.5 (± 0.15) |
77.5 (± 20) |
100 |
| MG1655 araB T7 RNAP |
pACYCDuet-1 pCas3-pACYC (arabinose) pCas3-pACYC (glucose) |
29.5 (± 2.8) 29.5 (± 2.8) 39.75 (± 11.7) |
1.7 (± 0.01) 1.46 (± 0.04) 1.75 (± 0.05) |
115 (± 35) 65 (± 7) 115 (± 21) |
100 100 100 |
| EB304 (Δcas3) |
pCascade |
25.4(± 4.0) |
0.94 (± 0.2) |
190 (± 16) |
1.0 |
| pUC19 | 47.66(± 7.0) | 2.0 (± 0.02) | 147 (± 10) | 100.0 |
a)Plasmid concentrations were measured in a Nanovue spectrophotometer (GE Healthcare). Values are means of three independent experiments with standard deviations given in parentheses. b)The percentage cells retaining plasmid was measured by comparing viable colony counts after plating on agar with, or without ampicillin or appropriate antibiotic. c)Viable cell number was from counting colonies after plating 100 µl of serially diluted bacteria on LB agar plates from at least two independent experiments.
In contrast to pCas3, pCascade had little effect on plasmid copy number compared with pUC19 (respectively, 25.6 +- 4.4 ng/μl; 68.4 +- 22.5 ng/μl). However, pCascade+ was unstable, being retained in only 0.1% of cells even though antibiotic selection was maintained. This retention rate was improved to 5% in cells expressing only the CasC subunit of Cascade (pCasC) under otherwise identical conditions (Table 2). The observed effect of plasmid yield and stability of pCascade was independent of Cas3, because results were the same in Δcas3 cells, lacking chromosomally encoded Cas3 (Table 2).
Cas3 does not stimulate copy number of plasmids with an RSF or p15A origin
Yields of non-ColE1 plasmids were also measured for any effect of Cas3 (Fig. 1B and Table 2). First, plasmids based on pRSF-1b were extracted from E. coli strain BL21 AI (Novagen), which has an arabinose inducible T7 RNA polymerase (ParaT7) needed for transcription of cas3 in pRSF-1b. Cas3-RSF constructs generate functional Cas3 in BL21 AI that is effective in defense against phage λvir.2,16 Cells grown in arabinose, to induce Cas3 expression, showed similar plasmid yield to cells grown in glucose to repress Cas3, or to cells harboring empty pRSF-1b. Next, further assays measured yields of pACYC-based plasmids from an E. coli MG1655 strain, IIB967 (Table 1), into which had been transduced ParaT7, inducible by arabinose as in BL21 AI, but preserving the same strain background as used with pUC plasmids in Figure 1. pACYC plasmids are low copy number in comparison to pRSF and pUC reflected in much lower yields of every pACYC construct (Fig. 1B). The presence of Cas3 had no effect on plasmid yield compared with empty pACYC control or if cells were grown in glucose or arabinose (Fig. 1B and Table 2), consistent with results from pRSF.
Observations thus far showed that Cas3 stimulates copy number of a ColE1 replicon, but not RSF or p15, and requiring Cas3 ATPase/helicase activity to do so. Cas3 exerted this effect independently of Cascade. RNaseHI is important for precise initiation of ColE1 replication from R-loops, and purified E. coli Cas3 can dissociate R-loops in vitro.16 Therefore, we tested for interplay in the effects of these two proteins in E. coli.
Cas3 and RNaseHI are antagonistic in E. coli cells
RNaseHI is required for precise initiation of ColE1 plasmid replication, by processing RNA in an R-loop formed between the RNAII pre-primer and plasmid ori.22 RNaseHI-deficient cells can support ColE1 replication, by mechanisms thought to involve multiple priming sites arising from hybridization of RNAII with other parts of plasmids.29 When we repeated plasmid copy number measurements from E. coli cells lacking RNaseHI (ΔrnhA, IIB796 Table 1) we observed that pCas3 yield was similar to pUC19 or pCas3K320L (Fig. 2A). This suggested that Cas3-stimulated plasmid copy number is dependent on RNaseHI and cannot rely on alternative methods of priming.
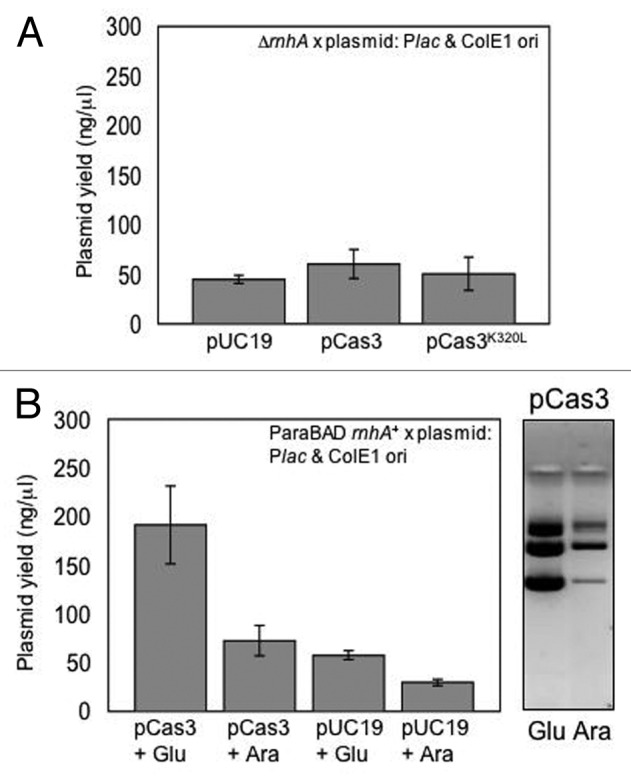
Figure 2. Interplay between Cas3 and RNaseHI affects plasmid copy number. (A) RNaseHI is needed for Cas3 to promote plasmid copy number. Experiments described in Figure 1A were repeated in MG1655 lacking RNaseHI (ΔrnhA) measuring yields of pUC19 as empty plasmid or expressing Cas3/Cas3K320L, as indicated. Results are means of three independent tests in each case, with standard deviation error bars. (B) In this experiment, in addition to chromosomal rnhA, RNaseHI can also be overexpressed from the chromosome by arabinose induction from ParaBAD. Cells were transformed with pUC empty vector or pCas3 and then grown in either glucose (+ glu, to repress RNaseHI) or arabinose (+ ara, to induce RNaseHI) as indicated. The panel shows representative plasmid yield from 4 μl of pCas3 (uncut) extracted from cells grown in arabinose or glucose, electrophoresed in an ethidium bromide-stained agarose gel (0.8%).
We then measured yields of pCas3 from E. coli cells expressing RNaseHI at high levels from the chromosome. These cells had an additional rnhA+ locus in the chromosome, under control of the arabinose inducible promoter ParaBAD, creating E. coli ParaBAD-rnhA+ (strain RCe442, Table 1). Cells grown in arabinose showed much-reduced yield of pCas3 comparable with empty pUC19 (Fig. 2B). In glucose, to repress ParaBAD RNaseHI expression, cells maintained a high yield of pCas3. Therefore, in these experiments, a surplus of RNaseHI induced from ParaBAD prevented a high yield of pCas3, despite RNaseHI being required to observe high yields of pCas3. We were unable to measure the abundance of RNaseHI molecules in cells using coomassie or silver-stained SDS-PAGE gels of total soluble cell proteins and, therefore, cannot accurately correlate RNaseHI protein expression with plasmid yields in these experiments. However, presumably in glucose media, RNaseHI expression from rnhA under its own promoter, rather than ParaBAD-rnhA+, is sufficient to support high yields of pCas3 that were lost in ΔrnhA cells.
As presented in Figure 1B and developed in the next section, the appearance of pCas3 concatemers is associated with cells lacking adequate RNaseHI activity.30 This is consistent with antagonistic effects of Cas3 against RNaseHI. As discussed later, this and the resulting stimulatory effect of Cas3 on pUC plasmid replication are more consistent with Cas3 acting at a site other than R-loops in this assay.
Cas3 influences plasmid topology: Stimulation of concatemers and removal of supercoiled plasmid
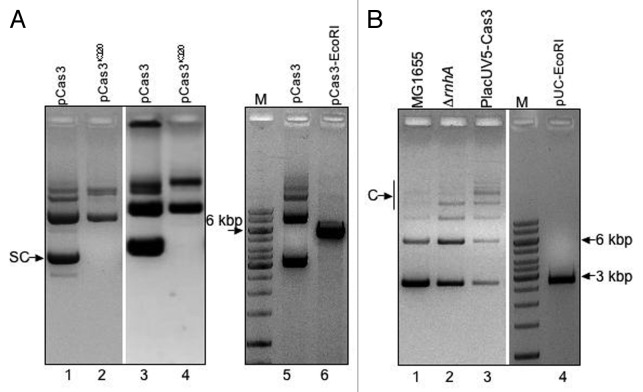
Cells lacking RNaseHI (ΔrnhA) can replicate ColE1 plasmids, characterized by formation of plasmid concatemers arising from homologous recombination and problems in plasmid segregation.29,30 Overexpression of RNaseHI (ParaBAD rnhA) antagonized the ability of Cas3 to promote pUC plasmid replication (Fig. 2B). Therefore, if in MG1655 cells, Cas3 from pCas3 was dominant to RNaseHI expressed from its own promoter, leading to a high yield of pCas3, then a phenotype similar to ΔrnhA cells might be observed, e.g., concatemer formation. pCas3+, but not pCas3K320L, reproducibly gave DNA product that was slowly migrating when analyzed uncut on agarose gels (Fig. 3A, lanes 1 and 2), and in some extractions, pCas3 DNA remained in gel wells (Fig. 3A, lanes 3 and 4). As in Figure 1, we noted that pCas3K320L failed to yield any detectable band corresponding to supercoiled plasmid (e.g., Fig. 3A, lanes 2 and 4). Digestion of pCas3 extracts with EcoRI restriction endonuclease, to introduce one cut, converted slowly migrating DNA bands into a single fragment (5.7 kbp), consistent with resolution of concatemers into linear DNA molecules (Fig. 3A, lanes 5 and 6). Evidence for Cas3-induced concatemers was also obtained by comparing pUC19 empty plasmid vector extracted from “wild-type” MG1655, ΔrnhA or cells expressing a chromosomal-engineered cas3 under control of IPTG inducible PlacUV5 instead of from pCas3 (Fig. 3B). As expected, pUC extracted from ΔrnhA cells gave concatemers compared with MG1655 (Fig. 3B, compare lanes 1 and 2), and these were even more prevalent in IPTG induced PlacUV5 Cas3 cells (Fig. 3B, lane 3). Again, the concatemers could be resolved into a single pUC-sized DNA band by treatment with EcoRI (Fig. 3B, lane 4). Therefore, increased ColE1 plasmid copy by Cas3 also provoked formation of plasmid species that migrated like concatemers, and which also seemed to require Cas3 helicase activity. This is discussed more below, with reference to what is known about Cas3 helicase activity and on measurements of plasmid copy number in cells lacking PcnB.
Figure 3. Cas3 provokes formation of plasmid concatemers. (A) Lanes 1–4 show ethidium bromide-stained agarose electrophoresis of uncut pCas3 or pCas3K320L from two independent plasmid extractions, to illustrate typical pCas3 concatemer formation including high molecular mass species that remain in gel wells (lane 3). Restriction digestion of uncut pCas3 converted multimeric plasmid (lane 5) into a single product consistent with linear pCas3 (5.4 kbp, lane 6). The DNA size marker is Fermentas GeneRuler. (B) Agarose gel showing concatemer (C) formation of pUC19 extracted from ΔrnhA cells (lane 2) compared with rnhA+ cells (lane 1). This effect is typical of cells lacking RNaseHI, as reported previously.30 IPTG induced expression of chromosomal Cas3 from PlacUV5 further increases concatemer formation of pUC19 (lane 3). All DNA species from PlacUV5-Cas3 were converted to single band of linear pUC19 after restriction digestion (lane 4).
Extractions of pCas3K320L consistently lacked detectable supercoiled DNA, summarized in Figure 4 (compare lanes 2 and 3). However, mutation of Cas3 nuclease HD domain in pCas3K78L restored the supercoiled species, even if the K320L mutation was also present (compare lanes 3–5). This is consistent with plasmid DNA nicking activity of E. coli Cas3 that requires the HD nuclease domain, as shown previously in work on E. coli CRISPR/Cas interference reactions.12 A possible nicking activity is intriguing in the assay we report, leading to the question; why does Cas3K320L have this effect on the supercoiled species but wild type Cas3 does not, even though both proteins have intact HD nuclease domains? This is discussed below.
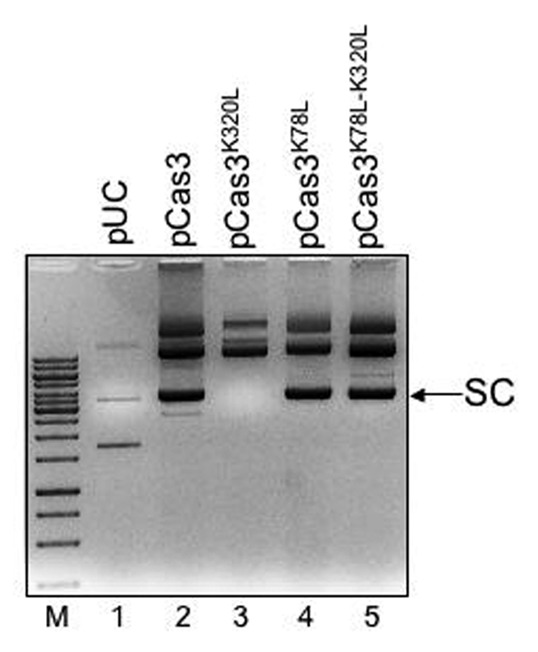
Figure 4. Potential for Cas3 nicking activity on supercoiled plasmid DNA. Ethidium bromide-stained agarose gel (0.8% w/v) showing 4 μl loadings of plasmid extractions, uncut, from E. coli MG1655. The position of supercoiled plasmid is labeled (SC), and is absent in lane 3 (pCas3K320L), but restored in lanes 4 and 5, representing nuclease defective Cas3 (Cas3K78L).
Discussion
We have shown that E. coli Cas3 promotes runaway replication of the ColE1 plasmid pUC19. This required Cas3 ATPase/helicase activity; mutation to the Walker A motif (Cas3K320L) reduced plasmid yield to levels of empty pUC19. A number of specific factors can increase ColE1 plasmid copy number, reviewed recently in reference 31. Many of these involve alterations to stability or relative levels of plasmid-encoded transcripts, RNAI and RNAII, which modulate ColE1 replication through RNA-DNA and RNA-RNA interactions. RNAII stimulates ColE1 replication by forming an R-loop with ori DNA, but this is blocked by RNAI binding to the 5′ end of RNA II, effectively decreasing plasmid copy number. It was unexpected that Cas3 would increase pUC copy number. Purified Cas3 can dissociate an R-loop in vitro,16 the same activity as previously reported for E. coli RecG helicase.27,28 However, RecG exerts the opposite effect to Cas3 in vivo, decreasing pUC plasmid copy number when measured in the same assay as we have used in this work.28 We noted that pCas3 was toxic in cells lacking RecG (ΔrecG), resulting in at least 30-fold less cell viability compared with pUC (Table 3). This was accompanied by little, if any, increase in plasmid yield in this strain, suggesting that the lethality of pCas3 in ΔrecG is unlikely to be caused by uncontrolled plasmid replication. Viability of ΔrecG cells was unaffected if Cas3 was expressed from pACYC, a low copy number non-ColE1 plasmid (Table 3). This is consistent with activities of Cas3 on ColE1 plasmids being responsible for reduced cell viability in the absence of RecG, possibly because RecG can counterbalance the effects of Cas3. In summary, in these assays it seems unlikely that Cas3 is unwinding R-loops. R-loop unwinding is consistent with reduced, not increased, ColE1 plasmid copy number, because it would dismantle RNAII-ori priming that initiates replication. What activity of Cas3 could increase ColE1 plasmid replication?
Table 3. Cas3 overexpression from ColE1 plasmid is toxic to cells lacking RecG.
| E. coli strain (MG1655) | Plasmid | Plasmid concentration (ng/µl) | OD600 | Viable cells per ml (×107) |
AmpR or ChlmR cells (% total) |
|---|---|---|---|---|---|
| N4256 ΔrecG |
pUC19 |
108.31 (± 15.4) |
1.9 (± 0.05) |
95 (± 30) |
100.0 |
| pCas3K320L |
91.38 (± 11.2) |
2.3 (± 0.01) |
248 (± 21) |
97.6 |
|
| pCas3 |
148.75 (± 24.9) |
2.0 (± 0.04) |
3 (± 3) |
100.0 |
|
| IIB968 ΔrecG araB::T7 RNAP) |
pACYCDuet-1 |
33.2 (± 3.9) |
1.7 (± 0.02) |
120 (± 14) |
100.0 |
| pCas3-pACYC (arabinose) |
29.5 (± 0.7) |
1.4 (± 0.03) |
70 (± 14) |
100.0 |
|
| pCas3-pACYC (glucose) |
46.7 (± 3.9) | 1.8 (± 0.13) | 100 (± 0) | 100.0 |
Details for measurement of cell viabilities and plasmid concentrations are the same as in footnotes to Table 1.
One possible explanation is that Cas3 may have regulatory roles on RNA that are distinct from CRISPR immunity. This is suggested by results showing interplay between Cas3 and RNaseHI: to achieve a high yield of pCas3 requires RNaseHI, but this is repressed if RNaseHI is overexpressed. Antagonism between activities of Cas3 and RNaseHI is a further indication that Cas3 does not target R-loops for unwinding, at least in this assay: if Cas3 were, then an overall effect o fdissociating R-loops would be the same as RNaseHI destroying R-loops, as reported for RecG. Our data are consistent with Cas3 stabilizing, promoting or generating R-loops for activation of ori ColE1. It is not likely that Cas3 is either forming or stabilizing R-loops in this context because its helicase activity is needed for increased plasmid copy number, an activity that dissociates R-loops.16 We propose instead that Cas3 helicase might promote pairing of RNAII with ori DNA, at the expense of RNAI-RNAII pairing, thus stimulating ColE1 replication. To do this, Cas3 helicase could dissociate RNAI from RNAII in a 3′ to 5′ direction to free up RNAII, indirectly promoting ColE1 replication. In support of this, ΔpcnB E. coli had no effect on pCas3 plasmid yield, but did reduce yields of pUC19 empty plasmid (Table S2). pcnB encodes poly(A)polymerase that ensures short half-life (< 2 min) of RNAI, thereby promoting ColE1 replication. RNAI persists when PcnB is absent, with the effect of reducing pUC copy number. Removal of RNAI from RNAII by another factor (e.g., Cas3) in the absence of PcnB would manifest as there being no major decreased plasmid copy number, as we have observed. However, future work would require testing this hypothesis in vitro using purified Cas3 and defined RNAI-RNAII substrate. It is interesting to note that RNAI-RNAII pairing creates several stem-loop or hairpin secondary structures in specific sequences, which could be targets for Cas3 for remodeling. There is currently some interest in manipulating host factors as a tool for upregulation of ColE1 plasmid replication.31 The mechanism by which Cas3 achieves it might uncover new possibilities for plasmid modifications.
Cas3 is a core CRISPR/Cas protein that was first studied as a requirement for effective defense of E. coli cells in assays against plaque formation by phage λvir, when expressed ectopically (from a plasmid) with Cascade.2 It is clear from that and later work, that E. coli Cas3-Cascade can be manipulated as an effective defense against phage.5,32 A crucial part of CRISPR defense is nicking and degradation of target DNA, catalyzed by Cas3 HD-nuclease when in complex with Cascade.12 In our assays, using a plasmid (pUC19) that lacked any PAM sequence and in the absence of Cascade, we observed a disappearance of supercoiled DNA that is consistent with nicking, but this was only clearly apparent from Cas3K320L, an ATPase defective mutant. After plasmid nicking, Cas3 did not processively degrade the plasmid, perhaps because Cascade is absent, and also because this pUC plasmid has not been engineered with PAM containing target DNA.12 As expected, nicking needed the active HD domain nuclease (e.g., Fig. 4). This intriguing observation suggests that correct binding and/or hydrolysis of ATP by Cas3 might be an important regulator of Cas3 nuclease activity. Given that very little is known about the structure or mechanism of ATPase catalysis by Cas3, this might be an interesting line of inquiry. It is possible that Cas3 nicking of plasmid DNA, only at a position corresponding to an R-loop, might offer a mechanism for ensuring Cas3 nuclease in CRISPR defense is targeted to the Cascade-crRNA R-loop formed after successful recognition of invasive DNA. We are also aware that, although speculative, such a nicking activity could direct rolling circle plasmid replication to occur. We suggest that slow migrating DNA species observed by expressing Cas3, especially in the absence of RNaseHI, result from R-loop provoked recombination. There is a possible alternative explanation that they are unresolved plasmid multimers from rolling circle replication, rather than products of R-loop provoked recombination. To distinguish between these possibilities will be addressed in future studies.
Detailed analyses of CRISPR loci across E. coli strains,33-35 and their relationship to coliphage, do not support a role for CRISPR/Cas solely in defense against plasmid or phage. It seems that E. coli CRISPR/Cas may be quite specialized and at least some components might be co-opted into other nucleic acid processing systems.36 It is thought that Cas3 and Cascade physically interact,12 which would provide a neat mechanism to target Cas3 to ssDNA for successful CRISPR interference. It will be interesting to see how this could interplay with recent evidence that Cas3 requires stabilization or chaperoning by HtpG.37 Very little is known about the Cas3 helicase/translocase mechanism. It is an intriguing possibility that Cas3 could recognize and translocate a common structural feature of both DNA and RNA, a 3′ single-stranded region, but this will require more detailed analysis in vitro using purified substrates.
Materials and Methods
Strains and plasmids
E. coli K-12 strains and plasmids used in this study are listed in the Table 1. Primers used are listed in Table S2. Deletion of the allele ygcB (cas3) in E. coli MG1655 was by the one-step gene disruption method.38 The substrate for this method was generated by PCR amplification of a cassette encoding resistance to apramycin (aprR). aprR PCR product had primer-generated extensions homologous to ygcB, encoding Cas3 in MG1655. Successful incorporation of aprR was designed to delete all of the ygcB except for flanking sequence of 10 codons, at both the beginning and at the end of the Cas3 coding sequence. aprR integrated into cas3 was verified by PCR using primers specific to ygcB flanking sequences (Table S2): Δcas3 giving a 0.9 kbp DNA fragment instead of 2.7 kbp from intact cas3. The 0.9 kbp aprR fragment was further verified by digestion with XhoI, to generate two DNA fragments from a single cleavage of aprR. aprR was then P1 transduced from the recombineering strain into MG1655, creating strain EB304 and again verified by PCR and XhoI. Table 1 lists the manipulations to create ΔrnhA and ΔpcnB in MG1655 background for consistency with other strain backgrounds, and strains obtained from other sources.
Cloning of E. coli ygcB (encoding Cas3), and K320L, K78L and K78L-K320L Cas3 variants, into pUC19 or pRSF-1b (pEB542) was by PCR amplification from existing cas3 plasmids that are described in reference 16 using primers in Table S1. Sub-cloning of ygcB into pACYC-Duet was from pEB542 via NcoI and KpnI, giving pEB574 (Table 1). Cascade was cloned into pUC19 (pCascade) as an operon (casA-casE /cse1-cas6e), or containing only CasC (pCasC) by PCR amplification from MG1655 genomic DNA, using primers listed in Table S2.
Media and general microbiology methods
LB broth and agar (10 g bacto-tryptone, 5 g yeast extract, 10 g NaCl to 1 L of water and sterilised) was used for all E. coli cell growths except during recombineering, as described in Datsenko and Wanner. Selection of ColE1 pUC plasmids was by ampicillin (100 μg/ml), kanamycin (50 μg/ml) for pRSF-1b plasmids and chloramphenicol (15 μg/ml) for pACYC-based plasmids. When required, arabinose or glucose was added to a final concentration of 13 mM, and IPTG to 0.2 mM. To extract plasmids, cell cultures were grown overnight with shaking at 37°C and cell density was measured at 600 nm (OD600, Table 2). Cell viabilities were calculated by plating onto agar plates 100 µl of bacteria cultures that had been serially diluted in 67 mM phosphate buffer (pH = 7.0). The fraction of plasmid-free cells was calculated by counting colonies on plates with and without ampicillin selection.
Agarose gel analysis of plasmid DNA yields and conformation
All plasmid DNA was extracted from 2 ml of culture grown as described above, using new, unused columns from a Wizard plus SV mini-prep kit (Promega). Plasmid DNA concentrations were measured from 2 µl aliquots in a Nanovue spectrophotometer (GE Healthcare). Typically, a 4 μl sample of plasmid DNA was analyzed after electrophoresis on a 1% agarose ethidium bromide gel. We also tested if an alternative method of plasmid extraction, gentle lysis with lysozyme followed by phenol extraction and ethanol precipitation, gave any difference to plasmid yields or concatamers compared with the mini-column kit method, but observed no difference. Restriction digestion of plasmid DNA used EcoRI (New England Biolabs) with digestion under standard conditions for this enzyme.
Supplementary Material
Acknowledgments
This work was supported by Grant 119-1191196-1201 from the Croatian Ministry of Science, Education and Sports and an award from Croatian Academy of Sciences and Arts and a UK BBSRC PhD studentship for J.L.H. We are grateful to Dr J. Vogel, Dr C. Rudolph, Prof Robert G Lloyd and Dr K. Datsenko for E. coli strains, and to two anonymous referees for interesting observations.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23876
References
- 1.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O’Toole GA. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol. 2012;194:5728–38. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, et al. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol. 2010;77:1367–79. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer S, Maier LK, Stoll B, Brendel J, Fischer E, Pfeiffer F, et al. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem. 2012;287:33351–63. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, et al. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–39. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 10.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 11.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–36. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 12.Westra ER, van Erp PB, Künne T, Wong SP, Staals RH, Seegers CL, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–40. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 14.Thomas M, White RL, Davis RW. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci USA. 1976;73:2294–8. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivančić-Baće I, Howard JA, Bolt EL. Tuning in to interference: R-loops and cascade complexes in CRISPR immunity. J Mol Biol. 2012;422:607–16. doi: 10.1016/j.jmb.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 16.Howard JA, Delmas S, Ivančić-Baće I, Bolt EL. Helicase dissociation and annealing of RNA-DNA hybrids by Escherichia coli Cas3 protein. Biochem J. 2011;439:85–95. doi: 10.1042/BJ20110901. [DOI] [PubMed] [Google Scholar]
- 17.Beloglazova N, Petit P, Flick R, Brown G, Savchenko A, Yakunin AF. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30:4616–27. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beloglazova N, Petit P, Flick R, Brown G, Savchenko A, Yakunin AF. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30:4616–27. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulepati S, Bailey S. Structural and biochemical analysis of nuclease domain of clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 3 (Cas3) J Biol Chem. 2011;286:31896–903. doi: 10.1074/jbc.M111.270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–42. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguilera A, García-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Itoh T, Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc Natl Acad Sci USA. 1980;77:2450–4. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogawa T, Okazaki T. Function of RNase H in DNA replication revealed by RNase H defective mutants of Escherichia coli. Mol Gen Genet. 1984;193:231–7. doi: 10.1007/BF00330673. [DOI] [PubMed] [Google Scholar]
- 24.Summers D. Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol Microbiol. 1998;29:1137–45. doi: 10.1046/j.1365-2958.1998.01012.x. [DOI] [PubMed] [Google Scholar]
- 25.Cesareni G, Muesing MA, Polisky B. Control of ColE1 DNA replication: the rop gene product negatively affects transcription from the replication primer promoter. Proc Natl Acad Sci USA. 1982;79:6313–7. doi: 10.1073/pnas.79.20.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polisky B. ColE1 replication control circuitry: sense from antisense. Cell. 1988;55:929–32. doi: 10.1016/0092-8674(88)90235-8. [DOI] [PubMed] [Google Scholar]
- 27.Fukuoh A, Iwasaki H, Ishioka K, Shinagawa H. ATP-dependent resolution of R-loops at the ColE1 replication origin by Escherichia coli RecG protein, a Holliday junction-specific helicase. EMBO J. 1997;16:203–9. doi: 10.1093/emboj/16.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vincent SD, Mahdi AA, Lloyd RG. The RecG branch migration protein of Escherichia coli dissociates R-loops. J Mol Biol. 1996;264:713–21. doi: 10.1006/jmbi.1996.0671. [DOI] [PubMed] [Google Scholar]
- 29.Dasgupta S, Masukata H, Tomizawa J. Multiple mechanisms for initiation of ColE1 DNA replication: DNA synthesis in the presence and absence of ribonuclease H. Cell. 1987;51:1113–22. doi: 10.1016/0092-8674(87)90597-6. [DOI] [PubMed] [Google Scholar]
- 30.Subia NL, Kogoma T. Concatemer formation of ColE1-type plasmids in mutants of Escherichia coli lacking RNase H activity. J Mol Biol. 1986;189:389–99. doi: 10.1016/0022-2836(86)90311-6. [DOI] [PubMed] [Google Scholar]
- 31.Camps M. Modulation of ColE1-like plasmid replication for recombinant gene expression. Recent Pat DNA Gene Seq. 2010;4:58–73. doi: 10.2174/187221510790410822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol. 2010;75:1495–512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 33.Díez-Villaseñor C, Almendros C, García-Martínez J, Mojica FJ. Diversity of CRISPR loci in Escherichia coli. Microbiology. 2010;156:1351–61. doi: 10.1099/mic.0.036046-0. [DOI] [PubMed] [Google Scholar]
- 34.Touchon M, Charpentier S, Pognard D, Picard B, Arlet G, Rocha EP, et al. Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology. 2012;158:2997–3004. doi: 10.1099/mic.0.060814-0. [DOI] [PubMed] [Google Scholar]
- 35.Touchon M, Rocha EP. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One. 2010;5:e11126. doi: 10.1371/journal.pone.0011126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol. 2011;79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yosef I, Goren MG, Kiro R, Edgar R, Qimron U. High-temperature protein G is essential for activity of the Escherichia coli clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Proc Natl Acad Sci USA. 2011;108:20136–41. doi: 10.1073/pnas.1113519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stockum A, Lloyd RG, Rudolph CJ. On the viability of Escherichia coli cells lacking DNA topoisomerase I. BMC Microbiol. 2012;12:26. doi: 10.1186/1471-2180-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 41.McGlynn P, Lloyd RG. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101:35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 42.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–68. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.