Abstract
The clustered regularly interspaced short palindromic repeats (CRISPR) system represents a highly adaptive and heritable defense system against foreign nucleic acids in bacteria and archaea. We analyzed the two CRISPR-Cas systems in Methanosarcina mazei strain Gö1. Although belonging to different subtypes (I-B and III-B), the leaders and repeats of both loci are nearly identical. Also, despite many point mutations in each array, a common hairpin motif was identified in the repeats by a bioinformatics analysis and in vitro structural probing. The expression and maturation of CRISPR-derived RNAs (crRNAs) were studied in vitro and in vivo. Both respective potential Cas6b-type endonucleases were purified and their activity tested in vitro. Each protein showed significant activity and could cleave both repeats at the same processing site. Cas6b of subtype III-B, however, was significantly more efficient in its cleavage activity compared with Cas6b of subtype I-B. Northern blot and differential RNAseq analyses were performed to investigate in vivo transcription and maturation of crRNAs, revealing generally very low expression of both systems, whereas significant induction at high NaCl concentrations was observed. crRNAs derived proximal to the leader were generally more abundant than distal ones and in vivo processing sites were clarified for both loci, confirming the previously well-established 8 nt 5′ repeat tags. The 3′-ends were more diverse, but generally ended in a prefix of the following repeat sequence (3′-tag). The analysis further revealed a 5′-hydroxy and 3′-phosphate termini architecture of small crRNAs specific for cleavage products of Cas6 endonucleases from type I-E and I-F and type III-B.
Keywords: methanoarchaea, CRISPR-Cas system, immunity of prokaryotes, regulatory RNA, phages, Methanosarcina mazei
Introduction
The CRISPR system has been discovered as a defense mechanism against exogenous nucleic acids (e.g., plasmids and phages) in bacteria and archaea (recently reviewed e.g., in refs. 1–8). In contrast to other gene transfer and phage defense mechanisms, the CRISPR system is highly adaptive and heritable as it incorporates sequences derived from foreign elements directly into its respective locus. The characteristic short palindromic repeats of a CRISPR locus are separated by short non-repetitive spacer sequences derived from previous encounters with foreign nucleic acids that specify the targets of CRISPR interference.9-13 The number of spacer and direct repeats in one CRISPR locus can vary considerably from one organism to another.14 The CRISPR loci are usually flanked on one site by a set of cas (CRISPR-associated) genes, encoding the protein machinery essential for the CRISPR defense system.6,14 Those Cas proteins are arranged in different combinatory sets that serve as a criteria to classify CRISPR-Cas systems into three different types (reviewed in Westra et al.7). The defense mechanism of all three CRISPR types relies on similar tasks that are grouped into the following phases: Acquisition of new spacers into the CRISPR arrays (adaptation phase), processing of the small interfering RNAs (crRNAs) (transcription and processing phase) and CRISPR interference with the invading foreign nucleic acids (interference phase).2,15-17 In general, little is known about the molecular mechanism of spacer acquisition,18-20 whereas insight into the molecular process of the two other phases has been recently obtained, unraveling three major types of RNA biogenesis and interference (types I–III) (reviewed in detail7,8), a classification which also takes phylogeny and composition of the cas genes into account.6,14 Transcription of CRISPR loci is initiated within the leader region resulting in long precursor CRISPR RNAs (pre-crRNA) containing the full set of CRISPR repeats including the embedded invader derived spacers.21-24 All pre-crRNAs are subsequently processed by specific endoribonucleases, CRISPR-specific Cas6-endoribonuclease (type I and type III) and RNase III (type II), generating the small interfering RNAs, called crRNAs.18,23,25-29 A general hallmark of CRISPR-based immunity is that pre-crRNAs are in general cleaved at a specific site within the repeat sequences followed by either 5′ trimming (type II), 3′ trimming (type III) or no trimming (certain type I subtypes).8
In types III and I, cleavage occurs in a sequence-specific manner at the 3′-end of a stem-loop structure resulting in well-defined 5′-ends of the crRNAs containing individual spacer sequences and a 5′ flanking part of the repeat sequence (at least 8 nucleotides, 5′ tag), which is considered to be important for binding to the Cas-complex. During the interference phase, the matured crRNAs are used to guide Cas protein complexes for recognition and cleavage of the invading nucleic acid in the context of a crRNA/Cas ribonucleoprotein complex, resulting in elimination of the invader. The Cas-protein complexes constitute to ribonucleoprotein effector complexes containing respective crRNAs and differ in their composition for type I, II and III.7,18,30-35 For a detailed description, see recent reviews.7,8
CRISPR loci are found in almost all sequenced archaeal genomes and ~40% of the bacterial genomes and show a surprising diversity of types and subtypes.6,14,31,36 In 2005, Haft et al.14 defined eight CRISPR subtypes; only recently Makarova et al.6 proposed a modified “polythetic” classification based on evolutionary relationship and the definition of cas genes characteristic for certain subtypes (called signature genes). Overall, Makarova et al.6 defined three major types of CRISPR-Cas systems with a further division of several subtypes. We use the more recent subtype classification from Makarova et al.6 In methanoarchaea and haloarchaea, examples for CRISPR-cas subtype I-B systems have been identified and analyzed only very recently.37,38 The recent investigation of the single CRISPR-Cas system (subtype I-B) and respective RNA processing in Methanococcus maripaludis C5 led to the identification of a novel Cas6 enzyme of subtype I-B designated Cas6b.38 In general, Cas6-like proteins are involved in processing the precursor crRNA and represent one of the most diverse Cas proteins in the different CRISPR-Cas subtypes. Initially identified in CRISPR-Cas subtype III-B in Pyrococcus furiosus,25,27,28,39 Cas6-like proteins have also been recognized in subtype I-F in Pseudomonas aeruginosa (Cas6f),26 Thermus thermophilus and Escherichia coli (Cas6e).29,40 Though all Cas6-like endoribonucleases are involved in the initial step of crRNA maturation and therefore appear to be functionally analogous, these Cas6 family endoribonucleases show only moderate sequence homology, differ in their RNA recognition and binding behavior (sRNA, ssRNA) as well as in the composition of their catalytic center (reviewed in Wiedenheft et al.8). In contrast to most Cas6-like proteins, which require one single-conserved histidine residue for activity besides lysine or tyrosine,26-29,40,41 Cas6b in M. maripaludis and Clostridium thermocellum contain two additional complementary histidine residues (H38 and H40) essential for the cleavage activity.38 Besides this work, Beloglazova et al.42 provide an insight into the structure and activity of the Cas3 HD nuclease of Methanocaldococcus jannaschii, which is most likely involved in the interference step. In the Methanosarcinales however, no functional analysis of CRISPR-Cas systems has been performed today.
Methanosarcina mazei strain Gö1 is a representative mesophilic archaeon of significant ecological importance due to its role in biogenic methane production in various anaerobic habitats on Earth.43 The original genome annotation published in 200244 did not include the information on potential CRISPR loci. However, re-analyzing the genome identified two CRISPR-Cas systems. In this work, we provide the first insights into the function and specific roles of these two CRISPR-Cas systems, belonging to CRISPR-Cas subtype I-B and subtype III-B, respectively (see Fig. 1). To elucidate CRISPR RNA (crRNA) processing in M. mazei, we performed an RNA sequencing approach and northern blot analyses under several stress conditions. In addition, both potential Cas6b endonucleases were characterized and analyzed for crRNA processing in vitro.
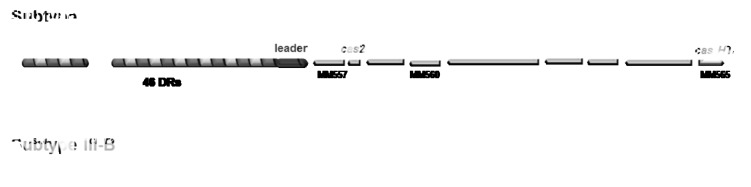
Figure 1. The two CRISPR loci present in Methanosarcina mazei. Organization of the subtype I-B and subtype III-B CRISPR locus. The respective ORF numbers are indicated according to Deppenmeier et al.44 Gene names are based on the nomenclature of Makarova et al.6
Results
M. mazei strain Gö1 encodes a subtype I-B and a subtype III-B CRISPR-Cas system
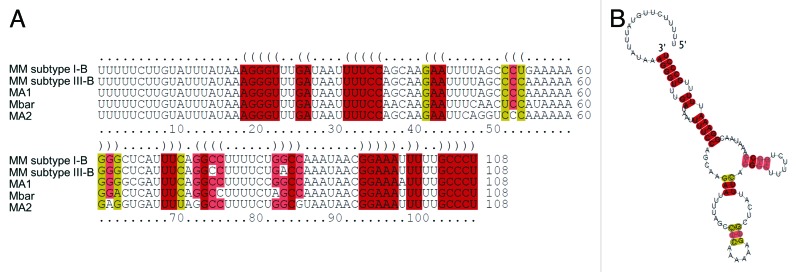
In addition to a CRISPR-Cas system of subtype I-B that posses 8 cas genes, a second system representing a CRISPR-Cas subtype III-B is encoded in the chromosome comprised of at least 6 cas genes (see Fig. 1). The CRISPR-Cas subtype I-B (Hmari subtype) contains 46 direct repeats with separating spacers (34–43 nt). The Cas proteins are encoded on the reverse strand in one long polycistronic operon (MM565-MM557) directly flanking the leader and CRISPR array as depicted in Figure 1. Overall, this CRISPR-Cas subtype I-B module shows a gene structure very similar to the single CRISPR-Cas module of M. maripaludis C538 and includes a Cas6-like endonuclease (MM560), with high sequence similarity to the novel Cas6b enzyme of M. maripaludis. The two systems differ, however, concerning their operon structure. By an RT-PCR analysis, we determined a single operon organization for the I-B cas-genes in M. mazei (data not shown). In contrast, the cas-genes in M. maripaludis are encoded in two separate operons in opposite orientations: Cas8b, Cas7, Cas5, Cas3 and Cas6b in one and Cas1, Cas2 and Cas4 in the other. The second CRISPR locus, subtype III-B, is also on the reverse strand, but these cas-genes are downstream of the CRISPR array. The array contains 81 direct repeats separated by spacers (32–48 nt) with a 152 nt leader. It is flanked by a potential polycistronic operon (MM3360-MM3353) encoding the RAMP module of Cas proteins (Cas6 like protein, Cmr1, Cmr6, Cas10, Cmr4 and Csm4) including two open reading frames (MM3360 and MM3354), for which no function could be assigned (Fig. 1). Strikingly, the CRISPR subtype III-B also contains a homolog of the novel Cas6b enzyme identified in M. maripaludis (MM3359). Both CRISPR loci contain a highly conserved direct repeat of 37 nucleotides (see below). Overall, the 5′-end of the direct repeats found in the CRISPR-Cas systems in methanoarchaea (see Fig. S1) and moreover across very many CRISPR repeats of different repeat clusters is highly conserved,31,36 potentially leading to a distinct and highly conserved 5′ tag of the respective downstream spacer during crRNA maturation. The leader sequence upstream of the CRISPR array in both different M. mazei subtypes is nearly identical, its predicted structure showed high conservation with several CRISPR leaders in methanosarcinales (see Fig. 2).
Figure 2. Structural comparison of the conserved leader sequences of CRISPR-Cas systems in Methanosarcinales. (A) Secondary structure alignment of leader sequences identified by computer-based searches of related Methanosarcina species performed with LocARNA.67 MM, M. mazei; Mbar, M. barkeri; MA, M. acetivorans. (B) Consensus secondary structure prediction by RNalifold.71
Characterizing Cas6b proteins demonstrated repeat specific endonucleolytic activity
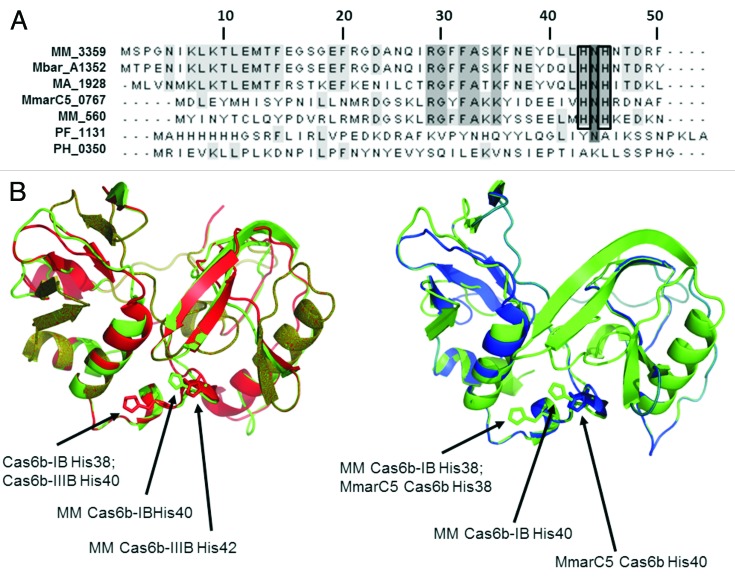
Both CRISPR I-B and III-B modules contained a homolog of the Cas6b endonuclease of M. maripaludis, which shows only 12% identity to the P. furiosus Cas6. The two M. mazei Cas6b homologs [Cas6b-IB (MM560) and Cas6b-IIIB (MM3359)] share 39% amino acid identity (63% amino acid similarity). In comparison to the M. maripaludis Cas6b, they showed 44% (Cas6b-IB) and 38% (Cas6b-IIIB) amino acid identity (69% and 64% amino acid similarity), including the two conserved histidines (H38 and H40) recently reported to be essential for catalytic activity in M. maripaludis38 (Fig. 3A). In addition, both M. mazei Cas6b proteins contained the typical Cas6 motif. Modeling the structure of the two M. mazei Cas6b endonucleases using the phyre 2 server demonstrated high structural conservation (Fig. 3B) and verified the conserved position of the two catalytic histidines, H38 and H40 (Cas6b-IB) and H40 and H42 (Cas6b-IIIB), respectively. A comparison based on the predicted structure of the M. maripaludis Cas6b revealed that the overall structure of all three proteins is highly conserved. Moreover, the structural location and steric orientation of the first conserved histidine of all three Cas6b proteins (MM560 H38, MM3359 H40 and M. maripaludis Cas6b H38) is predicted to be identical, whereas in case of the second histidine, the steric orientation in MM3359 and M. maripaludis is predicted to be identical, but differs from the orientation of MM560. Taking the high conservation of the two repeats in M. mazei into account as well, this finding indicates that the endonucleolytic activities of the M. mazei Cas6b proteins is similar.
Figure 3. Structural comparison of Cas6 proteins. (A) Alignment (ClustalW2) of the N-terminal part of Cas6 homologs shows the conserved histidines for all Cas6b homologs (black boxes). MM, M. mazei; Mbar, M. barkeri; MA, M. acetivorans; MmarC5, M. maripaludis C5; PF, P. furiosus; PH, P. horikoshii. (B) Structural models (Phyre2) of the whole proteins of M. mazei (MM) Cas6b-IB and Cas6b-IIIB and the M. maripaludis C5 (MmarC5) Cas6b were aligned (DaliLite). Depicted are comparisons of MM Cas6b-IB (green) and Cas6b-IIIB (red) and of MM Cas6b-IB and MmarC5 Cas6b (blue). Homolouge histidines are indicated through arrows.
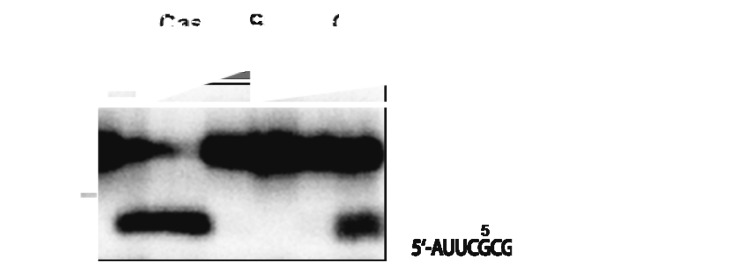
To test this assumption, both potential Cas6b endoribonucleases, Cas6b-IB and Cas6b-IIIB, were cloned as N-terminal His-tag fusions and heterologously overexpressed in E. coli, leading to a highly soluble Cas6b-IIIB protein fraction and a significant less soluble Cas6b-IB fraction (approx. 95% insoluble, see Fig. S2A). Both proteins were purified by affinity chromatography on Ni-NTA agarose leading to approx. 95% pure protein fractions (Fig. S2B). The purified proteins were analyzed concerning their in vitro cleavage activity using 5′ radioactively labeled single repeat RNAs (37 nt) derived from both CRISPR modules (consensus repeat I-B and repeat III-B, see Materials and Methods). Each Cas6b homolog was able to cleave both repeat variants and showed the same cleavage specificity, generating a 5′-labeled product of 29 nt and the respective non-labeled 3′ fragment of 8 nt (5′AUUGAAAC) (Fig. 4, repeat III-B not shown). Additional smaller 5′-labeled fragments were detected (e.g., 20 nt, 25 nt), some of them represent non-enzymatic cleavage products (control assay) or result from further unspecific processing of the remaining 29 nt repeat by Cas6b proteins (data not shown). In comparison, Cas6b-IIIB showed a significantly higher cleavage activity than Cas6b-IB (appr. 20-fold). This observation might be due to the optimal folding of Cas6b-IIIB; in case of Cas6b-IB, only a subpopulation might be folded correctly since the protein tended to precipitate. As a control, the repeat-IB with a dGTP at position 29 was used as a substrate for both Cas6b homologs, which resulted in both cases in the loss of cleavage activity verifying the processing site (data not shown). The cleavage activity of in vivo synthesized Cas6b homologs in M. mazei cell extracts showed significant but very low levels of cleavage activity (data not shown).
Figure 4. Endonuclease activity of purified Cas6b-IB and Cas6b-IIIB proteins. The cleavage assay was performed with 5′ radioactively labeled repeat 1 RNA in the presence of Cas6b-IB (10 µg, 15 µg, 20 µg) and Cas6b-IIIB (0.5 ng, 5 ng, 50 ng, 0.5 µg, 5 µg) protein. Cleavage patterns were analyzed on 8% polyacrylamide/7 M urea sequencing gel. On the right side, the identified repeat cleavage site is schematically depicted.
CRISPR-Cas subtypes I-B and III-B are transcribed and processed in vivo under high NaCl conditions
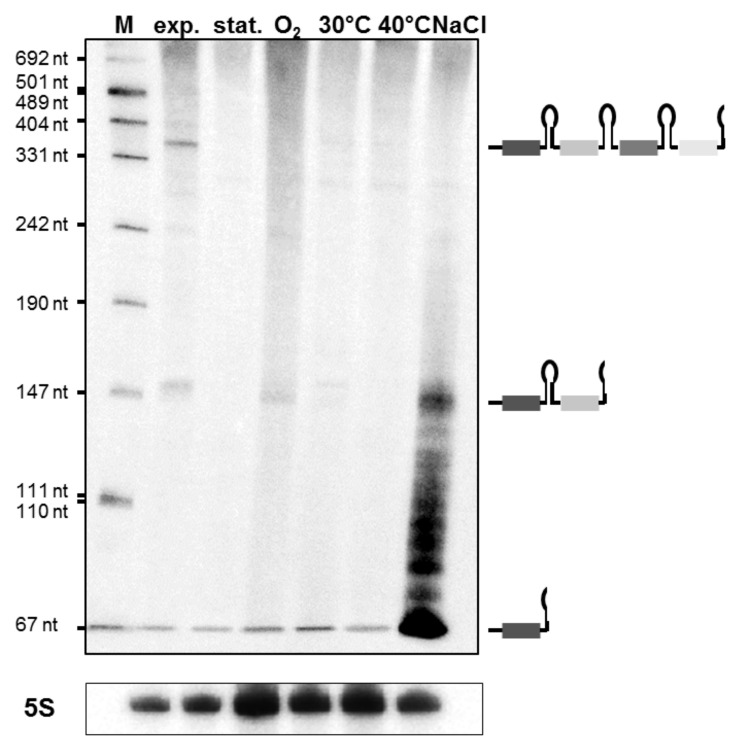
Previous transcriptome analyses using genome-wide DNA microarrays indicated that all genes encoding Cas proteins of the CRISPR-Cas subtype I-B and subtype III-B are transcribed in M. mazei.45-47 Studying in vivo transcription and maturation of precursor crRNA by a northern blot analysis using labeled repeat-IB probes demonstrated only very low expression and maturation of crRNAs under optimal growth conditions during both exponential and stationary growth, under oxygen or temperature stress (see Fig. 5) or UV-light exposure or nitrogen limitation (data not shown). In the presence of high NaCl concentrations (500 mM) however, transcription and maturation was strongly induced, leading to large amounts of distinct crRNAs with an approximate length of 67 nt; also, several longer fragments of precursor crRNAs were detected due to incomplete 5′ processing and unspecific processing at the 3′ end of crRNA precusors (see Fig. 5; Fig. S3).
Figure 5. crRNA abundance in M. mazei. Total RNA was isolated from M. mazei growing under optimal growth conditions and under various stress conditions (O2, 30°C, 40°C, NaCl) as described in Materials and Methods. Subsequent northern blot analysis using 5′ radioactively labeled repeat I-B (subtype I-B) was performed. In addition, the expression of 5S rRNA of the respective RNA preparation is shown. The pUC Mix 8 marker (Thermo Scientific, cat. no. SM0303) was used for designation of fragment size. On the right side, observed crRNA processing is depicted schematically.
Analyzing crRNA processing for CRISPR-Cas subtype I-B and subtype III-B by an RNAsequencing approach
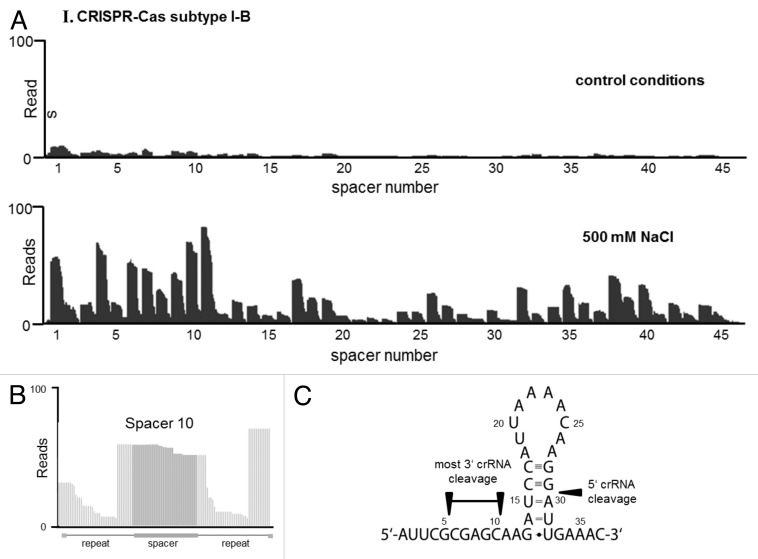
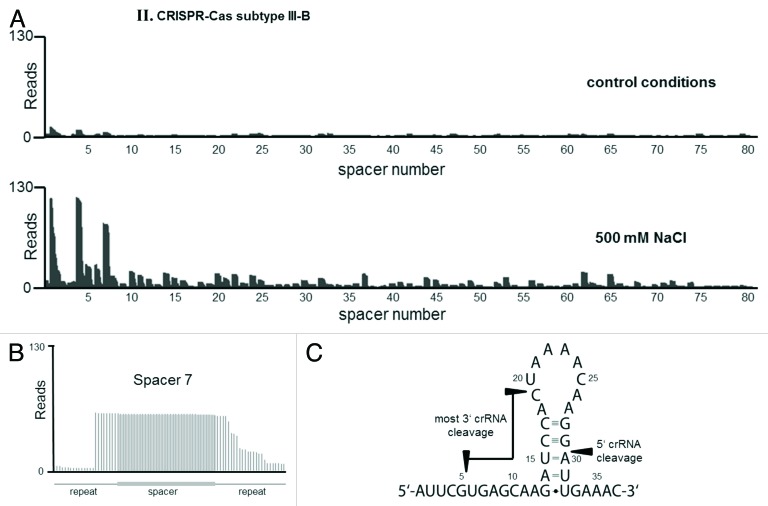
Total RNA from from exponentially growing cultures under optimal conditions and in the presence of high NaCl concentrations was isolated and freed of residual genomic DNA by DNase I treatment. At next, total RNA was divided into two parts. One was directly used (untreated library) for Solexa sequencing (see next part), whereas the second one was enriched for transcripts that possess 5′-hydroxy and 3′-phosphate termini by T4 polynucleotide kinase (PNK) to phosphorylate all 5′ ends. T4 polynucleotide kinase treatment also removes non-3′-OH ends comprising 2’ phosphate, 3′ phosphate or cyclic 2’,3′ phosphate. The four obtained cDNA libraries (salt +/− PNK and control +/− PNK) were sequenced using a HiSeq 2000 machine (Illumina) in single read mode (see Materials and Methods). Approximately 3.6 million sequence reads were obtained for each library, which were mapped to the M. mazei genome. The number of cDNA hits for each nucleotide position for both strands were calculated and the data visualized using the Integrated Genome Browser (IGB, Affimetrix) as recently described.48 Analyzing the reads of the respective CRISPR loci, the abundance of crRNAs and processing patterns confirmed active transcription and processing of both arrays under high NaCl conditions, whereas under control conditions, very low expression and processing was obtained. Comparing the respective differential libraries normalized against each other demonstrated that the PNK-treated cDNA libraries are in general enriched for crRNAs at both loci (see Fig. S4), strongly arguing that the mature crRNAs contain 5′-OH/ ends and 3′-P ends due to cleavage and processing. In the following, the analyses mainly focused on the PNK-treated libraries. Under control conditions, both CRISPR subtypes were expressed at very low (but comparable) levels, whereas under NaCl conditions, expression and maturation was approximately 5–10-fold higher (see Fig. 6A and 6B). As was expected, 46 mature crRNAs of subtype I-B and 81 of subtype III-B were detected and they showed different abundances with an overall tendency that crRNAs derived from the leader end are more abundant than the distal ones. This tendency was more pronounced in subtype III-B than in subtype I-B. In addition, a few crRNAs appear to be extremely high abundant in comparison with their respective flanking crRNAs (e.g., crRNA containing spacer 7 in subtype III-B or spacer 32 in subtype I-B), which might be due to direct effects of the respective flanking spacers on the formation of the repeat structure, potentially leading to a processing order for the biogenesis of crRNAs (see below), as well as on crRNA stability.
Figure 6A. crRNA processing for CRISPR-Cas subtype I-B by an RNA sequencing approach. (A) Sequencing reads for the CRISPR array are mapped to the M. mazei genome. Comparison of analyzed RNAseq reads derived from a culture under optimal growth condition and high salt condition. (B) The general processing of crRNA is shown exemplary for spacer 10. (C) Within the repeat structure, the distinct conserved 5‘-cleavage of crRNAs is shown, whereas the 3‘-ends of crRNAs are variable trimmed at marked region.
Figure 6B. crRNA processing for CRISPR-Cas subtype III-B by an RNA sequencing approach. (A) Sequencing reads for the CRISPR array are mapped to the M. mazei genome. Comparison of analyzed RNAseq reads derived from a culture under optimal growth condition and high salt condition. (B) The general processing of crRNA is shown exemplary for spacer 7. (C) Within the repeat structure, the distinct conserved 5‘-cleavage of crRNAs is shown, whereas the 3‘-ends of crRNAs are variable trimmed at marked region.
Zooming into the 5′- and 3′-ends of individual crRNAs as exemplarily depicted in Figure 6 IB and IIB for crRNA containing spacer 10 and 7, respectively, demonstrated that the 5′-ends of the crRNAs of both subtypes are distinct, whereas the 3′-ends are quite diverse, potentially due to further processing or exonuclease activities in the cell. In the case of subtype III-B, 3′-ends were even more diverse than for subtype I-B ranging from position 5–19 of the repeat (see Fig. 6 IIC). The distinct 5′-ends of the crRNAs further established the specific in vivo cleavage site of both Cas6b endonucleases within the 3′-end of the direct repeat between position 29 and 30 (see Fig. 6 IC and IIC), confirming the cleavage site of both Cas6b homologs determined in vitro. These specific cleavage sites were also detectable when analyzing the respective – PKN cDNA libraries. In general, we found no antisense transcription (asRNA) as has been extensively obtained in M. maripaludis,38 with the exception of a single spacer in the subtype III-B (spacer 18, see Fig. S4), for which a low number of the respective asRNA was detected. However, this did not correlate with a significantly reduced abundance of the respective sense crRNA in comparison to the flanking crRNAs.
Direct repeats form a five base pair hairpin structure despite multiple mutations
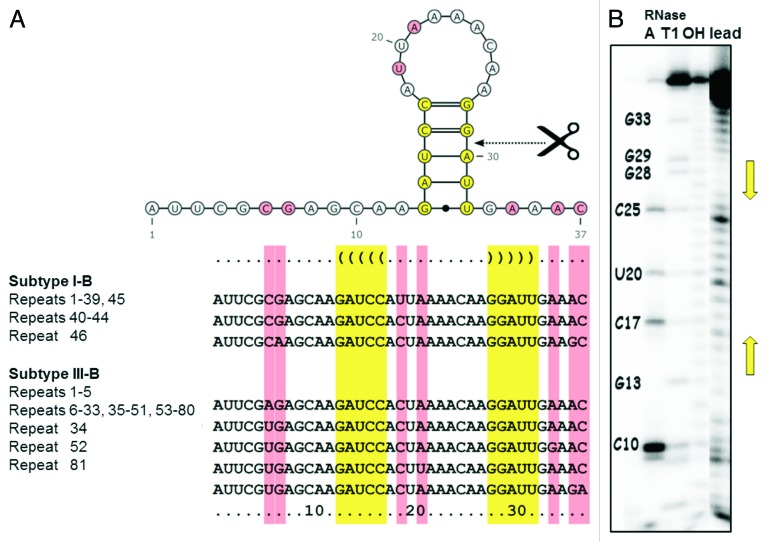
Comprehensive comparative sequence analysis demonstrated that both CRISPR loci contain multiple mutations in the repeat sequences. The minimum free energy structure of these variations, however, is the same hairpin containing five base pairs (Fig. 7A). The experimentally identified crRNA processing site that results in the 8 nt 5′ tag is just below the two C-G base pairs in the middle of the stem; no mutations occur in this stem. Further, the influence of surrounding spacer sequences on each repeat instance in the array was analyzed. A variation in the hairpin stability is detectable in the respective spacer context; however, the majority of repeat instance show a high probability of forming this structure (i.e., have a base pair accuracy above 0.4) as depicted in Figure S5. Mutations in the repeat seem not to effect the formation of the structure and in the processing of mature crRNAs, only a very slight effect might occur for spacers 1–3 at the 3′-end in subtype III-B (see RNaseq data, Fig. 6). In addition, we verified the formation of the hairpin structure by in vitro structure probing of the repeat (Fig. 7B)
Figure 7. (A) RNA secondary structure of the CRISPR repeat sequences in M. mazei. For both loci, the different variants of the repeats and their relative positions in the array are depicted. Yellow indicates the conserved base pairs of the hairpin structure and pink indicates positions where mutations have occurred. The sequence in the RNA structure is depicted according to the majority of repeats in subtype I-B. The scissors show the processing site in the generation of mature crRNAs that results in the 8 nt 5′ tag. (B) Structure probing of 5′-end-labeled CRISPR repeat RNA (subtype I-B). Cleavage pattern of RNase A and T1 treatment with identified G and C nucleotides, OH ladder and lead cleavage is pictured. Base pairing regions are indicated through yellow arrows.
Analyzing CRISPR loci from different M. mazei isolates indicates low CRISPR activity
Re-sequencing the I-B CRISPR locus demonstrated that no new spacer has been acquired in the M. mazei strain Gö1 during the past 10 y of culturing M. mazei in our laboratory. Thus, the CRISPR subtype I-B locus was PCR-cloned and sequenced from 3 different M. mazei isolates from three different continents, which are commercially available (DSMZ 4556, DSMZ6300, DSMZ2244). The analysis showed that all isolates share the identical organization of the respective cas-genes, and contain the same leader sequence. Analyzing each spacer sequence of the different isolates demonstrated that isolate DSMZ2244 contains exactly the same CRISPR array as M. mazei strain Gö1, including all 46 spacers. Furthermore, the first two spacers are identical in all isolates analyzed. In M. mazei DSMZ 4556 however, the four spacers following spacer 1 and 2 are identical to spacer 28–31 of strain Gö1 followed by spacer identical to spacer 14–31 of strain Gö1. Sequence analysis of the further downstream sequence was not successful. In M. mazei DSMZ6300, the spacer following spacer 1 and 2 are identical to spacer 7–46 of strain Gö1 with an additional inserted spacer 15, 16 and 22 (see Fig. S6). This pattern of spacers demonstrate that no new spacers have been acquired in the different environments, where the strains were originally isolated. The duplications of several spacers in DSM6300 and DSMZ4556 argue for rearrangements within the CRIPSR array by homologous recombination including deletions and duplications.
Discussion
In this first report on CRISPR-Cas systems in M. mazei, we discovered two different CRISPR loci of subtypes I-B and III-B. The latter one is also known for Cmr effector protein complexes, another class of multisubunit ribonucleoprotein complexes most common in archaea and well-studied in P. furiosus.14,30 Based on in vitro experiments, this type III-B CRISPR system is assumed to recognize and target foreign RNA in the interference phase.25,27,40 Only very recently direct in vivo evidence was obtained for RNA targeting and, thus, these subtype III-B CRISPR systems might be capable to also defend against viruses that have an RNA-based genome. However, it is not known so far which protein is responsible for RNA cleavage. It has been suggested to be Cmr2, but an impressive study by Zhang et al.35 showed that Cmr2 is unlikely to be the catalytic subunit of the Cmr complex for target RNA cleavage. The requirement to identify the nuclease responsible for target RNA cleavage is accelerating the importance to study type III systems in different species. In addition, the presence of an HD domain in Cas10 that is usually located in Cas3 proteins and posses nuclease activity suggests that type III-B could also be active against foreign DNA.49,50
Cas6b endonucleases are present in both subtypes of CRISPR systems
Evidence was obtained that in both CRISPR-Cas subtypes present in M. mazei, subtypes I-B and III-B containing considerably different cas genes, a Cas6b class endonuclease involved in crRNA processing is present. Since the different subtypes have almost identical repeats they can be recognized by Cas6-like proteins encoded by two different CRISPR subtypes, as shown in vitro. In most cases, the type III-B is often linked to a type I system and cannot be found independently since it is missing its own cas1 gene that is required for the adaptaion process.6
Both Cas6b endonucleases of M. mazei show significant homology to the Cas6b protein recently identified in M. maripaludis.38 Comparative analysis demonstrated the presence of further Cas6b homologs in the CRISPR-Cas locus in Methanosarcina acetivorans (MA1928) and Methanosarcina barkeri strain fusaro (Mbar A1352)51,52 (see Fig. 3). All of these Cas6b homologs show around 38% identity to the M. mazei homologs and the overall structure and the respective catalytic histidines most likely involved in the cleavage reaction are highly conserved. Besides the Cas6b homologs, the direct repeats of the two different CRISPR subtypes in M. mazei are highly conserved and share the same RNA hairpin structure. The Cas6-like protein cleavage site from the different CRISPR-subtypes was identical, resulting in the generation of a 8 nt 5′ tag of the mature crRNAs (Figs. 4 and 6). Unexpectedly, the identified cleavage site is located within the predicted hairpin structure of the repeat after the second base pair of the 5 bp stem (see Fig. 7). Thus, the hairpin structure, which has been verified in vitro but appears to be rather unstable upon the thermodynamically unstable base pairs (A/U and G/U), might be required for recognition and opened upon binding and processing by the Cas6b proteins. Alternatively, it is also possible that under in vivo conditions, the repeats in the context of the spacer in the crRNA precursor do not form a hairpin structure resulting in unstructured repeats. The repeat sequence in M. maripaludis shows some homology to the M. mazei repeat, however the structure proposed for the M. maripaludis repeat differs significantly, with a hairpin located nearly in the center of the repeat just upstream of the Cas6b unstructured cleavage site at the 3′ end.38 Processing however results in the exact same 8 nt-long crRNA 5′-tag. For CRISPR-specific endoribonuclease Cas6e (type I-E) and Cas6f (type I-F), it has been shown that the endoribonucleases remain tightly associated with the crRNAs after pre-crRNA cleavage and serve as nucleation point for assembling the remainder of the ribonucleoprotein complex.41,53 The extreme high substrate-specificity of those endoribonucleases for stem-loop structures of pre-crRNA repeat sequences ensures a correct processing of pre-crRNAs but limits their function as single-turnover enzymes to CRISPR-Cas activities. In contrast, Cas6 from type I-A CRISPR shows only a loose association with the archaeal CASCADE complexes.54 Similar observations were obtained for Cas6 of P. furiosus that is absent from Cmr effector protein complexes.30 It gives rise to the speculation that in those case, Cas6 activity can be exchanged between different CRISPR systems as well and might function in cleavage of non-cognate pre-crRNA RNAs.
Strikingly, the leader sequence upstream of the CRISPR array in both different M. mazei subtypes is nearly identical; moreover, the predicted respective structure of the leader is highly conserved in several Methanosarcinales (see Fig. 2), which might indicate that a potentially conserved leader-binding protein is binding to the leader. These findings, together with the fact that both Cas6b homologs could cleave either repeat in vitro (repeat I-B and repeat III-B) with the identical cleavage specificity, might argue for a complementary mode of action of both M. mazei Cas6b proteins during precursor processing and crRNA maturation of both subtypes (representing a genetic backup). A cross-cleavage activity of the Cas6b homologs of both different CRISPR subtypes might occur in vivo as well, although all other cas genes of the two CRISPR loci are significantly different and are classified to be members of two different subtypes. In order to address this query, a genetic approach generating both single Cas6b deletion mutants could be performed with a subsequent analysis of the remaining processing and maturation of crRNAs of both loci. Alternatively, it is attractive to speculate that both Cas6b proteins might form active heterodimers, which process the precursor crRNA of both loci. In order to experimentally verify this hypothesis, the formation and activity of heterodimers need to be studied. Moreover, it has to be elucidated whether the Cmr complex of type III-B system in M. mazei can use crRNAs from the type I-B system and vice versa. For P. furiosus, it has been reported that indeed, different crRNA can be bound by different effector complexes.49
Overall, it is tempting to study Cas proteins in molecular detail as e.g., Cas6 endonucleseases due to valuable characteristics provide new tools for application in biotechnology. Many of the Cas proteins have specific kinds of new enzymatic activities and are highly substrate-specific and overexpression of those proteins is not detrimental to the cell. Furthermore, sequence motives that serve as recognition motivs have been determined already. First examples of CRISPR-based RNA processing mediated by Cas6f endoribonuclease that enabeles predictable programming of gene expression has been recently shown.55
Evolving CRISPR systems in M. mazei?
The crRNA maturation of the subtypes I-B and III-B observed in M. mazei, both resulting in a distinct 8 nt long 5′ tag and less well-defined 3′ tag of the crRNA derived from the repeat is in accord with crRNA maturation reported for other subtypes and prokaryotes summarized in Westra et al.7 In contrast to most other (methano)archaeal CRISPR systems studied until today, e.g., in M. maripaludis38 or P. furiosus,23 however, both M. mazei loci appear not to be active under optimal growth conditions with methanol as sole energy and carbon source (see Fig. 6). In the presence of high salt concentrations, a significant induction of expression and maturation of the crRNAs was obtained for both loci, although still low in comparison to the single CRISPR system in M. maripaludis. Whereas the cas genes are all constitutively expressed as observed by the expression levels of cas genes in the cDNA sequencing data sets, the abundance of the leader transcript and matured crRNAs is significantly higher in the presence of high salt conditions (see Fig. S4, +PKN libraries). Thus, it is most likely that transcription of the crRNA precursor, including the leader, is induced in response to the NaCl stress. This might be due to the perception of potential disintegration of DNA upon NaCl stress, which to some extent might mimic phage infection. Analyzing both promoter regions upstream of the highly conserved leader allowed the identification of a BRE- and TATA box, binding sites for the general transcription factors TFB and TBP in archaea (see Fig. S4), however no clear putative regulator (repressor) binding site was identified. It is tempting to speculate that the DNA binding protein encoded upstream of the cas genes in subtype I-B (MM565) (see Fig. 1) functions as repressor for the promoter of the CRISPR arrays, until a yet-unknown signal is perceived potentially leading to reduced DNA-binding abilities of the repressor and, thus, dissociation from the promoter. A comparable regulatory mechanism has been shown for example for the CRISPR-Cas system present in E. coli K12, where the global transcriptional repressor H-NS is involved in repressing the CRISPR-Cas promoter, and is antagonized by the transcriptional activator LeuO.24,56 If the transcriptional regulator is indeed repressing the transcription of the both CRISPR arrays, this might be a feature to maintain the CRISPR system in the M. mazei genome and induce the system in case of exposure to foreign nucleic acids or generally under stress conditions.
The finding of an overall very low activity of the CRISPR system under optimal growth conditions might argue for low exposures of M. mazei to foreign nucleic acids. This is in accordance with the fact that M. mazei is not natural competent,57 and until today, no M. mazei specific phage has been identified. Further, no phage complementarity to M. mazei spacers has been identified by standard Blast searches of the common databases; and analyzing spacer contents and patterns in the subtype I-B array of four different isolates of M. mazei demonstrated that no new spacers have been aquired. Moreover, the spacer content showed a pattern of spacers in a mainly conserved order, however with several missing spacers and some spacer duplications (see Fig. S6). This pattern suggests that the CRISPR subtype I-B system in M. mazei is evolutionarily old and no new spacers have been acquired from different environments located at different continents (USA, USSR and Israel), from which the different strains were originally isolated (see Table 1). The duplications and deletions of several spacers, however, are most likely based on rearrangements within the CRISPR array by homologous recombination. Overall, these findings indicate that the two CRISPR arrays in M. mazei evolve very slowly and over a long time, which would be supported by the finding that under normal growth conditions the expression of the CRISPR array is repressed. Examples for long evolution times for CRISPR-loci have been recently reported and demonstrated for several strains of E. coli and Salmonella by Touchon and Rocha1 analyzing 51 complete genomes.
Table 1. Strains and plasmids.
| Strain or plasmid | Genotype or description |
Source or reference |
|---|---|---|
|
Strain |
|
|
|
Methanosarcina mazei strain Gö1 |
wild-type |
DSM Z No. 3647 |
|
Methanosarcina mazei |
Methanosarcina mazei (Barker 1936),1 isolated from cow dung, USSR |
DSMZ No. 2244 |
|
Methanosarcina mazei |
Methanosarcina mazei (Barker 1936),1 isolated from alkaline mud at an oil exploration drilling site; Israel, Hula swamp area |
DSMZ No. 4556 |
|
Methanosarcina mazei |
Methanosarcina mazei (Barker 1936),1 isolated from biomethanation granules, USA |
DSMZ No. 6300 |
|
E. coli Rosetta™ |
F-ompT hsdSB(rB- mB-) gal dcm(DE3) pRARE (CamR) |
Novagen, Cat. No.70954 |
|
E. coli DH5α |
general cloning strain |
2
|
|
Plasmid |
|
|
|
pRS714 |
MM560 in pET28a |
This study |
|
pRS833 |
MM3359 in pET28a |
This study |
|
pRS834 |
spacer 1–31 of CRISPR/Cas subtype I-B in DSM No. 4556 in TopoPCRII-vector |
This study |
|
pRS835 |
spacer of CRISPR/Cas subtype I-B DSMZ No. 6300 in TopoPCRII-vector |
This study |
| pRS836 | spacer of CRISPR/Cas subtype I-B of DSMZ No.2244 in TopoPCRII-vector | This study |
1 Mah RA, Kuhn DA. Transfer of the Type Species of the Genus Methanococcus to the Genus Methanosarcina, Naming it Methanosarcina mazei (Barker 1936) comb. nov. et emend. and Conservation of the Genus Methanococcus (Approved Lists 1980) with Methanococcus vannielii (Approved Lists 1980) as the Type Species. International Journal of Systematic Bacteriology 1984; 34:263-5.
2 Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol 1983; 166:557-80.
Materials and Methods
Strains and plasmids
Strains and plasmids used in this study are listed in Table 1. The DNA sequence from the cas6b genes (MM560 and MM3359) were PCR-amplified from chromosomal DNA from M. mazei Gö1. Both PCR products containing additional primer-generated NdeI and SacI restriction sites were cloned into the pET28a vector. pRS835 and pRS836 were constructed as follows: The CRISPR-Cas subtype I-B array (MM_0552 to MM_0557) was amplified using chromosomal M. mazei DNA (M. mazei DSMZ6300 for pRS835 and M. mazei DSMZ2244 for pRS836 construction) as a template and the following primers: C1for 5′-TGCGTAGATTGCTGTTACCGG-3′ and C1rev2 5′-TCCCCTGTTTTCCAGATACCG-3′. The obtained 4130 bp PCR product was cloned into TopoPCRII-vector (Invitrogen, Cat. No. 450641) as described by the manufactures instructions. For pRS834, the first 31 spacers of the CRISPR-Cas subtype I-B locus were amplified using chromosomal DNA from M. mazei 4556 and the primers C1for 5′-TGCGTAGATTGCTGTTACCGG-3′ and C1rev 5′-GCTCGCGAATCCCAATTTCTC-3′. The 2400 bp PCR-product was cloned into TopoPCRII-vector and the resulting plasmids were sequenced using the primers listed in Table S1. Plasmid DNA was in general transformed into E. coli according to the method of Inoue et al.58
Growth of M. mazei
M. mazei strain Gö 1 was in general grown under anaerobic conditions at 37°C with an atmosphere of 80% N2 plus 20% CO2 in 70-mL closed bottles in minimal medium that contained 150 mM methanol as sole energy and C-source according to Ehlers et al.,59 and was supplemented with 100 μg/mL ampicillin to prevent bacterial contamination. Growth under nitrogen starvation was performed in the absence of ammonium in the medium with molecular nitrogen of the gas phase as the sole nitrogen source as described by Ehlers et al.59 Oxygen stress was performed by sterile injection of 20 ml air into a 70 ml exponential growing culture (corresponding to 2.9% oxygen concentration in the total gas volume of cultivation flask) at turbidity at 600 nm (T600) of 0.5, followed by further incubation at 37°C for 30 min. For setting temperature stresses after inoculation, the cultures were incubated at 30°C or 40°C. For high salt stress, an additional 500 mM NaCl was added to the minimal medium right after inoculation. Cultures were grown until cells reached T600 of 0.3 (high salt) or 0.5 and were harvested by 4°C via centrifugation. Cultures were grown until cells reached T600 of 0.3 (high salt, nitrogen limitation) or 0.5 and were harvested by 4°C via centrifugation. UV-light exposure was performed as described before in Weidenbach et al.47
RNA extraction
Total RNA was isolated by phenol extraction as described before60 but using the Isol-RNA Lysis Reagent (5′PRIME GmbH, Cat. No. 2302700) and was followed by DNase I treatment.
Northern blot analyses
Northern blot analyses of total RNA with (γ-32P)-ATP end-labeled oligodeoxynucleotide 5′-CTTGTTTTAATGGATCTTGCTCGC-3′ directed against the repeat were performed as described before.61
Reverse transcriptase PCR
Reverse transcriptase PCR was performed using the One Step RT-PCR Kit (Qiagen, Cat. No. 210210) according to manufactures instructions. Two µg total RNA from M. mazei 3647 (wild-type) and the listed primers in Table S2 were used.
PCR for localization of the cas genes
For analyzing the genomic organization and localization of the cas genes in various M. mazei isolates. PCR amplification was performed using chromosomal DNA from M. mazei strain Gö1, DSMZ 3647 (wild-type); DSMZ 2244; DSMZ 4556 and DSMZ 6300 as well as primers listed in Table S2.
Isolation of chromosomal DNA
Chromosomal DNAs from the M. mazei isolates DSMZ 2244, DSMZ 4556, DSMZ 6300 and wild-type (DSMZ 3647) were isolated with Wizard® Genomic DNA Purification Kit (Promega, Cat. No. A1620).
Purification of heterologously expressed Cas6 proteins in E. coli
The Cas6b proteins were expressed, fused to an N-terminal His-tag from plasmid pRS714 and pRS833 in E. coli Rosetta™ (Novagen, Cat. No.70954) growing in LB-medium in the presence of 30 µg/ml Kanamycin. The protein expression was induced at a T600 of 0.6 with 30 µM isopropyl-β-D-thiogalactopyranoside for 16 h at 18°C. The cells were harvested at 4°C and resuspended in His-tag puffer (250 mM NaH2PO4, 1.5 M NaCl, pH 8). After DNaseI and RNaseA was added, cell disruption was performed using a French pressure cell at 800 psi. The His-tagged recombinant fusion proteins were purified from the respective cell-free cell extract by affinity chromatography using Nickel-NTA agarose (Qiagen, Cat. No. 30210). After wash steps, the Cas6b proteins were eluted in the presence of 100 mM, 250 mM and 500 mM imidazole; the wash and elution fractions were analyzed by sodium dodecyl sulfate-PAGE (SDS-PAGE) for quality control.
In vitro structure probing and endonuclease activity assay
Secondary structure probing and endonuclease activity assay was conducted on 5′-end-labeled CRISPR repeat RNA (type I-B repeat 5′-AUUCGCGAGCAAGAUCCAUUAAAACAAGGAUUGAAC-3′, type III-B repeat 5′-AUUCGUGAGCAAGAUCCACUAAAACAAGGAUUGAAC-3′ and the type I-B deoxy variant repeat 5′-AUUCGCGAGCAAGAUCCAUUAAAACAA(dG)GAUUGAAC-3′). An OH ladder and structure probing with RNase T1 was performed in a total volume of 10 µl as described before.62 Additionally, lead cleavage was conducted. In this case, RNA was denatured 1 min at 95°C and chilled on ice for 5 min and was incubated for 2, 3 and 5 min at 37°C with 2.5 mM lead (II) acetate. Endonucleases activity assays were performed in total volume of 20 µl. Initially, RNA was denatured 1 min at 95°C, chilled on ice for 5 min and incubated with different indicated concentrations of heterologously expressed and purified Cas6b-IB and Cas6b-IIIB protein in reaction puffer (20 mM Hepes pH8, 1 mM DTT, 250 mM KCl, 1.875 mM MgCl2,) for 5 min at 37°C. All reactions were stopped and precipitated with 70% ethanol. Samples were denatured for 5 min at 95°C prior to separation on 8% polyacrylamide/7 M urea sequencing gels in 1 × TBE buffer. Gels were analyzed using a PhosphorImager (FLA-3000 Series, Fuji) and AIDA software (Raytest).
Construction of cDNA libraries for dRNA-seq and Illumina sequencing
Libraries for Solexa sequencing (HiSeq) of cDNA were constructed by vertis Biotechnology AG, Germany (www.vertis-biotech.com/), as described previously for eukaryotic microRNA63 but omitting the RNA size-fractionation step prior to cDNA synthesis. In brief, after PNK treatment, equal amounts of RNA samples from treated and untreated RNA samples were poly(A)-tailed using poly(A) polymerase. Then, the 5′PPP structures were removed using tobacco acid pyrophosphatase (TAP). Afterwards, an RNA adaptor was ligated to the 5′-phosphate of the RNA. First-strand cDNA was synthesis by an oligo(dT)-adaptor primer and the M-MLV reverse transcriptase. In a PCR-based amplification step using a high-fidelity DNA polymerase, the cDNA concentration was increased to 20–30 ng/μl. A library-specific barcode for sequencing multiplexing was part of a 3′-sequencing adaptor. The resulting cDNA libraries were sequenced using a HiSeq 2000 machine (Illumina) in single read mode at the Max Planck Genome Centre Cologne.
Repeat structure prediction analysis
Single repeat sequences for each locus were folded with RNAfold from the Vienna Package 1.8.4.64 (options-noLP-d2-p). Every repeat variation folded into the same minimum free energy structure as presented in Figure 6. No stable sub-optimal structures were determined. The influence of surrounding spacer sequences on each repeat position in the CRISPR array was determined as follows: The entire repeat-spacer array was folded using a local folding algorithm called RNAplfold,65 also from the same Vienna Package (options-noLP-W 150-L 100). The stability of the hairpin structure from Figure 6 (at each repeat instance in the array) was measured using the base pair accuracy (i.e., mean base pair probability). The base pair accuracy for measuring the stability of local structures and the parameters for RNAplfold were taken from previous work.66
Leader conservation
The conserved 108 nt region of the leaders of both CRISPR loci was identified by a local pairwise smith waterman sequence alignment. Further leader sequences similar to the ones found in M. mazei were identified in a Blast search against all available nucleotide sequences. The sequence and structure alignment and consensus structure was generated using the LocARNA web server67 with default parameters.
Protein structure prediction
The protein structures from both Cas6b proteins were determined using the phyre2 server68 with the Pyrococcus horikoshii Cas6 (PDB ID 3QJJ) as the top template for structure prediction. Cas6 sequences were aligned using ClustalW269 and homologous structureswere aligned with DaiLite.70Table 2.
Table 2. Primer pairs used for cas gene localization, RT-PCR and sequencing.
| Primer designations | 5`→3` |
|---|---|
|
csh1-csh2 for |
GGTGACTTGGAAGTTCAGCC |
|
csh1-csh2 rev |
GTGAACTGTCGAACGATAGCG |
|
csh2-cas5h for |
TCTTCAAGCTCATCAAGTTTGTCAG |
|
csh2-cas5h rev |
GTGACGGCACAAACATATACGC |
|
cas5h-cas3 for |
TTCTGGAAGCCACCAGTAAGC |
|
cas5h-cas3 rev |
GTTCATAGTAGGAGAGACTTCG |
|
cas3-cas6 for |
CAGTAGTGCCAAAGAAAGTAGATTC |
|
cas3-cas6 rev |
CGCCTTCTATAAGCCATAGCTTA |
|
cas6-cas1 for |
CCGCATCTCATCGAGTTTTCC |
|
cas6-cas1 rev |
GGTTAAGGTTACATTAGTCCCTCT |
|
cas1-cas2 for |
ACCTGTTTAAAAGACTGTTCATCCA |
|
cas1-cas2 rev |
CAAGGACGGGGACATCGAATA |
|
cas2-cas4 for |
TTCAAGAGGAGCTGCTGTTCC |
|
cas2-cas4 rev |
ACATAGTTAGGTGAGAAGTGTAAGT |
|
cas4-crRNA-Spacer1 for |
CCGAAAAGCGGTGTTAAGTCAG |
|
cas4-crRNA-Spacer1 rev |
GACCTTCAGAACCCTATATTCGG |
| Gö1–42for |
AAAGCCAGTCGGACCCAC |
| Gö1–21for |
TTCCTCCGCGTTCTTTCGTTG |
| Gö1–9for |
AGGATCAGGACGACGGC |
| Spacer8_4556f |
TCATCTTATCGCCATTCGCG |
| Spacer 22 wt r |
GTATGTGTACACATGGTG |
| Spacer 30 wt f |
CCACCTCTTCCTGATTAT |
| Spacer 30 r | CTGATAATCAGGAAGAGGTGG |
Supplementary Material
Acknowledgments
We thank the members the Schmitz laboratory for useful discussions, as well as Claudia Kießling and Jutta Kock for technical assistance. This work was supported by the German Research Council (DFG) Forschergruppe FOR1680 “Unravelling the prokaryotic immune system” (Schm1052/12-1 to R.S. and BA 2168/5-1 to R.B.).
Glossary
Abbreviations:
- sRNA
small RNA
- HTS
high throughput sequencing
- crRNA
CRISPR RNA
- pre-crRNAs
precursor CRISPR RNA
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cas
CRISPR associated
- nt
nucleotide
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/23928
References
- 1.Touchon M, Rocha EP. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One. 2010;5:e11126. doi: 10.1371/journal.pone.0011126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 3.Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem. 2011;392:277–89. doi: 10.1515/bc.2011.042. [DOI] [PubMed] [Google Scholar]
- 6.Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–39. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 8.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 9.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–63. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 10.Lillestøl RK, Redder P, Garrett RA, Brügger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–6. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 12.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 14.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin EV, Makarova KS. CRISPR-Cas: an adaptive immunity system in prokaryotes. F1000 Biol Rep. 2009;1:95. doi: 10.3410/B1-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrangou R, Horvath P. CRISPR: new horizons in phage resistance and strain identification. Annu Rev Food Sci Technol. 2012;3:143–62. doi: 10.1146/annurev-food-022811-101134. [DOI] [PubMed] [Google Scholar]
- 17.Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–40. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 20.Fineran PC, Charpentier E. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology. 2012;434:202–9. doi: 10.1016/j.virol.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 21.Lillestøl RK, Shah SA, Brügger K, Redder P, Phan H, Christiansen J, et al. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009;72:259–72. doi: 10.1111/j.1365-2958.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- 22.Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, et al. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–81. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 23.Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–9. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol. 2010;75:1495–512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 25.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–96. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–8. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–8. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–64. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–7. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 30.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard JA, Delmas S, Ivančić-Baće I, Bolt EL. Helicase dissociation and annealing of RNA-DNA hybrids by Escherichia coli Cas3 protein. Biochem J. 2011;439:85–95. doi: 10.1042/BJ20110901. [DOI] [PubMed] [Google Scholar]
- 33.Plagens A, Tjaden B, Hagemann A, Randau L, Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J Bacteriol. 2012;194:2491–500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–82. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell. 2012;45:303–13. doi: 10.1016/j.molcel.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–29. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- 37.Fischer S, Maier LK, Stoll B, Brendel J, Fischer E, Pfeiffer F, et al. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem. 2012;287:33351–63. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richter H, Zoephel J, Schermuly J, Maticzka D, Backofen R, Randau L. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012;40:9887–96. doi: 10.1093/nar/gks737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang R, Zheng H, Preamplume G, Shao Y, Li H. The impact of CRISPR repeat sequence on structures of a Cas6 protein-RNA complex. Protein Sci. 2012;21:405–17. doi: 10.1002/pro.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–92. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 41.Haurwitz RE, Sternberg SH, Doudna JA. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J. 2012;31:2824–32. doi: 10.1038/emboj.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beloglazova N, Petit P, Flick R, Brown G, Savchenko A, Yakunin AF. Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference. EMBO J. 2011;30:4616–27. doi: 10.1038/emboj.2011.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogers JE, Whitman WB, eds. Microbial production and consumption of greenhouse gases: methane, nitrogen oxides and halomethanes. ASM Press, Washington DC 1991. [Google Scholar]
- 44.Deppenmeier U, Johann A, Hartsch T, Merkl R, Schmitz RA, Martinez-Arias R, et al. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J Mol Microbiol Biotechnol. 2002;4:453–61. [PubMed] [Google Scholar]
- 45.Veit K, Ehlers C, Ehrenreich A, Salmon K, Hovey R, Gunsalus RP, et al. Global transcriptional analysis of Methanosarcina mazei strain Gö1 under different nitrogen availabilities. Mol Genet Genomics. 2006;276:41–55. doi: 10.1007/s00438-006-0117-9. [DOI] [PubMed] [Google Scholar]
- 46.Weidenbach K, Ehlers C, Kock J, Ehrenreich A, Schmitz RA. Insights into the NrpR regulon in Methanosarcina mazei Gö1. Arch Microbiol. 2008;190:319–32. doi: 10.1007/s00203-008-0369-3. [DOI] [PubMed] [Google Scholar]
- 47.Weidenbach K, Glöer J, Ehlers C, Sandman K, Reeve JN, Schmitz RA. Deletion of the archaeal histone in Methanosarcina mazei Gö1 results in reduced growth and genomic transcription. Mol Microbiol. 2008;67:662–71. doi: 10.1111/j.1365-2958.2007.06076.x. [DOI] [PubMed] [Google Scholar]
- 48.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–5. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 49.Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–42. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, et al. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 2002;12:532–42. doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS, et al. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J Bacteriol. 2006;188:7922–31. doi: 10.1128/JB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–72. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–56. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi L, Haurwitz RE, Shao W, Doudna JA, Arkin AP. RNA processing enables predictable programming of gene expression. Nat Biotechnol. 2012;30:1002–6. doi: 10.1038/nbt.2355. [DOI] [PubMed] [Google Scholar]
- 56.Westra ER, Pul U, Heidrich N, Jore MM, Lundgren M, Stratmann T, et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol. 2010;77:1380–93. doi: 10.1111/j.1365-2958.2010.07315.x. [DOI] [PubMed] [Google Scholar]
- 57.Metcalf WW, Zhang JK, Apolinario E, Sowers KR, Wolfe RS. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc Natl Acad Sci USA. 1997;94:2626–31. doi: 10.1073/pnas.94.6.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–8. doi: 10.1016/0378-1119(90)90336-P. [DOI] [PubMed] [Google Scholar]
- 59.Ehlers C, Weidenbach K, Veit K, Deppenmeier U, Metcalf WW, Schmitz RA. Development of genetic methods and construction of a chromosomal glnK1 mutant in Methanosarcina mazei strain Gö1. Mol Genet Genomics. 2005;273:290–8. doi: 10.1007/s00438-005-1128-7. [DOI] [PubMed] [Google Scholar]
- 60.Veit K, Ehlers C, Schmitz RA. Effects of nitrogen and carbon sources on transcription of soluble methyltransferases in Methanosarcina mazei strain Go1. J Bacteriol. 2005;187:6147–54. doi: 10.1128/JB.187.17.6147-6154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jäger D, Sharma CM, Thomsen J, Ehlers C, Vogel J, Schmitz RA. Deep sequencing analysis of the Methanosarcina mazei Gö1 transcriptome in response to nitrogen availability. Proc Natl Acad Sci USA. 2009;106:21878–82. doi: 10.1073/pnas.0909051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jäger D, Pernitzsch SR, Richter AS, Backofen R, Sharma CM, Schmitz RA. An archaeal sRNA targeting cis- and trans-encoded mRNAs via two distinct domains. Nucleic Acids Res. 2012;40:10964–79. doi: 10.1093/nar/gks847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berezikov E, Thuemmler F, van Laake LW, Kondova I, Bontrop R, Cuppen E, et al. Diversity of microRNAs in human and chimpanzee brain. Nat Genet. 2006;38:1375–7. doi: 10.1038/ng1914. [DOI] [PubMed] [Google Scholar]
- 64.Hofacker I, Fontana W, Stadler P, Bonhoeffer L, Tacker M, et al. Fast folding and comparison of RNA secondary structures. Monatshefte für Chemie - Chemical Monthly. 1994;125:167–88. doi: 10.1007/BF00818163. [DOI] [Google Scholar]
- 65.Bernhart SH, Hofacker IL, Stadler PF. Local RNA base pairing probabilities in large sequences. Bioinformatics. 2006;22:614–5. doi: 10.1093/bioinformatics/btk014. [DOI] [PubMed] [Google Scholar]
- 66.Lange SJ, Maticzka D, Möhl M, Gagnon JN, Brown CM, Backofen R. Global or local? Predicting secondary structure and accessibility in mRNAs. Nucleic Acids Res. 2012;40:5215–26. doi: 10.1093/nar/gks181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38(Web Server issue):W373-7. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 69.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 70.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–7. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 71.Bernhart SH, Hofacker IL, Will S, Gruber AR, Stadler PF. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.