Abstract
An RNA-based screen was performed to reveal a possible evolutionary scenario for the CRISPR-Cas systems in two cyanobacterial model strains. Following the analysis of a draft genome sequence of Synechocystis sp PCC6714, three different CRISPR-Cas systems were characterized that have different degrees of relatedness to another three CRISPR-Cas systems in Synechocystis sp PCC6803. A subtype III-B system was identified that is extremely conserved between both strains. Strong signals in northern hybridizations and the presence of different spacers (but identical repeats) indicated this system to be active, despite the absence of a known endonuclease candidate gene involved in the maturation of its crRNAs in the two strains. The other two systems were found to differ significantly from each other, with different sets of repeat-spacer arrays and different Cas genes. In view of the otherwise very close relatedness of the two analyzed strains, this is suggestive of an unknown mechanism involved in the replacement of CRISPR-Cas cassettes as a whole. Further RNA analyses revealed the accumulation of crRNAs to be impacted by environmental conditions critical for photoautotropic growth. All six systems are associated with a gene for a possible transcriptional repressor. Indeed, we identified one of these genes, sll7009, as encoding a negative regulator specific for the CRISPR1 subtype I-D system in Synechocystis sp PCC6803.
Keywords: comparative genomics, CRISPR, cyanobacteria, defense mechanisms, crRNA maturation, transcriptional regulator
Introduction
The CRISPR-Cas (clustered regularly interspaced short palindromic repeats, CRISPR associated proteins) system provides adaptive immunity in archaea and bacteria.1-8 In addition, alternative functions in DNA repair were demonstrated of at least some components of the CRISPR-Cas system.9
Ribonucleoprotein complexes consisting of Cas proteins and properly processed short CRISPR RNAs, the crRNAs, target foreign DNA or RNA molecules for cleavage and degradation.1,4,10-13 CRISPR-Cas systems are extremely diverse across different organisms and can be exchanged via horizontal gene transfer,14 contributing to the observed high diversity and variability among the CRISPR-Cas loci from different organisms.
Previously, 45 families of Cas proteins were identified,15 of which subsets make up the diverse modules of Cas proteins. These modules function independently and highly specific with their respective crRNAs. Characterized examples include the Cmr [Cas module RAMP (repeat-associated mysterious proteins)] and the CASCADE (CRISPR-associated complex for antiviral defense) complexes of Pyrococcus furiosus and E. coli, respectively.16,17 By comparing phylogenies of common cas genes, repeat sequences and the architecture of CRISPR-cas loci, CRISPR-Cas systems were further categorized into three major types of CRISPR-Cas systems, I, II and III. These can be further divided into at least 10 subtypes and some chimeric variants.13,18
A hitherto only marginally studied group are the CRISPR-Cas systems of oxygenic phototrophic cyanobacteria.19,20 In the model cyanobacterium Synechocystis sp PCC6803 (from here: Synechocystis 6803), three fundamentally different CRISPR-Cas systems were found, called CRISPR1, CRISPR2 and CRISPR3, which are encoded together on pSYSA, a single large plasmid.21 From these, CRISPR1 was assigned to the subtype I-D,13,18 whereas CRISPR2 and CRISPR3 are type III systems. All three systems were found to be associated after maturation with abundant transcripts.21 In addition to the CRISPR systems, the plasmid pSYSA encodes at least six different type II toxin-antitoxin systems.22 These were shown to be simultaneously active and are likely involved in post-segregational killing, suggesting that the maintenance of pSYSA and of the three CRISPR systems located on it has been highly selected for.22
A hallmark of CRISPR-Cas systems is the involvement of short crRNAs that guide associated ribonucleoprotein complexes in the destruction of invading DNA or RNA. The precise maturation of these crRNAs is therefore functionally important for the function of these systems. Endoribonucleases from various type I and type III CRISPR-Cas systems have been characterized as key players in crRNA maturation. Based on genetic, transcriptomic and bioinformatic approaches we previously identified in Synechocystis 6803 distinct processing pathways for each of the three CRISPR-Cas systems. It is well known that the CRISPR maturation endoribonucleases cleave repeats at distinct sequence and structure motifs leaving a 8 nt 5′ repeat handle (5′ tag): Cas6,23-25 Cse3,16,26,27 recently renamed to Cas6e,13 and Csy4,28,29 recently renamed to Cas6f.13 After cleavage, the crRNAs are trimmed further by a poorly characterized ruler mechanism to fixed lengths.30 In Synechocystis 6803, the maturation of crRNAs was found to include at least one maturation step dependent on the endonucleases Cas6-1 (CRISPR1) or Cas6-2a (CRISPR2). However, the maturation of CRISPR3 crRNAs was found to be Cas6-independent,21 which is unusual for a type III system. Instead, a Cmr2 protein (CRISPR module RAMP protein no. 2) was identified as involved in the maturation, regulation of expression, Cmr complex formation or stabilization of transcripts from the CRISPR3 spacer-repeat locus, as in its absence no CRISPR3-derived crRNAs were detected.21 Cmr2 proteins contain a GGDEF domain, an RNA recognition motif (RRM)-fold31,32 and have some similarity to the Palm/Cyclase polymerase domain.18,33 In Pyrococcus furiosus Cmr2 is a component of the CRISPR-Cas effector complex that destroys complementary RNAs, was suggested as the subunit responsible for target RNA cleavage,17,34 which, however, was not supported by structural analysis.35 In view of these findings, a role of Cmr2 in the maturation of Synechocystis 6803 CRISPR3 crRNAs appeared surprising and furthermore, it appeared unclear if CRISPR3 is an active system at all. However, a resequencing analysis of the used substrain, Synechocystis 6803 “PCC-M,” revealed two deletions in the repeat-spacer arrays of CRISPR1 and CRISPR2, whereas CRISPR3 appeared unchanged.36
To reveal a possible evolutionary scenario for these cyanobacterial CRISPR-Cas systems, here we performed an RNA-based screen of four other Synechocystis strains available in the Pasteur Culture Collection. We identified one closely related strain, Synechocystis sp PCC6714 (from here: Synechocystis 6714), with signals in northern hybridizations clearly indicating the presence of a CRISPR3 system similar to the one in Synechocystis 6803. Both strains were isolated by R. Kunisawa from freshwater in California USA, in 1967 (Synechocystis 6714) and 1968 (Synechocystis 6803), i.e., from very similar environments, suggesting a possible close relationship. Following draft genome analysis, we identified three distinct CRISPR-Cas systems also in Synechocystis 6714, one of which is indeed very close to the CRISPR3 of Synechocystis 6803. However, the other two are very distinct despite the generally very close relatedness of the two analyzed strains. A striking similarity among the six systems is the presence of genes encoding possible transcriptional regulators. We tested their functionality by genetic knockout experiments and identified one as a repressor of the subtype I-D CRISPR1 locus in Synechocystis 6803.
Results
Identification of a strain carrying a CRISPR-Cas system closely related to CRISPR3 of Synechocystis 6803
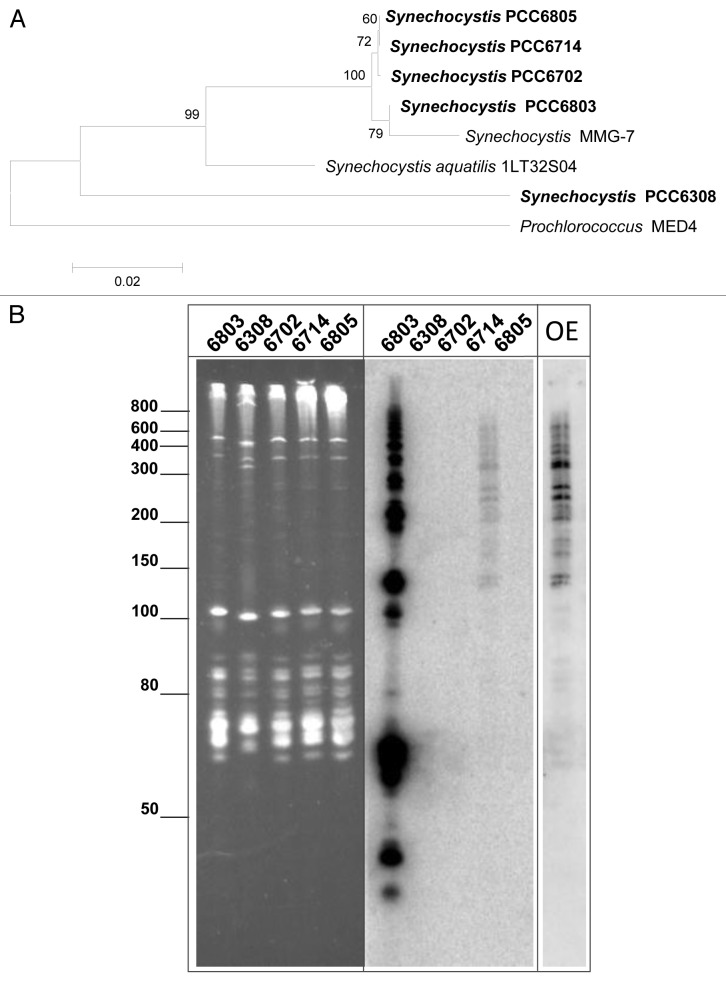
In our previous work, we demonstrated the very high transcript abundance for CRISPR3-derived precursor transcripts, maturation intermediates and crRNAs in Synechocystis 6803 despite the lack of a known endonuclease gene involved in these steps.21 Therefore, here we employed an RNA-based screen for possible homologs of Synechocystis 6803 CRISPR3 in four further Synechocystis strains. These strains were chosen based on their published 16S rRNA sequences (Fig. 1A) as very closely related (< 1% sequence divergence, strains PCC6714, PCC6702 and PCC6805), or only moderately related (PCC6308, 89% 16S rRNA sequence identity with Synechocystis 6803), and were obtained as axenic cultures from the Pasteur Culture Collection. Following cultivation under standard growth conditions, total RNA was extracted, electrophoretically separated and hybridized to a transcript probe encompassing the four 5′ terminal repeat-spacer units of the repeat-spacer array of Synechocystis 6803 CRISPR3 (Fig. 1B). As expected, a very strong signal was observed for the total RNA from Synechocystis 6803, but there was a second lane with signal, corresponding to the total RNA from Synechocystis 6714 (Fig. 1B). Overexposure of the latter revealed a signal pattern of comparable complexity for both strains. Upon successful PCR amplification and sequence analysis of a 1,231 nt fragment, the presence of a Synechocystis 6803 CRISPR3-homologous repeat-spacer array in Synechocystis 6714 was confirmed (Fig. 2). However, whereas the direct repeat sequences were identical, the spacer sequences were not, consistent with the lowered hybridization signal intensity in the lower molecular weight range observed for Synechocystis 6714 RNA compared with Synechocystis 6803 RNA (Fig. 1B), as with decreasing length these signals are increasingly spacer-related. The CRISPR repeat-spacer array shown in Figure 2 was named CRISPR3*, because of its similarity to CRISPR3 in Synechocystis 6803. The two homologous arrays CRISPR3/3* are preceded by a highly conserved leader region as can be concluded from the 95% sequence identity (78/82 nt) of the sequence between the 3′ end of the PCR primer used for amplification and the first transcribed nucleotide (transcription start site, TSS; Fig. 2).
Figure 1. Identification of a locus homologous to the Synechocystis 6803 CRISPR3 array in closely related cyanobacteria. (A) Phylogenetic relatedness among analyzed (in boldface letters) Synechocystis strains, based on 16S rRNA sequences. The sequence of the unicellular cyanobacterium Prochlorococcus sp MED4 served as outgroup. The numbers at nodes refer to bootstrap support values (10,000 repetitions). The phylogenetic tree was generated using the minimum evolution method within MEGA5.53 The optimal tree with the sum of branch length = 0.2726 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree and are given in the number of base substitutions per site. There were a total of 1441 positions in the final data set. The GenBank accession (GI) numbers are 14625357, 226844852, 298103759, 16215697, 16215698 and 16215699 for Synechocystis strain PCC6308, SMMG-7, aquatilis 1LT32S04, PCC6702, PCC6714 and PCC6805. For Synechocystis 6803 and Prochlorococcus MED4, data were obtained from the whole genome sequences (CP003265 and BX548174). (B) Northern analysis of the selected strains. Left side: Denaturing gel electrophoresis of total RNA; Right side: hybridization with a single-stranded RNA probe for the 5′-terminal section of the Synechocystis 6803 CRISPR3 array (exposure time 1 h). The lane with Synechocystis 6714 total RNA was overexposed for 6 d (very right, lane “OE”). The positions of selected size marker bands are indicated.
Figure 2. Comparison of the PCR-amplified CRISPR3* repeat-spacer sequence from Synechocystis 6714 (6714_C3*) compared with CRISPR3 from Synechocystis 6803 (6803_C3). A sequence alignment of the complete PCR-amplified and resequenced CRISPR3* and of the first 14 direct repeat, spacer units of CRISPR3 is shown. The highly conserved leader region in the 5` region of both CRISPR arrays (position 1–107) is marked by a dotted line. The oligonucleotide primers used for amplification are indicated by long horizontal arrows in black. The bent arrows indicate mapped TSS and the spacers are labeled by roman numerals.
Genome analysis identifies three CRISPR-Cas loci in Synechocystis 6714
To obtain a more comprehensive view on the CRISPR-Cas systems in Synechocystis 6714, we determined its total genome sequence based on a whole genome shotgun sequencing approach, yielding five scaffolds. The sequence information has been deposited at DDBJ/EMBL/GenBank and can be accessed under the accession AMZV01000000.
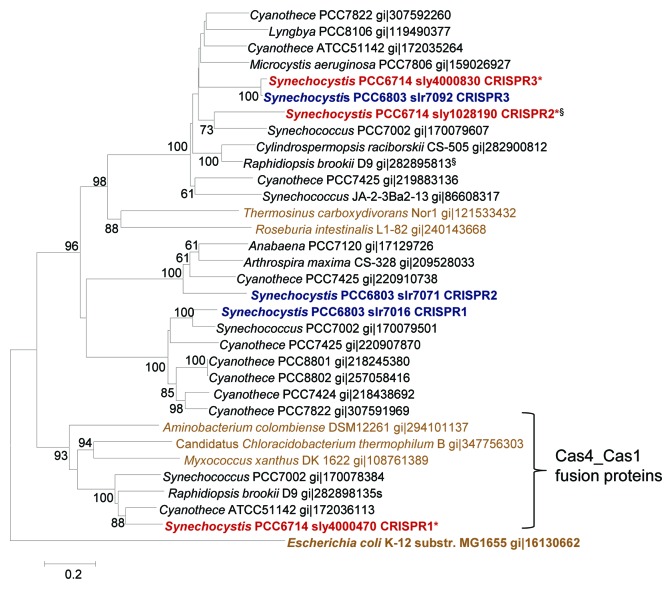
The 16S rRNA sequence identities between Synechocystis 6714 and Synechocystis 6803 are 1429/1437 residues, corresponding to 99.4%. Accordingly, the gene content and predicted proteome between both strains is very highly conserved. However, with regard to the CRISPR-Cas systems, a different picture was obtained. Three cas1 genes were identified, located on two different scaffolds and indicating the presence of three distinct CRISPR-Cas systems also in Synechocystis 6714. From these cas1 genes, one (sly4000830) encodes a protein that is very closely related to Slr7092 in the CRISPR3 array of Synechocystis 6803 (97% identical residues). However, phylogenetic analysis revealed the other two Cas1 proteins to be only remotely related to the other two Cas1 proteins of Synechocystis 6803, Slr7016 and Slr7071 (Fig. 3). This is further substantiated by the fact that the other two proteins possess a very distinct domain architecture, one (Sly4000470) consisting of a Cas4-Cas1 protein fusion of 564 amino acids, whereas the other (Sly1028190) is a truncated form of only 129 residues (compared with ~300 amino acids for most Cas1 proteins). Accordingly, both are found in very distinct phylogenetic clades. Cas4-Cas1 fusion proteins exist also in the CRISPR-Cas systems of many other cyanobacteria and non-cyanobacteria, including Myxococcus xanthus DK 1622 and gram-positive bacteria (Fig. 3), whereas the truncated form might have been affected by a recombination with the CRISPR3 system and might be non-functional.
Figure 3. Phylogenetic relationship among Cas1 proteins of Synechocystis and of selected other bacteria. Gene identification numbers (gi) are given for all compared sequences expect for the Cas1 proteins from Synechocystis 6803 and Synechocystis 6714. For Synechocystis sequences the gene names are indicated together with the designation of the respective CRISPR-Cas system they are associated with (Fig. 5). §Truncated sequences (< 200 amino acids). Non-cyanobacterial sequences are in lighter color and the sequence of the E. coli K12 Cas1 protein was used as outgroup. The evolutionary history was inferred using the minimum evolution method as implemented in MEGA5.53 The optimal tree with the sum of branch length = 9.7587 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) are shown next to the branches if > 60. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. All ambiguous positions were removed for each sequence pair. There were a total of 607 positions in the final data set.
Detailed comparison of CRISPR repeat-spacer arrays
All three cas1 genes in Synechocystis 6714 are associated with an extended set of Cas genes and a transcribed repeat-spacer array, here called CRISPR1*, CRISPR2* and CRISPR3* in analogy to CRISPR1-3 in Synechocystis 6803.
General features of the CRISPR arrays from both Synechocystis strains are summarized in Table 1. The direct repeat sequences are identical within the CRISPR1* or CRISPR2* repeat-spacer arrays and differ substantially from the CRISPR1 and CRISPR2 repeats. In contrast, the CRISPR3* repeats are identical with those of Synechocystis 6803 CRISPR3, except a degeneration of 4 nt, which can be detected within the final repeat of CRISPR3*.
Table 1. Details of the three repeat-spacer arrays CRISPR1*-CRISPR3* present in Synechocystis 6714 compared with the three loci CRISPR1-CRISPR3 in Synechocystis 6803 substrain ‘PCC-M’.36.
| CRISPR | Subtype13,18 | Repeat sequence | length | No. of spacers | Spacer length (nt) | Identical spacer-repeat units |
|---|---|---|---|---|---|---|
| CRISPR1* |
I-A or I-B |
GTGTCCAAACCATTGATGCCGTAAGGCGTTGAGCAC GTGTCCAAACCATTGATGCCATAAGGCGTTGAGCAC |
36 |
11 |
35–37 |
none# |
| CRISPR1 |
I-D |
CTTTCCTTCTACTAATCCCGGCGATCGGGACTGAAAC |
37 |
49 |
31–41 |
10–11 |
| CRISPR2* |
III-B |
CCCTACCGATTGGATTAAATCGGATTAGTTGGAAAC CCCTACCGATTGGATTAAATCGGATTAGTCGGAAAC ACCTATCAATGGAATTAAATCAGATTAGTTAGAAAC |
36 |
8 |
32–44 |
none |
| CRISPR2 |
III |
GTTCAACACCCTCTTTTCCCCGTCAGGGGACTGAAAC GTTCAACACCCTCTTTTCCCCGTTAGGGGACTGAAAC |
37 |
56 |
34–46 |
6/7–8/9, 37–39 |
| CRISPR3* |
III-B |
GTCTCCACTCGTAGGAGAAATTAATTGATTGGAAAC GTCTCGACTCGTAGGAGAAATTAACTGATTTAAAAC |
36 |
13 |
35–47 |
6–7 |
| CRISPR3 | III-B | GTCTCCACTCGTAGGAGAAATTAATTGATTGGAAAC GTCTCCACTCGCAGGAGAAATTAATTGATTGGAAAC |
36 | 38 | 35–47 | none |
For each CRISPR, the repeat sequence and single occurring mutant variants in the terminal repeats (array 3′ end) are given underneath (the mutated nucleotide is in boldface). Nucleotide positions identical to the widely conserved 3′ repeat motif are underlined. The numbers and lengths of spacers are given, together with information on spacer-repeat unit duplications (consecutive identical spacer-repeat units are separated by a dash, duplicated unit pairs are united by a slash). #A very similar sequence (27/36 residues) is shared between spacer 10 of CRISPR1* and CRISPR1.
CRISPR direct repeats frequently contain imperfect palindromes. Thus, after transcription, these elements are subject to the formation of secondary structures and these structures are important determinants of RNA maturation by RNA endonucleases.27,28,37,38 The general practice in the search for the functional CRISPR repeat structure is to compute the minimum free energy (MFE) structure of a single repeat sequence. Such MFE predictions are frequently correct due to highly stable stem-loop structures with many GC base-pairs, for example in E. coli.16 In Figure 4, the predicted secondary structures for CRISPR1*, CRISPR2* and CRISPR3* are shown. These structures are similar for CRISPR2* and CRISPR3*, suggesting some similarity between these two systems. Moreover, taking the entire array sequence into account, we discussed previously a likely improved similarity to the native structures for the Synechocystis 6803 CRISPR3 repeats and identified by transcriptome analysis the endonuclease cleavage site within this context, leaving the unusual 13 nt repeat handle 5′-AUUGAUUGGAAAC remaining at the processed spacer 5′ ends,21 as indicated in Figure 4. As the CRISPR3 and CRISPR3* repeats are identical, this should apply to CRISPR3* as well. Interestingly, also for CRISPR2* a long unpaired section in the repeat’s 3′ half is predicted, which is except one substitution (5′-AUUAGUUGGAAAC) identical to the experimentally identified 13 nt repeat handle of CRISPR3/3*, reinforcing the possible relationship between CRISPR2* and CRISPR3*.
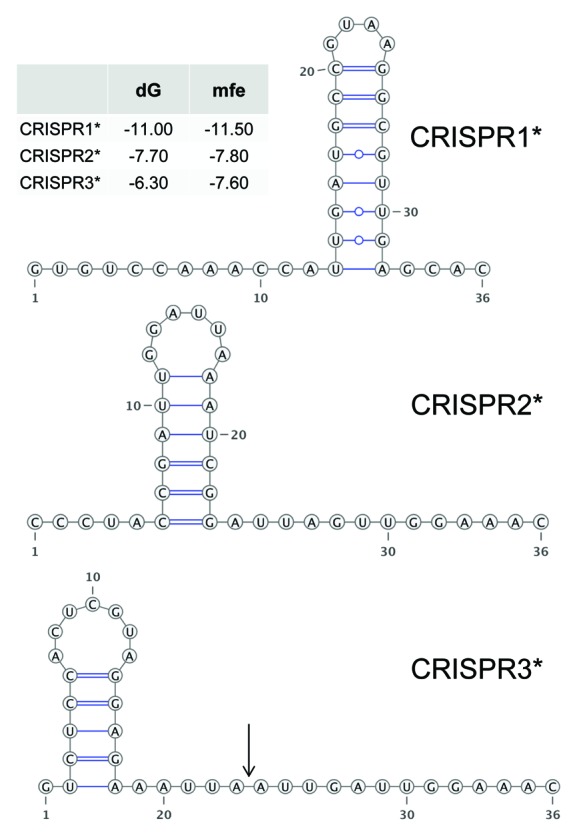
Figure 4. Predicted secondary structures of CRISPR repeats in Synechocystis 6714. The energy values (mfe, minimal folding energy and dG, energy of the selected structure) are given in the inset. The CRISPR3* structure is identical to the one published for CRISPR3 of Synechocystis 680321 and is characterized by a 13 nt tag remaining at the 5′ end of the mature spacers after maturation (arrow).
The sequence comparison of different repeats revealed a widespread conserved 3′ end, consisting of 5 nt (GAAA-C/G).14 This is also observed for CRISPR2* and CRISPR3* repeats in Synechocystis 6714 (and all Synechocystis 6803 CRISPRs), which end in GAAAC. In contrast, CRISPR1* repeats finish with the unusual GAGCAC (Table 1). A CRISPR array with identical repeat sequences exists in the marine strain Synechococcus PCC7002, which is also associated with closely related Cas proteins, such as the fused Cas4-Cas1 protein (Fig. 3). These facts suggest the presence of a CRISPR1*-like CRISPR-Cas system in Synechococcus PCC7002.
CRISPR1*-CRISPR3* consist of 11, 8 and 13 repeat-spacer units per locus (each with an additional final repeat). The spacer sequences in Synechocystis 6714 differ in lengths from 32–47 nt and are all unique, except spacer 6 and 7 of C3*, which is different from the situation in Synechocystis 6803, where duplicate spacers exist only in CRISPR1 and 2 (Table 1). Despite the very substantial differences in cas-gene content and CRISPR subtype, a very similar sequence (27/36 residues) is shared between spacer 10 of Synechocystis 6714 CRISPR1* and spacer 10 of Synechocystis 6803 CRISPR1, indicating a past challenge by a closely related invader DNA in the progenitor of both strains at about the same time, as the order of spacers can be considered to be chronologically. All other Synechocystis 6714 spacer sequences differ from those in Synechocystis 6803.
Analysis of Cas genes indicates swap of two CRISPR-Cas cassettes but conservation of the third
The arrangement and types of CRISPR1*-associated Cas genes are only remotely related to those accompanying CRISPR1 of Synechocystis 6803, and differ even more from CRISPR2 and CRISPR3. CRISPR1* possesses in addition to the fused cas4-cas1 gene sly4000470 a cas5/devS gene sly4000490, which indicates the CRISPR subtype MYXAN (GenProp0922, named after Myxococcus xanthus).39 Another gene (sly4000520) is a homolog of cas3. All these genes are compatible with CRISPR subtypes I-A and I-B.13 Consequently, although other subtype-specific markers such as a cas8 gene were not identified, CRISPR1* belongs into subtype class I-A or I-B. Thus, the CRISPR1* subtype is clearly distinct from Synechocystis 6803 CRISPR1 that was classified as subtype I-D21 due to the presence of signature genes csc1, csc2 and csc3.13 Among the CRISPR1*-associated Cas genes is furthermore a putative cas6 endonuclease gene (sly4000530). However, this is not closely related to the cas6-1, cas6-2a and cas6-2b genes of Synechocystis 6803 but belongs to the MYXAN subtype.39
In contrast, CRISPR2* and CRISPR3* resemble type III CRISPRs, indicated by the presence of cmr2/cas10 homologs. Although the subtype-specific marker Cmr513 could not be identified for either of them, the presence of cmr2, cmr3 and cmr4 genes associates CRISPR2* and CRISPR3* with subtype III-B rather than with III-A, for which csm-type genes would have been expected.18 However, a single subtype III-A superfamily Csm6 protein is associated with CRISPR2* (gene sly1028100), as well as CRISPR2 (sll7062). Thus, CRISPR2* and CRISPR3* are of the same subtype as CRISPR3 of Synechocystis 6803, including the additional presence of a Csm6 homolog.
In fact, CRISPR3* and CRISPR3 loci appear as fully syntenic with very high percentages of identical residues in pairwise protein sequence alignments (Fig. 5), reaching 100% in case of the Cas2 protein. This high conservation is furthermore consistent with the identical repeat sequence for CRISPR3/3* and is in the range of what might have been expected for the genome sequences of two Synechocystis strains as closely related as PCC6714 and PCC6803. A difference is that the two separate genes slr7081 and slr7082 in Synechocystis 6803 are fused into a single gene (sly4000910) in Synechocystis 6714 (Fig. 5). A closer inspection showed that the DNA sequence in the critical region at the end of slr7081 and between the genes slr7081 and slr7082 is almost fully conserved between the two strains but that in Synechocystis 6714 a “TAT” codon is found instead of the “TAG” stop codon in Synechocystis 6803. Because the analysis was conducted with a Synechocystis 6714 draft genome sequence, we checked the possibility of sequencing error by sequence analysis of a PCR product from this region. However, both sequences were identical, providing independent verification for this difference (Fig. S1).
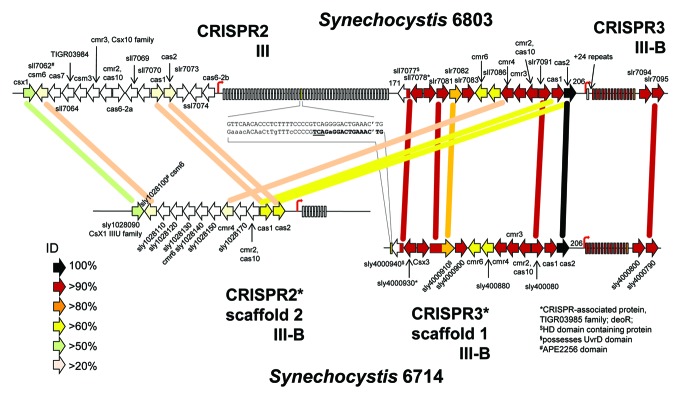
Figure 5. Organization of the two Type III CRISPR-cas systems in Synechocystis 6714 compared with Synechocystis 6803. Several cas genes are associated with each of these CRISPR arrays in both organisms, indicated by the arrows (genes not drawn to scale). Rectangles symbolize the direct repeats. For genes that could be annotated, the nomenclature introduced by Makarova et al. is followed,13,18 otherwise the systematic gene IDs are given. The percentage of sequence identity between predicted gene products (blastP) is coded as indicated. Start sites of transcription (TSS) are marked by thin arrows. For better orientation, selected genes are connected by lines that correspond to their pairwise percentage identity. The synteny between CRISPR3* and CRISPR3 begins within the sll7077/sly4000940 genes and includes the repeat spacer arrays plus immediately following genes. The cas1 and cas2 gene products of CRISPR2* have a substantially higher percentage of identical residues to their counterparts in CRISPR3 than in CRISPR2. Gene sly4000940 in CRISPR3* of Synechocystis 6714 terminates with a sequence motif aligned in the center of the figure to a direct repeat of CRISPR2 of Synechocystis 6803. The final nucleotides of the sly4000940 reading frame are in boldface letters and the stop codon is underlined (reverse orientation). Note that this sequence points to a particular set of repeats that are followed by a TG dinucleotide from the following spacer.
The CRISPR3 and CRISPR2 loci form in Synechocystis 6803 a contiguous stretch of 43 kb on plasmid pSYSA and encompass at least the genes slr7061-slr7095. Therefore, a corresponding degree of similarity as for CRISPR3/3* might also be expected for CRISPR2/2*. However, that is not the case. The high sequence conservation finishes within the sll7077 gene, encoding a protein with a predicted HD domain (metal dependent phosphohydrolases),40 whose first 65 codons are still 98% conserved (64/65 amino acids), whereas the remaining 128 residues (Sll7077) or 87 residues (Sly4000940) are so distant that they can hardly be aligned any more. In Synechocystis 6803 this gene (sll7077) is directly adjacent to the repeat-spacer array of CRISPR2 (Fig. 5). This is not the case in Synechocystis 6714 but, interestingly, a single direct repeat-like sequence element is present at the end of the sly4000940 ORF (Fig. 5). The correctness of this genomic arrangement was independently verified by PCR (Fig. S2).
Such metal dependent phosphohydrolases can very well play a role in nucleic acid metabolism. However, it should be noted that in addition to Sll7077/Sly4000940, there are several additional homologs in both genomes, these are sll1660 in Synechocystis 6803, genes sly1002080 and sly1028310 in Synechocystis 6714.
Expression of the CRISPR1* repeat-spacer array is modulated by environmental conditions
We noticed that in all six studied CRISPR systems possible transcriptional regulators are part of the cas gene cassettes. These genes are sll7009, sll7062 and sll7078 in case of Synechocystis 6803. In Synechocystis 6714, the respective genes are sly4000560, sly1028100 and sly4000930, which appear associated with CRISPR1*, 2* and 3*. Four of these six proteins are WYL domain proteins and all four respective genes belong into the same Cluster of Orthologous Genes 2378 (COG2378). The WYL domain is around 170 amino acids in length and typically found to the C terminus of a DNA-binding helix-turn-helix domain. WYL domain proteins encompass mainly proteins classified as transcriptional regulators, including several DeoR family proteins. Consistent with the WYL domain annotation is the COG2378, which encompasses proteins classified as transcriptional regulators, including many DeoR family proteins as well. The other two (Sly1028100 and Sll7062) of these six proteins belong into the Csm6_III-A family of proteins. Members of this family are often fused to an HTH domain and are also known as APE2256 family proteins. One representative of the Csm6 protein family, the Sulfolobus solfataricus protein Sso1445, has been structurally characterized;41 it contains an HTH domain and was suggested as a regulatory protein.18
It is known that in enterobacteria the expression of the CRISPR system is under transcriptional control.42-45 Therefore, we wondered if expression of the Synechocystis CRISPRs would be regulated in some way and what role, if any, these possible regulators would play in it. Nine different conditions relevant for photoautotrophic organisms were tested for their impact on the CRISPR expression; (1) exponential phase (standard conditions), (2) stationary phase (standard conditions), (3) cold stress, (4) heat stress, (5) high light stress, (6) darkness, (7) CO2 limitation, (8) nitrogen and (9) iron limitation.
Northern hybridizations were performed with an oligonucleotide probe against the direct repeat sequence of the respective CRISPR. For CRISPR1*, we observed a reduced amount of mature crRNA and processing intermediates in conditions with temperature or light stress, compared with standard growth conditions (exponential phase; Fig. 6). In contrast, nitrogen and iron limitation seemed to enhance the expression of the CRISPR1* array.
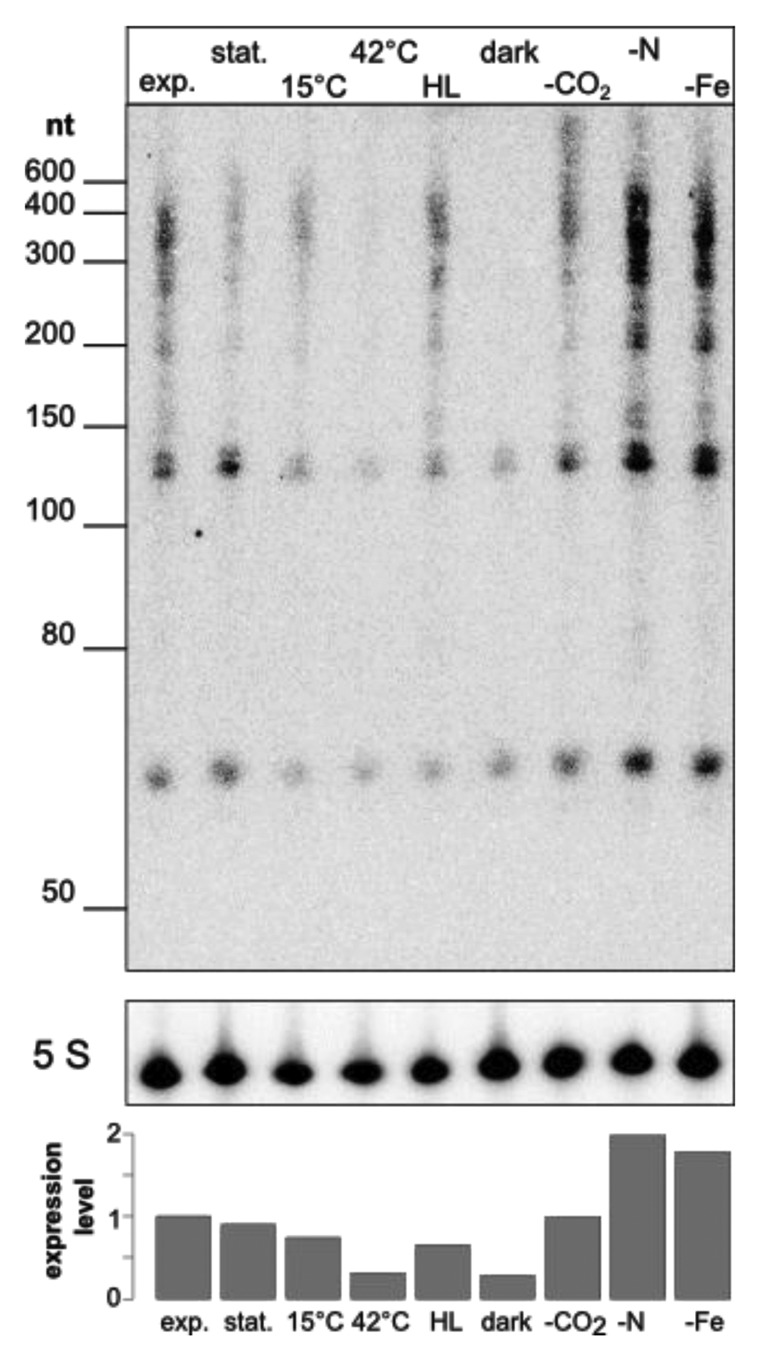
Figure 6. Environmental conditions affect expression levels of CRISPR1*. Nine different environmentally relevant conditions were tested for the accumulation of CRISPR1*-derived transcripts by hybridization using a synthetic oligonucleotide against the direct repeats. Labels are used as follows, exp., exponential phase (standard conditions); stat., stationary phase (standard conditions); 15°C, cold stress; 42°C, heat stress; HL, high light stress; dark, darkness; -CO2, CO2 limitation; -N, nitrogen limitation; -Fe, iron limitation. Five S, 5S rRNA was hybridized as a loading control and after densitometric scanning was used for normalization of CRISPR1* band intensities (lower panel).
A transcriptional regulator involved in the regulation of CRISPR expression
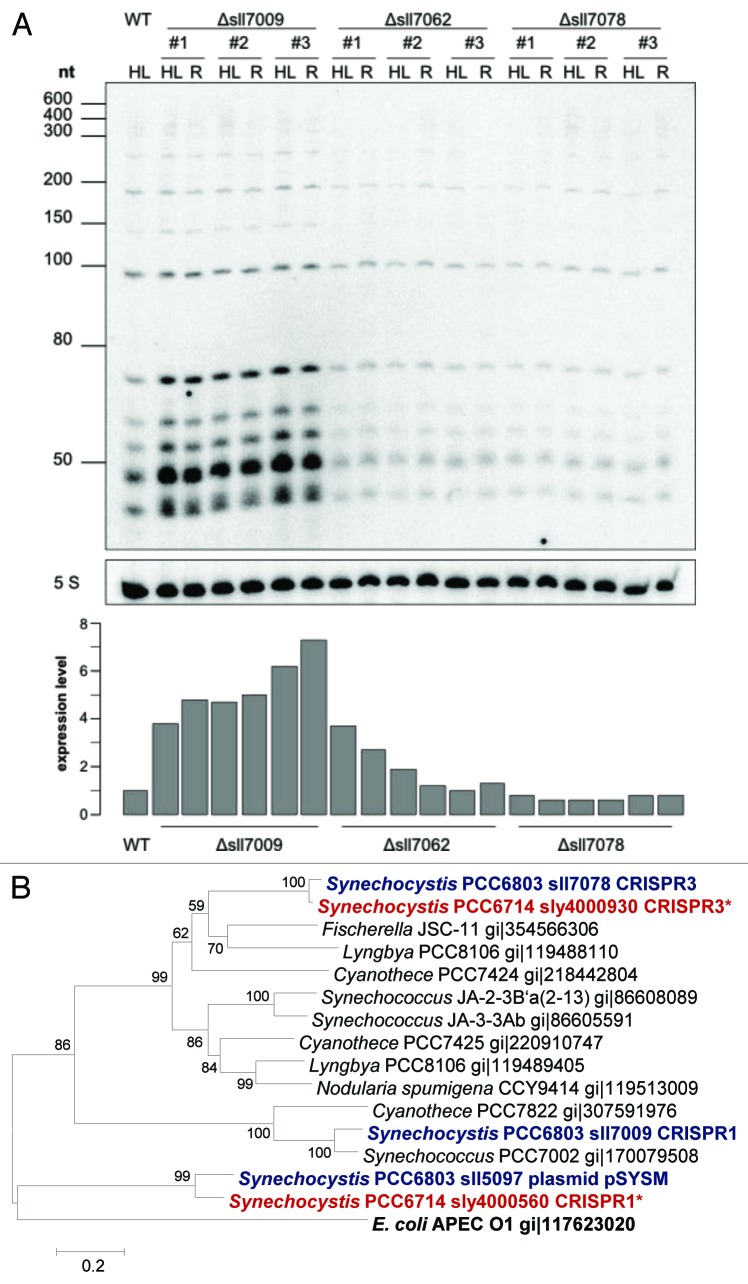
The observed altered expression of CRISPR1* in Synechcocystis 6714 under a variety of stress conditions (Fig. 6) and the presence of genes encoding possible transcriptional regulators as part of the CRISPR loci in both Synechcocystis strains led us to analyze these genes. Here, we chose the genes sll7009, sll7062 and sll7078 of Synechcocystis 6803 because of its well-characterized system for genetic manipulation. We tested respective single knockout mutants after 30 min of high light stress and 30 min high light stress followed by a 30 min recovery phase under standard light conditions, respectively. In northern hybridization using an oligonucleotide probe against spacer one of CRISPR1 an increased amount of small CRISPR-derived transcripts (mature crRNA) of about 50 nt length can be seen in the deletion background of sll7009. No such drastic effect is seen for the knockouts of sll7062 nor sll7078 (Fig. 7). This result if fully consistent with the association of sII7009 with the CRISPR1 array, whereas sll7062 and sll7078 rather appear associated with the CRISPR2 and CRISPR3 arrays.21
Figure 7. A transcription factor acting as repressor is involved in the regulation of CRISPR1 expression. (A) Knockout mutants of putative transcription factor genes were tested for the abundance of CRISPR1 transcripts. Chosen conditions were 30 min high light (HL) and 30 min high light followed by a 30 min recovery phase under standard light conditions (R). The positions of selected size marker bands are indicated. Five S, 5S rRNA was hybridized as a loading control and after densitometric scanning was used for normalization of CRISPR1 band intensities (lower panel). (B) Phylogenetic relationships among CRISPR-associated transcription factor proteins of Synechocystis and of selected other bacteria using the minimum evolution method. Gene identification numbers (gi) are given for all compared sequences expect for the proteins from Synechocystis 6803 and Synechocystis 6714. For Synechocystis sequences the gene names are indicated together with the designation of the respective CRISPR-Cas system they are associated with (compare Fig. 5). We noticed the plasmid pSYSM-encoded Synechocystis 6803 protein Sll5097 to be very similar to Synechocystis 6714 protein Sly4000560. However, the relation of Sll5097 to the CRISPR system, if any, is unknown. The sequence of the E. coli APEC O1 DeoR protein was used as outgroup. The optimal tree with the sum of branch length = 6.15816 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. The multiple sequence alignment was obtained using T-Coffee.54 All positions containing gaps and missing data were eliminated. There were a total of 105 positions in the final data set. Evolutionary analyses were conducted in MEGA5.53
Discussion
Recently, we have described three distinct CRISPR systems, named CRISPR1, CRISPR2 and CRISPR3 in the widely used cyanobacterial model strain Synechocystis 6803.21 The CRISPR3-associated Cas genes in Synechocystis 6803 lack a gene with a known function as maturation endonuclease. Moreover, we found in genetic analyses the maturation and/or accumulation of crRNAs from the CRISPR3 system to be independent of the Cas6 activities associated with CRISPR1 and CRISPR2 of Synechocystis 6803.21 Instead, we identified a cmr2 gene that upon inactivation led to a loss of CRISPR3-derived crRNAs, a phenotype that was reversed upon re-introduction of the gene. This result suggested Cmr2 (Sll7090) as involved in the maturation, regulation of expression, Cmr complex formation or stabilization of transcripts. There are several facts known about Cmr2 proteins,31,32 and the structure of Cmr2 from Pyrococcus furiosus has recently been solved.35,46 However, none of these data did point directly toward a possible role of Cmr2 in the maturation of crRNAs. Therefore, the function of Cmr2 in Synechocystis 6803 CRISPR3 crRNA accumulation appeared enigmatic and it appeared even possible that CRISPR3 is not a functionally active system at all.
Therefore, here we addressed the evolutionary history of these cyanobacterial CRISPR-Cas systems and set out to identify a possibly Cmr2-dependent system closely related to CRISPR3 of Synechocystis 6803. Indeed, we identified one closely related strain, Synechocystis 6714, which possesses a CRISPR3 system similar to the one in Synechocystis 6803 (called here CRISPR3*). The direct repeat sequences of CRISPR3* and CRISPR3 are identical, the Cas gene clusters are perfectly syntenic to each other (Fig. 5), and the predicted Cas proteins are also very close, with up to 100% sequence identity (Cas2). Like in Synechocystis 6803, a cas6 gene is lacking, whereas a well-conserved cmr2 gene (94% amino acid identity) is present. The only noteworthy difference between the CRISPR3 and CRISPR3* systems of both strains are the spacer sequences. Hence, these systems must have been active in spacer acquisition in both strains at least until very recently. In addition, the complex pattern of transcript accumulation detected for both (Fig. 1B and Scholz et al.21) is indicative of a well-working maturation apparatus. We conclude that Cmr2 is indeed a candidate factor involved in the maturation of crRNAs.
In the course of our draft genome analysis, we identified two additional CRISPR-Cas systems in Synechocystis 6714. In contrast to CRISPR3*/CRISPR3, these two are very different, despite the generally very close relatedness of the two analyzed strains (99.4% sequence identity in 16S rRNA). Hence, this observation suggested substitution of the two complete CRISPR-Cas cassettes had occurred. In an alternative scenario, it could be that CRISPR3/3* were introduced to their current respective hosts via horizontal gene transfer, in which CRISPR1/1* and 2/2* were present prior to this transfer.
Whereas extensive recombination events and horizontal gene transfer are known to play a role in the genetic modification of CRISPR-Cas cassettes, the exact mechanisms are not. In Synechocystis 6803, the CRISPR2 locus is directly adjacent to the CRISPR3 locus. Because of the high conservation of CRISPR3*/3, it was possible to search for the possible break point, where sequence identity would drop. We identified this region within the sll7077/sly4000940 gene pair (Fig. 5). Within this region we identified a direct repeat-like sequence in Synechocystis 6714, similar to the sequence of Synechocystis 6803 CRISPR2 direct repeats. This is relevant as there is no CRISPR2 in the Synechocystis 6714 genome, the possible substitute is not only at another location but also possesses an entirely different repeat (compare CRISPR2* and CRSIPR2 repeats in Table 1). Therefore, the CRISPR2 direct repeat-like element in gene sly4000940 (Fig. 5) is indicative of a previous Synechocystis 6803 CRISPR2-like CRISPR-Cas system that was present at this region some while ago.
A striking feature of the six systems compared here is their association to genes encoding possible transcriptional regulators. We noticed a remote similarity to DeoR-type regulators for four of these six proteins. The archetypical transcriptional repressor DeoR (“Deoxyribose Regulator”) is involved in the negative regulation of genes that are involved in nucleotide catabolism.47 What makes this association so interesting is our finding that certain environmental conditions impact accumulation of crRNAs (Fig. 6). Proteins of this family of transcriptional regulators consist of two domains, a potential helix-turn-helix DNA-binding motif and a domain involved in the oligomerization and the recognition of a possible co-inducer in the C-terminal part [WYL (pfam13280) domain in the proteins Sll7009, Sll7078, Sly4000560 and Sly4000930 compared in Fig. 7B].48 It is tempting to speculate that this domain could mediate the response to an environmental cue to the CRISPR system.
We tested the functionality of these possible regulators by genetic knockout experiments and identified one (Sll7009) as a repressor of the subtype I-D CRISPR1 locus in Synechocystis 6803 (Fig. 7A). This association is specific as knockout mutagenesis of the other two possible regulators, Sll7062 and Sll7078, had no effect on the accumulation of CRISPR1-derived transcripts. Transcriptional control over the CRISPR system has been demonstrated for E. coli and Salmonella enterica.42-45 However, our finding identifies for the first time a transcriptional regulator of CRISPR expression outside the enterobacteria and it is to our knowledge the first observation of transcriptional control of CRISPR expression by a WYL domain-containing protein of the COG2378-type transcription factor family.
Materials and Methods
Strains, culture media and growth conditions
The Synechocystis PCC6803 “PCC-M” strain36 was obtained from A. Wilde, University of Freiburg (originally from S. Shestakov, Moscow State University). Synechocystis strains PCC6308, PCC6702, PCC6714 and PCC6805 were purchased from the Pasteur Culture Collection (PCC) in Paris, France.
Liquid cultures were grown at 30°C in BG11 medium49 under continuous white light illumination of 50 µmol quanta m−2s−1 and a continuous stream of air to the desired growth phase (OD750 = 0.6 – 0.8). For the expression analysis, cultures of Synechocystis 6714 were initially grown under standard conditions and then transferred to nine different conditions: (1) exponential growth until an OD750 of 0.67; (2) stationary phase until an OD750 of 4.1; (3) cold stress, 15°C for 30 min; (4) heat stress, 42°C for 30 min; (5) high light, 470 µMol q s−1 m−2 for 30 min; (6) dark, no light for 12 h; (7) Ci depletion, 150 mL of culture were washed three times with 100 mL of carbon-free BG11 and cultivated further for 20 h; (8) N depletion, 150 mL of culture were washed three times with 100 mM of nitrogen-free BG11 and cultivated further for 12 h; (9) Fe stress, addition of DFB chelator for 24 h.
For the expression analysis of the transcription factor knockout mutants, cultures were initially grown under standard conditions an OD750 of 0.85 and then exposed to high light, 600 µMol q s−1 m−2 for 30 min (HL) and recovered for 30 min at standard illumination of 50 µmol quanta m−2s−1.
Knockout experiments and transformation of Synechocystis 6803
To analyze gene functions, putative transcription factor genes were knocked out by replacing the gene with a resistance cassette through homologous recombination. The upstream and downstream flanking regions of the corresponding gene were amplified via PCR (for primer sequences, see Table 2) and ligated with the resistance cassette, resulting in following construct: upstream flanking region, resistance cassette, downstream flanking region. Three different resistance cassettes were used, providing resistance against the antibiotics kanamycin (from vector puc4K), streptomycin (from vector pRL692) or chloramphenicol (from vector pACYC184). These constructs were ligated into the multiple cloning site of pJet1.2 and the resulting vectors were subsequently used to transform cells of Synechocystis 6803. For each transformation, 10 ml of Synechocystis 6803 culture (OD750 = 0.5 - 1.0) were centrifuged (4,000 rpm, 20°C, 10 min) and the pellet resuspended in 200 µl BG11 medium. After addition of 1–3 µg plasmid (vector pJet1.2 with adequate insert which should be integrated into the pSYSA plasmid via homologous recombination) the sample was incubated at room temperature for 1 h and then plated on BG11 agar plates without antibiotics. Slightly shaded plates were incubated for 1–2 d at 30°C. For subsequent selection, kanamycin (10 µg/ml) was added to the plates underneath the agar layer. After 3–4 wk, single colonies were picked and cultivation on plates continued with increasing concentration of antibiotics until full segregation was achieved.
Table 2. Desoxyoligonucleotide primers used in this study.
| Oligonucleotide | sequence (5‘→3‘) | |
|---|---|---|
|
Knockout constructs | ||
| ∆sll7009_I_fw |
CGATCGCCTCATGTCTGTTTTAAC |
|
| ∆sll7009_I_rev |
CTAGGCCGGCCACAAAATAATTAGGTCTGA |
|
| ∆sll7009_II_fw |
GACCGGTATTGAAATTTCATACTAGTCTTCAAAC |
|
| ∆sll7009_II_rev |
CCTGTGGTGCATTAAATGCTGTTTC |
|
| ∆sll7062_I_fw |
CCAAAATTCCTAGGAGGAATACCAAG |
|
| ∆sll7062_I_rev |
GATAACCGGTTCATTATTGATAAATTGGGGCTG |
|
| ∆sll7062_II_fw |
GGCCGGCCACAAGGGGAAACAATAACG |
|
| ∆sll7062_II_rev |
GAGGAAACTTCTGCTATTGGCG |
|
| ∆sll7078_I_fw |
GTTGCCAGTTTTGCCGTTTTTGCTG |
|
| ∆sll7078_I_rev_B |
GTACCGGTCATGGCAGCCCTTTAC |
|
| ∆sll7078_II_fw |
CATCGGCCGGCCTACTTTGGGCC |
|
| ∆sll7078_II_rev |
GCAATGGCCACCGACTGGG |
|
| Km_AgeI_fw |
GGCTTTACCGGTTATATGGGAATTCCG |
|
| Km_FseI_rev |
GGCCGGCCTTGTCGGGAAGATG |
|
| Cm_AgeI_pACYC184_fw |
GATACCGGTAAGCCCTGGGC |
|
| Cm_FseI_pACYC184_rev |
CCGACGGCCGGCCCGAATTTGC |
|
| Strep_AgeI_pRL692_fw |
CAGTAGACCGGTGTACAGAGTGATGTC |
|
| Strep_FseI_pRL692_rev |
GTAATTGGCCGGCCTATTATCGTAGTTGCTC |
|
|
Northern hybridization | ||
| C1S1 |
GCATTGAAAGCGACCGCCAGGGGCAC |
|
| DR1_6714 |
GTGCTCAACGCCTTACGGCATCAATGG |
|
| CRISPR3_TS_fw |
GGATGCCTTGTCCCGTAAGG |
|
| CRISPR3_TS_rev |
TAATACGACTCACTATAGGGAGAGACAGGCAGTGCGCTAACAC |
|
| 5S_6803 |
CGCTGTCACATTTCACAACCGAGTG |
|
|
PCR amplification |
|
|
| 6714_C3_FW |
TCAGCCGCTATTTTTACATTGAGC |
|
| 6714_C3_RV |
ACCGTATACTCCTGGTGATCCAAGG |
|
| 6714_4–910_FW |
AGTAATGTTCAATGGTAGTTGACTC |
|
| 6714_4–910_int_FW |
TTGTCACCATTGGCAAACGGAAGC |
|
| 6714_4–910_RV |
ACTTGTCATGGCTTTACCAGATTG |
|
| 40930–6714_FW |
TATCGCCACCAAGACAATAACG |
|
| dws40950_6714_RV | GTCGTAATATTCGTCTGCTATGC | |
RNA analysis and hybridization conditions
One hundred milliliters of Synechocystis cultures (samples Fig. 6: 50 ml) were harvested through centrifugation (10,000 rpm, 20°C, 8 min). The pellet was resuspended in 1 ml of PGTX50 and immediately frozen in liquid nitrogen. Samples were then incubated for 5 min at 95°C and put on ice for 5 min. After addition of 1 ml of chloroform/isoamylalcohol (24:1) and thorough agitation samples were incubated at room temperature for 10 min. Samples were centrifuged with a swing out rotor at 6,000 rpm, 15°C for 15 min. The upper aqueous phase was transferred into a new vial and the same volume of chloroform/isoamylalcohol (24:1) was added and mixed. Samples were centrifuged as described above and the aqueous phase removed again and combined with an equal volume of isopropanol. After gently inverting the tube, RNA was allowed to precipitate over night at -20°C. RNA was pelleted through centrifugation (13,000 rpm, 4°C, 30 min). The pellet was washed with 1 ml of 70% ethanol (13,000 rpm, 20°C, 5 min), allowed to air dry for approximately 10 min and resuspended in 30 µl H2O.
Eight µg of total RNA per lane were separated on 10% polyacrylamide-urea gels and electroblotted onto Hybond-N+ membranes from Amersham. Membranes were prehybridized for at least 30 min at 45°C with hybridization buffer (50% deionized formamide, 7% SDS, 250 mM NaCl, 120 mM NaPi buffer, pH 7.2) in glass tubes under continuous rotation. For northern hybridization, synthetic oligonucleotide probes (Table 2) labeled by (32P) ATP were used. For 5′ labeling of oligonucleotides with 30 µCi (32P) ATP, T4 polynucleotide kinase (Fermentas) was used: 5 µl oligonucleotide (10 pmol/µl) and 9 µl H2O were mixed and incubated for 5 min at 95°C and put on ice. After addition of 2 µl 10 x buffer A, 3 µl (32P) ATP (10 mCi/ml) and 1 µl PNK (10 U/µl) the probe was incubated for 30 min at 37°C and the reaction stopped at 95°C for 5 min. The probe was put on ice and then added to the prehybridized membrane for hybridization at 45°C overnight. Subsequent washing of the membrane was performed at 40°C with washing solution I (2 × SSC, 1% SDS), II (1 × SSC, 0.5% SDS) and III (0.1 × SSC, 0.1% SDS) for 10 min each. Signals were detected with a storage phosphor screen (Kodak), read on a BIO-RAD Molecular Imager FW system and analyzed with the Quantity One software (BIO-RAD). Signals were normalized by 5S rRNA.
Analysis of genomic DNA
The preparation of genomic DNA for deep sequencing analysis followed a recently published protocol.36 In short, DNA was isolated from 80 ml cultures harvested on a glass microfiber filter by vacuum filtration, followed by cell lysis in the presence of 2% SDS and 1.5 mg proteinase K at 50°C overnight. Following phenol/chloroform extraction and 2-propanol precipitation, the DNA was resuspended in 50 µl H2O. One µl of RNase A (Sigma) was added and the tube incubated at 37°C and 260 rpm overnight. RNase was removed by another round of phenol/chloroform extraction and 2-propanol precipitation.
Sequencing of genomic DNA was performed on an Illumina Genome Analyzer IIx system. We performed paired-end sequencing for a library with approx. Three hundred-bp long fragments and for a mate-paired library created for 3 kb long fragments. Velvet was used to assemble the resulting data sets into five scaffolds with lengths ranging from 46,504–2,984,476 nt. A preliminary annotation was performed with RAST.51
Sequence data
All sequence analyses with Synechocystis 6803 were done using the publicly available sequence for the “Kazusa” strain as reported in RefSeq (NC_005230.1) or GenBank (AP004311.1) and for the “PCC-M” strain in CP003267. The sequence information for Synechocystis 6714 can be accessed at DDBJ/EMBL/GenBank under the accession AMZV01000000.
RNA structure prediction
Putative secondary structures of the repeat sequences were predicted using RNAHeliCes.52 We inspected the low-energy structures and selected the ones with the highest similarity to the structures presented in Scholz, et al.21 Structure drawings were generated with VARNA.
Supplementary Material
Acknowledgments
We thank Gisle Vestergaard, Copenhagen, for discussion in the initial work leading to this manuscript, and Matthias Kopf, Freiburg, for support in the final phase of the genome assembly. Financial support for this work by the German Research Foundation (DFG) program FOR1680 “Unravelling the Prokaryotic Immune System” (grant HE 2544/8-1) to WRH is greatly acknowledged.
Glossary
Abbreviations:
- Cas
CRISPR-associated proteins
- CASCADE
CRISPR-associated complex for antiviral defense
- Cmr
Cas module RAMP
- CRISPR
clustered regularly interspaced short palindromic repeats
- MFE
minimum free energy
- nt
nucleotides
- RAMP
repeat-associated mysterious protein
- RNP
ribonucleoprotein
- TSS
transcription start site
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
The authors have made the following declarations about their contributions: Conceived and designed the experiments: W.R.H., I.S., S.H. Performed the experiments: I.S., S.H. Analyzed the data: B.V., W.R.H. Wrote the paper: W.R.H., S.H., B.V.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24160
References
- 1.Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem. 2011;392:277–89. doi: 10.1515/bc.2011.042. [DOI] [PubMed] [Google Scholar]
- 2.Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64:475–93. doi: 10.1146/annurev.micro.112408.134123. [DOI] [PubMed] [Google Scholar]
- 3.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 4.Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37:7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–90. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–7. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–8. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 8.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–39. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 9.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol. 2011;79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–6. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 11.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci. 2009;34:401–7. doi: 10.1016/j.tibs.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–29. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- 15.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heidelberg JF, Nelson WC, Schoenfeld T, Bhaya D. Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS One. 2009;4:e4169. doi: 10.1371/journal.pone.0004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuno S, Yoshida T, Kaneko T, Sako Y. Intricate interactions between the bloom-forming cyanobacterium Microcystis aeruginosa and foreign genetic elements, revealed by diversified clustered regularly interspaced short palindromic repeat (CRISPR) signatures. Appl Environ Microbiol. 2012;78:5353–60. doi: 10.1128/AEM.00626-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scholz I, Lange SJ, Hein S, Hess WR, Backofen R. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PLoS One. 2013;8:e56470. doi: 10.1371/journal.pone.0056470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopfmann S, Hess WR. Toxin antitoxin systems on the large defense plasmid pSYSA of Synechocystis sp. PCC 6803. J Biol Chem. 2013 doi: 10.1074/jbc.M112.434100. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–8. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–96. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–64. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–92. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 27.Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–7. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 28.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–8. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Przybilski R, Richter C, Gristwood T, Clulow JS, Vercoe RB, Fineran PC. Csy4 is responsible for CRISPR RNA processing in Pectobacterium atrosepticum. RNA Biol. 2011;8:517–28. doi: 10.4161/rna.8.3.15190. [DOI] [PubMed] [Google Scholar]
- 30.Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc Natl Acad Sci USA. 2011;108:21218–22. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anantharaman V, Iyer LM, Aravind L. Presence of a classical RRM-fold palm domain in Thg1-type 3′- 5’nucleic acid polymerases and the origin of the GGDEF and CRISPR polymerase domains. Biol Direct. 2010;5:43. doi: 10.1186/1745-6150-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pei J, Grishin NV. GGDEF domain is homologous to adenylyl cyclase. Proteins. 2001;42:210–6. doi: 10.1002/1097-0134(20010201)42:2<210::AID-PROT80>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 33.Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–96. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cocozaki AI, Ramia NF, Shao Y, Hale CR, Terns RM, Terns MP, et al. Structure of the Cmr2 subunit of the CRISPR-Cas RNA silencing complex. Structure. 2012;20:545–53. doi: 10.1016/j.str.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trautmann D, Voss B, Wilde A, Al-Babili S, Hess WR. Microevolution in cyanobacteria: re-sequencing a motile substrain of Synechocystis sp. PCC 6803. DNA Res. 2012;19:435–48. doi: 10.1093/dnares/dss024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juranek S, Eban T, Altuvia Y, Brown M, Morozov P, Tuschl T, et al. A genome-wide view of the expression and processing patterns of Thermus thermophilus HB8 CRISPR RNAs. RNA. 2012;18:783–94. doi: 10.1261/rna.031468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–36. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 39.Haft DH, Selengut JD, Richter RA, Harkins D, Basu MK, Beck E. TIGRFAMs and Genome Properties in 2013. Nucleic Acids Res. 2013;41(Database issue):D387–95. doi: 10.1093/nar/gks1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23:469–72. doi: 10.1016/S0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 41.Lintner NG, Frankel KA, Tsutakawa SE, Alsbury DL, Copié V, Young MJ, et al. The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system. J Mol Biol. 2011;405:939–55. doi: 10.1016/j.jmb.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medina-Aparicio L, Rebollar-Flores JE, Gallego-Hernández AL, Vázquez A, Olvera L, Gutiérrez-Ríos RM, et al. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J Bacteriol. 2011;193:2396–407. doi: 10.1128/JB.01480-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Rodriguez R, Haitjema C, Huang Q, Nam KH, Bernardis S, Ke A, et al. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol Microbiol. 2011;79:584–99. doi: 10.1111/j.1365-2958.2010.07482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol. 2010;75:1495–512. doi: 10.1111/j.1365-2958.2010.07073.x. [DOI] [PubMed] [Google Scholar]
- 45.Westra ER, Pul U, Heidrich N, Jore MM, Lundgren M, Stratmann T, et al. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol. 2010;77:1380–93. doi: 10.1111/j.1365-2958.2010.07315.x. [DOI] [PubMed] [Google Scholar]
- 46.Zhu X, Ye K. Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR-Cas systems. FEBS Lett. 2012;586:939–45. doi: 10.1016/j.febslet.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 47.Valentin-Hansen P, Albrechtsen B, Løve Larsen JE. DNA-protein recognition: demonstration of three genetically separated operator elements that are required for repression of the Escherichia coli deoCABD promoters by the DeoR repressor. EMBO J. 1986;5:2015–21. doi: 10.1002/j.1460-2075.1986.tb04458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valentin-Hansen P, Højrup P, Short S. The primary structure of the DeoR repressor from Escherichia coli K-12. Nucleic Acids Res. 1985;13:5927–36. doi: 10.1093/nar/13.16.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rippka R, Deruelles J. B. WJ, Herdmann M, Stanier RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- 50.Pinto FL, Thapper A, Sontheim W, Lindblad P. Analysis of current and alternative phenol based RNA extraction methodologies for cyanobacteria. BMC Mol Biol. 2009;10:79. doi: 10.1186/1471-2199-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang J, Backofen R, Voß B. Abstract folding space analysis based on helices. RNA. 2012;18:2135–47. doi: 10.1261/rna.033548.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39(Web Server issue):W13-7. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.