Abstract
To fend off foreign genetic elements, prokaryotes have developed several defense systems. The most recently discovered defense system, CRISPR/Cas, is sequence-specific, adaptive and heritable. The two central components of this system are the Cas proteins and the CRISPR RNA. The latter consists of repeat sequences that are interspersed with spacer sequences. The CRISPR locus is transcribed into a precursor RNA that is subsequently processed into short crRNAs. CRISPR/Cas systems have been identified in bacteria and archaea, and data show that many variations of this system exist. We analyzed the requirements for a successful defense reaction in the halophilic archaeon Haloferax volcanii. Haloferax encodes a CRISPR/Cas system of the I-B subtype, about which very little is known. Analysis of the mature crRNAs revealed that they contain a spacer as their central element, which is preceded by an eight-nucleotide-long 5′ handle that originates from the upstream repeat. The repeat sequences have the potential to fold into a minimal stem loop. Sequencing of the crRNA population indicated that not all of the spacers that are encoded by the three CRISPR loci are present in the same abundance. By challenging Haloferax with an invader plasmid, we demonstrated that the interaction of the crRNA with the invader DNA requires a 10-nucleotide-long seed sequence. In addition, we found that not all of the crRNAs from the three CRISPR loci are effective at triggering the degradation of invader plasmids. The interference does not seem to be influenced by the copy number of the invader plasmid.
Keywords: archaea, Haloferax volcanii, CRISPR/Cas, crRNA, PAM, seed sequence
Introduction
Every living organism must defend itself against foreign genetic elements. Prokaryotes use a variety of defense mechanisms, one of which is the recently discovered prokaryotic immune system called the CRISPR/Cas system.1-6 The system comes in various forms, which have been classified into three major types (I–III) and a minimum of 10 subtypes.7 In all of the CRISPR/Cas types, the central elements are the crRNAs and a set of proteins, called the Cas proteins. The defense reaction consists of three steps: (1) adaptation to the invader, (2) the expression of the crRNAs and (3) the degradation of the invader DNA (or RNA). In the first stage, the adaptation step, the cell recognizes a new invader as it enters the cell and degrades its DNA (or, in subtype III-B, its RNA, from now on we will only mention DNA as target, but as stated here in subtype III-B RNA is the target.).8-11 A piece of the invader DNA (known as the protospacer) is subsequently selected to be integrated into the CRISPR locus of the host (note that once the protospacer has been integrated into the CRISPR locus, it is renamed as a spacer). In the type I and type II CRISPR/Cas systems, an important distinguishing characteristic for the selection of a protospacer is the protospacer adjacent motif (PAM).7,12 This motif is located in the invader DNA, directly adjacent to the protospacer. The PAM sequence is important, not only for its selection as a spacer but also for the third step of the process, the interference reaction.7,12-15 In the second stage of the defense reaction, the expression step, the CRISPR RNA is synthesized as a precursor RNA and is subsequently processed into crRNAs, which are essential for the system to function. In the last stage of the process, the interference reaction, the crRNA and the Cas proteins form a ribonucleoprotein interference complex, which recognizes the DNA of an attacking invader in a sequence-specific manner (by base-pairing with the target sequence). The invader’s DNA is subsequently cleaved by the Cas proteins. In the CRISPR/Cas systems I and II, the invader is only recognized and degraded if it carries the correct PAM sequence.10
The crRNA is elemental in base-pairing with the invader DNA and, thereby, identifying it in a sequence-specific manner. In the type I and type III systems, the crRNA is generated by a member of the Cas6 protein family.7 After processing the crRNA, Cas6 remains bound to the 3′ handle,16,17 while the 5′ handle of the crRNA is protected by the binding of another Cas protein (e.g., Cse1, Cas7 or Cas5 for type I systems).17 The central part of the crRNA is the spacer sequence (Fig. 1B), which is flanked by parts of the repeat sequences. Because the spacer is an exact copy of the protospacer, there is initially a 100% match between the crRNA and the DNA of the invader. However, the invader could escape this immune system by mutating the protospacer; the success of the escape depends on the degree and the location of these mutations.4 In Escherichia coli,14 (CRISPR/Cas type I-E) and Pseudomonas aeruginosa17 (CRISPR/Cas type I-F), it has been shown that the interaction between the crRNA and the invader DNA is initiated and controlled by a seed region. A non-contiguous seven-nucleotide match proximal to the PAM sequence is required for the defense reaction to occur in E. coli.14 Mutations in the invader DNA that are distal to the PAM do not disable the defense.14
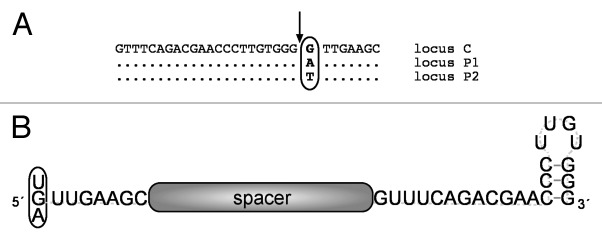
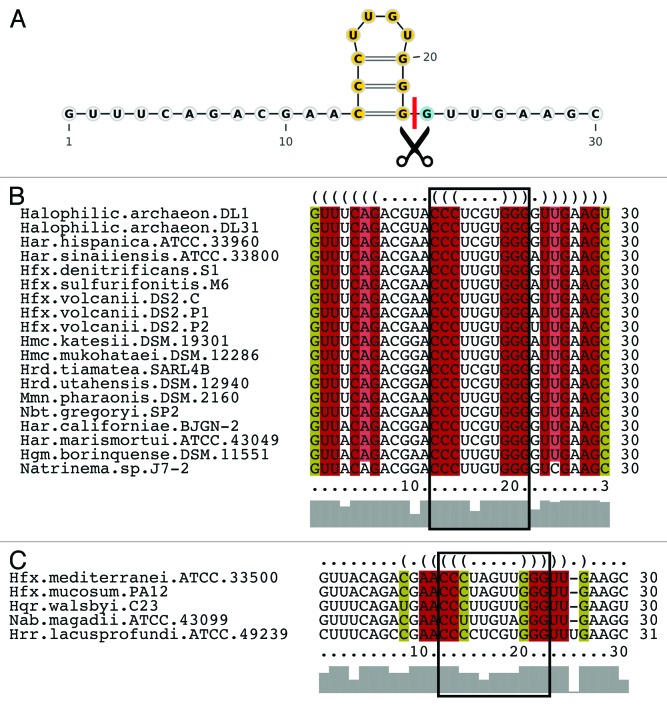
Figure 1. (A)Repeat sequences of the three CRISPR loci in H. volcanii. The repeat sequences of the three different CRISPR RNAs differ by one nucleotide (at position 23). The sequence of the CRISPR locus C repeat is shown. Positions that are identical in the P1 and P2 locus repeats are indicated with a dot. The pre-crRNA is processed in the repeat region directly upstream of nucleotide 23 (marked by the arrow). (B) Structure of the crRNA in Haloferax. The product of the pre-crRNA processing reaction is shown. It contains the spacer sequence and an eight-nucleotide 5′ handle, which originates from the upstream repeat. Because position 23 varies between the three CRISPR repeats, the first nucleotide of the 5′ handle is either a G (locus C), an A (locus P1) or a U (locus P2). Downstream of the spacer are the remaining 22 nucleotides of the downstream repeat, which can fold into a minimal stem loop.
Although the CRISPR/Cas systems have been classified into three major groups (I–III),7 data indicate that there are profound differences between the subtypes of each class. For instance, the subtype III-A targets invader DNA, whereas the subtype III-B attacks invader RNA.18 The CRISPR/Cas type I is the most diverse type, and it has been classified into six different subtypes. Subtypes I-A, I-C, I-E and I-F have been studied to some extent, but very little is known about the CRISPR/Cas types I-B and I-D.10
We have investigated the CRISPR/Cas type I-B in the halophilic archaeon Haloferax volcanii. This defense system in Haloferax consists of eight Cas proteins (Cas1-Cas8b) (Fig. S1), which are encoded as a gene cluster that is flanked by two of the three CRISPR loci15 on the minichromosome pHV4; the third CRISPR locus is located on the main chromosome. Recently, we have shown that the CRISPR system remains active in this archaeon and that the Haloferax defense system recognizes six different PAM motifs.15 The CRISPR RNAs are constitutively expressed and processed to crRNAs. Here, we analyzed the crRNA population of the CRISPR/Cas system of Haloferax and the details of the interference reaction, which depends on the crRNA-invader interaction. We show that the CRISPR/Cas type I-B also requires a seed sequence to interact with the invader.
Results
H. volcanii encodes three CRISPR RNAs with nearly identical repeat sequences
H. volcanii H119 contains three CRISPR loci with almost identical repeat sequences, which differ only at position 23 (Fig. 1A). The repeat sequences of all three of the CRISPR loci are 30 nucleotides long, and the last one or two repeats of each locus are mutated. The last repeat of locus C is shortened by 13 nucleotides at its 3′ end, and in locus P1, the last repeat is shortened by one nucleotide. In locus P2, the next-to-last repeat has a G instead of a T at position 23, thereby resembling the repeat of locus C. In addition, the last repeat in the P2 locus is shortened by 23 nucleotides. The spacer lengths vary and are between 34 and 39 nucleotides, with an average length of 36 nucleotides. To determine the exact 5′ ends of the crRNAs, we isolated the fraction of RNA from H. volcanii ranging in size from 55–80 nucleotides. After generating cDNA from this fraction, high-throughput sequencing was performed (for details, see Materials and Methods). Analysis of the reads obtained confirmed that all three of the loci are expressed, as had been suggested earlier by northern blot analysis.15 The crRNAs contain the spacer sequence, preceded by eight nucleotides of the upstream repeat (Fig. 1), the sequence known as the 5′ handle.19 Because the repeats of the three loci differ at position 23, we found three types of handles (Fig. 1B): GTTGAAGC (52% of the crRNAs, locus C), ATTGAAGC (41% of the crRNAs, locus P1), TTTGAAGC (7% of the crRNAs, locus P2). Accordingly, processing of the crRNAs occurs between nucleotides 22 and 23 of the repeat (Fig. 2). The individual crRNAs derived from the three CRISPR loci are not present in the same abundance; some of the crRNAs are overrepresented, while others are completely absent (Fig. 3). However, the previously reported trend of higher levels proximal to the leader and decreasing levels toward the 3′ end of the CRISPR locus20,21 was not observed here. In summary, we were able to demonstrate that the crRNAs of all three loci possess an eight-nucleotide long 5′ handle and differ in their abundance.
Figure 2. A small stem loop structural motif is conserved across 22 haloarchaeal species. (A) Part of the predicted structure for the repeat from locus C is conserved throughout the haloarchaeal species (highlighted in yellow). The red line corresponds to the cleavage site just upstream of the 5′ crRNA tag. The G nucleotide that is cyan in color corresponds to the 23rd nucleotide, which is different in the P1 (A) and P2 (U) loci. (B and C) Multiple sequence alignments generated by LocARNA; the red columns correspond to conserved base pairs and the mustard yellow columns correspond to the presence of a compensatory base pair that conserves the consensus structure. The conserved structural motif from (A) is surrounded by the black box. (B) The larger group of haloarchaea with the conserved motif and a 4-nucleotide hairpin loop. (C) The smaller group with a 5-nucleotide hairpin loop. The conserved CG stem loop motif is surrounded by stabilizing base pairs in both groups.
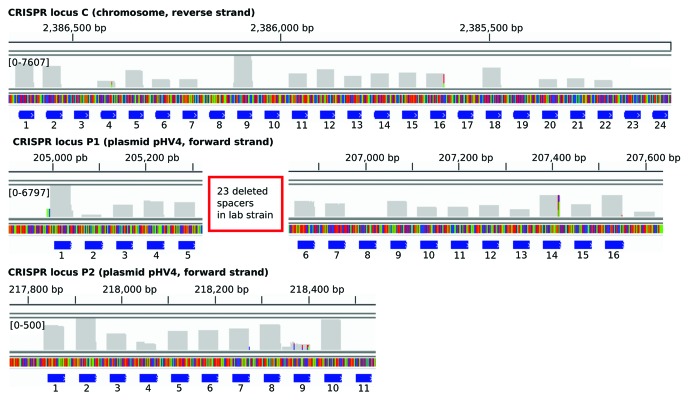
Figure 3. Profile of reads across each CRISPR locus: crRNAs accumulate to varying degrees. The reads of the extracted crRNA are mapped to the three CRISPR RNA loci and the variation in their abundance is shown. The blue bars correspond to the spacer number. The number of reads (y-axis) is given on a log-scale. In locus P1, a large deletion of 23 spacers occurred in our lab strain, in comparison to the sequenced genome in the database.15 Due to the internal priming of the 3′ oligonucleotide used in the sequencing protocol, the 3′ ends generated do not represent the in vivo 3′ ends. For mapping, the reads that started with the 5′ repeat tag were extracted to a maximum length of 35 nucleotides.
A small stem loop is conserved in haloarchaeal repeats
Kunin et al. grouped the CRISPR repeat sequences into 12 large clusters22 based on sequence similarity. These clusters were classified as “unfolded” or “folded” based on their ability to form a consensus structure. According to these analyses, the repeats from H. volcanii (Hmari23 CRISPR/Cas subtype) belong to two unfolded clusters (bacterial repeat cluster 1 and archaeal repeat cluster 9).22 However, we identified stable structures for each of the H. volcanii repeats using the thermodynamic structure prediction programs RNAfold24 and mfold25 (Fig. S3). Therefore, we analyzed the folding potential of each single repeat in all three of the CRISPR RNAs as a function of the surrounding spacer sequences. According to these analyses, all of the repeats share a minimal three base-pair stem loop, and some repeats have the potential to fold into a longer stem loop structure (repeats C5 and C9, for instance, Fig. S2). Variations in the folding potentials of individual repeats indicate that the structure of the repeat depends on the neighboring spacer sequences. A comparative approach, using CRISPR repeat sequences from other haloarchaeal genomes (most with type I-B cas genes), confirmed the individual structure predictions (Fig. S2). The minimal stem loop is conserved in all of the analyzed haloarchaea; it contains three C-G base pairs and is situated directly upstream of the cleavage site that generates the 8-nucleotide 5′ crRNA tag (Fig. 2). This conserved structural motif is generally surrounded by additional stabilizing base pairs within the repeat. The haloarchaea can be separated into two groups: the larger group is made up of haloarchaea that form a 4-nucleotide hairpin loop, and the other, smaller, group includes haloarchaea that form a 5-nucleotide hairpin loop. However, the sequences of both groups contain the three consecutive C-G base pairs (Fig. 2B and C).
In addition to the in silico analyses, we employed two different experimental approaches to identify base-pairing regions in the repeats from the P1 and P2 CRISPR loci. The experimental analyses did not reveal any structures in the repeat sequences under the conditions tested. 1D NMR analysis suggested that neither repeat folded into a stable structure, as only a few very broad signals were observed in the imino-proton 1D spectra, and these completely disappeared at temperatures higher than 10°C (Fig. S4). This lack of signal indicates the absence of stable base-pairing interactions in the putative stem loops because imino proton signals are only observed when their respective nucleotides are part of well-defined base-pairing interactions. In the second approach, we probed the structure of the repeat RNA using the ribonucleases T1, V1 and A. Again, no defined structure was found, confirming the results that had been previously obtained using NMR (data not shown).
In summary, we find that a minimal stem loop structure is conserved in haloarchaeal repeats; this structure is not stable at room temperature in vitro but might be stabilized in vivo by salt or proteins.
A seed region is essential for interference
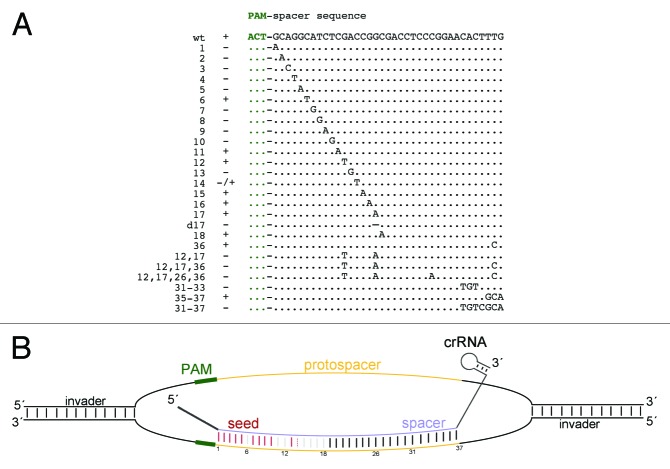
In E. coli14 and P. aeruginosa,17 a “seed region” was found to be essential for the recognition of the protospacer by the CRISPR/Cas system. The seed region must exactly match the corresponding sequence of the spacer to ensure successful interference.14,17 According to our previous analysis, the H. volcanii defense system recognizes a broad range of PAM sequences, suggesting that the defense system is quite flexible. To analyze how many protospacer mutations (based on spacer 1 of CRISPR locus P1: P1.1) can be tolerated by the H. volcanii CRISPR/Cas system, we systematically inserted single-point mutations in the potential seed region (5′ region) of the protospacer and exchanged nucleotides in the 3′ region of the protospacer (Fig. 4). Point mutations at positions 1–5 and 7–10 of the protospacer sequence resulted in transformation rates that were similar to those obtained when the pTA409 vector alone was transformed (Table 1). Thus, the mutated invader plasmid was no longer recognized as foreign. However, single nucleotide exchanges at positions 6, 11, 12, 15–18 or 36 of the 37-bp long protospacer and the combined mutation of the last three positions (35–37) did not affect the recognition of the plasmid by the CRISPR/Cas system or the efficiency of the interference reaction. A deletion of the nucleotide at position 17 and the combined mutation of positions 17 and 36, as well as 12 and 36, resulted in a protospacer variant that could no longer trigger the defense of H. volcanii CRISPR/Cas system. The same is true of a protospacer variant in which the last seven positions of the protospacer (31–37) are mutated. Mutations at position 14 gave ambiguous results; in some transformations, the mutation of position 14 resulted in interference, in others, it did not. This is the position at which the transition between the nucleotides that are essential for interference and the nucleotides that are non-essential seems to occur, and this transition is reflected by the results that we obtained. Taken together, our results suggest that the defense system requires a 10-nucleotide non-contiguous seed sequence but can tolerate a mismatch at position 6.
Figure 4. A seed sequence is required to trigger an interference reaction. (A) To investigate the positions that are essential for successful interference, nucleotides were mutated or deleted (marked by a d) in the protospacer sequence. The PAM sequence is indicated in green. Variants that were recognized by the CRISPR/Cas machinery are marked with a “+,” variants that were not recognized as invader are labeled with a “–“. (B) The interaction between the crRNA and the invader plasmid is shown schematically. The crRNA and the plasmid form an R loop, in which the crRNA is able to base pair with the protospacer sequence. The PAM sequence is shown in green, the protospacer sequence in yellow. Base pairs that are essential for the interference reaction are shown in red, while non-essential base pairs are shown in grey.
Table 1. Mutations in the target sequence that do not prevent successful interference.
| Plasmid | Position changed | Reduction in transformation rate by factor |
|---|---|---|
| pTA409-P1.1 |
— |
1 x 10−3 |
| pTA409-SEED6 |
6 |
2 x 10−3 |
| pTA409-SEED11 |
11 |
5 x 10−3 |
| pTA409-SEED12 |
12 |
1 x 10−3 |
| pTA409-SEED15 |
15 |
1 x 10−3 |
| pTA409-SEED16 |
16 |
1 x 10−3 |
| pTA409-SEED17 |
17 |
1 x 10−3 |
| pTA409-SEED18 |
18 |
4 x 10−4 |
| pTA409-SEED36 |
36 |
4 x 10−4 |
| pTA409-SEED35–37 | 35–37 | 6 x 10−3 |
The different positions mutated in the target sequence are shown (“position changed” column). The transformation rate for the vector pTA409 (carrying no insert) was set to 1. The reductions in transformation rate for the invader plasmid pTA409-P1.1 (without mutations) and the pTA409 plasmids with mutations in the target sequence are shown (“reduction in transformation rate by factor” column).
Not all crRNAs can trigger successful interference
The observed differences in the abundance of the crRNAs from each CRISPR locus (Fig. 3) prompted us to investigate whether each crRNA can trigger a successful interference reaction. We constructed a set of invader plasmids, each of which each carried a different protospacer that is identical to the spacers encoded in the CRISPR loci. In our previous experiments, we used only one protospacer, which was derived from the spacer 1 that is encoded by CRISPR locus P1 (P1.1). This protospacer was effectively recognized by its corresponding crRNA and by the CRISPR/Cas system when it was preceded by a functional PAM sequence. Here, we cloned spacer sequences from all three of the CRISPR loci: from each locus a spacer from the 5′ end, the central part and the 3′ end was cloned: spacers 6 and 16 from locus P1 (P1.6, P1.16); spacers 1, 6, 8 and 10 from locus P2 (P2.1, P2.6, P2.8 and P2.10) and spacers 1, 9, 10, 14 and 24 from locus C (C.1, C.9, C.10, C.14 and C.24). All 11 of the protospacers were cloned into plasmid pTA409 downstream of the previously identified functional PAM sequences TTC (PAM3) and ACT (PAM9).15 The transformation of Haloferax with these constructs demonstrated that four of these sequences triggered a defense reaction: P1.1, P1.6, P2.6 and C.1 (Table 2). The remaining seven protospacers (P1.16, P2.1, P2.8, P2.10, C.9, C.10 and C.24) were not recognized as invaders. While the P1.1 and P2.6 spacers were active in triggering a response with both of the PAM sequences tested (PAM3 and PAM9), spacers P1.6 and C.1 were active only with the PAM sequence TTC (PAM3) but not with the ACT sequence (PAM9). In summary, we could show that not all of the crRNAs can trigger a successful interference.
Table 2. Spacers have different abilities to trigger the defense reaction.
| spacer | PAM3 (reduction in transformation rate by factor) |
PAM9 (reduction in transformation rate by factor) |
Spacer length |
AT % |
|---|---|---|---|---|
| P1.1 |
1 x 10−3 |
1 x 10−3 |
37 |
35 |
| P1.6 |
6 x 10−3 |
no |
36 |
52 |
| P1.16 |
no |
no |
39 |
33 |
| P2.1 |
no |
no |
38 |
45 |
| P2.6 |
3 x 10−3 |
3 x 10−3 |
35 |
37 |
| P2.8 |
no |
no |
35 |
54 |
| P2.10 |
no |
no |
37 |
32 |
| C.1 |
2 x 10−3 |
no |
36 |
50 |
| C.9 |
no |
no |
35 |
34 |
| C.10 |
no |
no |
36 |
58 |
| C.24 | no | no | 36 | 39 |
The different spacers used as the invader sequence are shown (spacer column). Their ability to trigger the defense reaction, together with the PAM sequences TTC (PAM3) (PAM3 column) and ACT (PAM9) (PAM9 column), are shown, as well as their length (spacer length column, shown in nucleotides) and AT content (AT% column). The transformation rate for the vector pTA409 (carrying no insert) was set to 1. The reductions in transformation rate for the invader plasmid pTA409-P1.1 and the pTA409 plasmids with other spacers as the target sequence are shown.
Interference depends on the type of the origin of replication
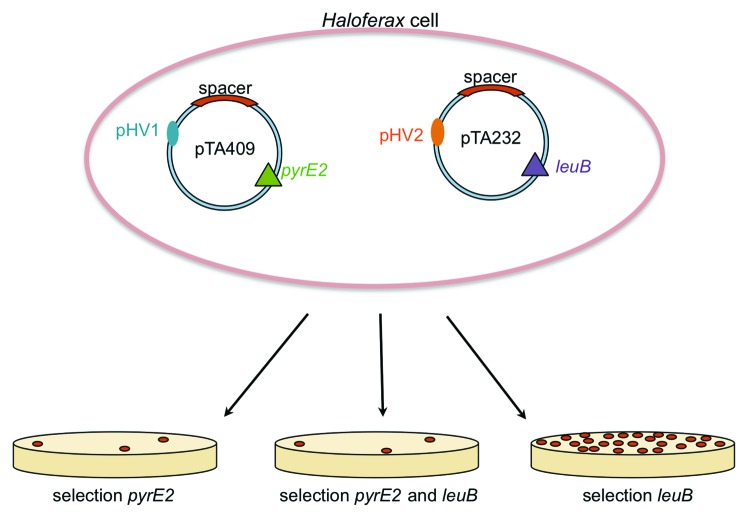
To investigate whether the copy number of the invader is relevant to a successful defense reaction, we cloned spacer P1.1 as a protospacer flanked by a functional PAM (PAM3 and PAM9) into various Haloferax vectors with different origins. The invader plasmid used originally was generated in vector pTA409, which is a low-copy vector that contains the Haloferax pHV1 ori.26,27 To test an additional low-copy vector with the same origin of replication but another selection marker (leuB instead of pyrE2), we used pTA352,26 which also triggered the defense reaction. To test whether a high-copy plasmid would behave differently, we used vector pTA232, which contains the ori pHV2.26,28,29 Upon transformation of Haloferax cells with this plasmid, no interference was observed; i.e., the plasmid was not recognized as an invader. To investigate whether this is due to the nature of the origin or due to the copy number, we transformed Haloferax cells with both the pTA409-PAM3 and pTA232-PAM3 plasmids simultaneously (the same experiment was performed with pTA409-PAM9 and pTA232-PAM9) (Fig. 5). If a high-copy number, resulting in too many copies of invader DNA for the restricted number of crRNAs available, overwhelms the defense system and renders it inoperable, we would expect normal transformation rates. Instead, we observed a selective interference reaction directed at only one type of plasmid, specifically, the pTA409 invader plasmid (Fig. 4). We observed interference if the transformants were plated on medium that was selective only for the pTA409-type invader plasmid; when transformants were plated on medium that was selective only for the pTA232 invader plasmid, we observed normal transformation rates. Thus, only the pTA409 plasmid is destroyed, while the pTA232 plasmid is not degraded. These results suggest that for successful interference, the copy number of the invader is not important but that the type of origin of replication is crucial.
Figure 5. Interference as a function of the invader plasmid origin. To analyze whether the efficiency of the interference reaction depends on the copy number, different types of vectors were used to clone the protospacer and PAM sequences. Haloferax cells were transformed with pTA409-PAM3-P1.1 (pyrE2 marker) and pTA232-PAM3-P1.1 (leuB marker) simultaneously. Plating on medium that was selective for both plasmids (ura- and leu-) resulted in very low transformation rates. Plating on medium selective only for plasmid pTA409-PAM3-P1.1 (ura-) resulted, likewise, in very low transformation rates, indicating that in both cases, the interference reaction was successful. In contrast, plating on medium that was selective only for plasmid pTA232-PAM3-P1.1 (leu-) resulted in full plates, indicating that the defense reaction was not active. Taken together, these data demonstrate that plasmid pTA409-PAM3-P1.1 is selectively degraded.
Discussion
Haloferax CRISPR repeats contain a conserved minimal stem loop
According to our bioinformatic analyses, the Haloferax CRISPR repeats have the potential to fold into a minimal stem loop structure. Because it contains only three base pairs, this potential structure is not very stable; it would unfold even at lower temperatures, which explains why the structure was not detected by NMR analysis and structure probing. The minimal stem loop might be stabilized in the cell by protein binding; in addition, the high salt concentration inside the Haloferax cell (up to 2 M KCl30) could stabilize this stem. The identified structural motif is in good agreement with previously published CRISPR RNA structures, which also contain multiple CG base pairs, and the Gs are predominantly on the right side of the stem. This G side of the stem has been suggested to be important for recognition and cleavage in the type III system of Staphylococcus epidermidis, based on the results of mutational analyses.31 The CRISPR precursor is cleaved 3′ to the final CG base pair in the identified structure,19,31-34 generating an eight-nucleotide 5′ handle.19,21,31,33-36 The length of the remaining repeat sequence at the 3′ end of the crRNA differs from organism to organism and consists of either the remaining repeat sequence or a shortened sequence, sometimes all but the spacer is removed.16,17,31,33-37 We could demonstrate here that the Haloferax crRNA contains an 8-nucleotide-long handle. According to northern blots, the Haloferax crRNAs are approximately 65–70 nucleotides long,15 which would imply that the 3′ handle consists of the remaining 22-nucleotide repeat sequence. However, further experiments will be needed to determine the exact nature of the 3′ end.
The Cas6 protein has been shown to interact with unstructured RNA, as has been found in Pyrococcus furiosus,38 as well as with structured RNA, as has been observed in P. aeruginosa32 (Cas6f) and Thermus thermophilus33 (Cas6e). It will be interesting to see whether the Haloferax Cas6 protein binds to the minimal loop structure that is predicted for the Haloferax repeats or to the unfolded Haloferax repeat.
Mature crRNAs are present in different concentrations
The sequencing data revealed that the crRNAs were not present in equal concentrations; a similar observation was made in Sulfolobus solfataricus,39 P. furiosus,40 Clostridium thermocellum21 and Methanococcus maripaludis.21 One reason for this unequal distribution might be technical biases; for instance, reverse transcription could be prematurely terminated by stable RNA structures. Two additional reasons for this unequal distribution have been discussed in previously published reports: transcription from the opposite strand could produce anti-crRNAs, which could base pair with the crRNAs, thereby reducing the detection of crRNAs, or there could be internal promoters present. To investigate whether any promoters are located in the spacer sequences, we analyzed the regions upstream of spacers, which accumulated to very high levels, and in two upstream spacers (C.8 and P2.1), a potential promoter was found, which could initiate transcription of the downstream spacers C.9 and P2.2, respectively. Additional experiments will show whether these potential promoter motifs are indeed active. With our approach to sequencing RNA in the size range of 55–80 nucleotides, we did not detect any antisense transcripts to the crRNAs.
A seed sequence is required for an effective interaction
Our data demonstrate that, for the defense reaction, a non-contiguous 10-nucleotide seed sequence is essential. Base pairing at nucleotides one to five and seven to 10 is essential for effective interference. Base pairing at position 11 and 12 is not essential, but position 13 must base pair. Notably, a point mutation at position 17 is tolerated, but a deletion of position 17 is not. Single mutations at positions 12, 17 or 36 are tolerated, but double mutations at positions 12 and 36, as well as at positions 17 and 36, result in the loss of interference. The observed seed sequence is similar to the observed seed sequence prerequisites of other organisms14,17,41 and to the eukaryotic RNAi seeds, which require a six- to seven-nucleotide long contiguous seed.42,43 The Haloferax seed sequence is, at 10 nucleotides, three nucleotides longer than the seven-nucleotide seed sequence that has been reported for E. coli. As discussed by Semenova et al.,14 the requirement for a seed sequence suggests that the interference complex (consisting of the Cas proteins and crRNA) scans the invader DNA for the initial identification of the target sequence to allow the subsequent base-pairing of the crRNA.
Not all crRNAs are effective at triggering the interference reaction
Analysis of the efficiency of the various protospacers as invaders revealed a complex picture. Interference is triggered if the crRNA contains any of four different spacers: P1.1, P1.6, P2.6 or C.1. The seven other spacers tested (P1.16, P2.1, P2.8, P2.10, C.9, C.10 and C.24) were not active in triggering the defense mechanism. The invader plasmid is efficient with both of the PAM sequences that were tested (PAM3 and PAM9), if it is carrying spacer P1.1 or spacer P2.6, while spacers P1.6 and C.1 are only active with the PAM sequence PAM3. The PAM sequences we initially identified were all selected for their efficient activity with spacer P1.1.15 So, for each spacer, one might need to find the optimal PAM sequence. Another factor contributing to efficient target recognition might be the AT content of the spacer sequence. The sequences of spacers P2.1 and P2.8 possess AT contents of 45% and 54%, respectively, which is considerably higher than average, when compared with the sequence of the Haloferax genome (35% AT in average30). The increased AT content might result in less stable base pairing between the crRNA and the invader DNA, reducing the efficiency of the interference. Spacers that are part of crRNAs that are present in high concentrations are not necessarily active in triggering the defense system, as protospacers C.9 and P2.10 are not recognized. Because spacers from all three of the CRISPR loci are active (P1, P2 and C), the difference in the repeat sequence at position 23 does not seem to affect activity. The Haloferax genome encodes only one set of Cas proteins, which must process all three types of CRISPR repeats. Although we observed that the spacers that were effective in triggering the defense reaction had lengths of 35, 36 and 37 nucleotides, other spacers of the same lengths did not work. Thus, spacer length alone is not important for recognition. It seems that different factors act together to make a certain crRNA effective.
Interference does not depend on the copy number of the invader plasmid
According to the observed results, we believe that the defense reaction is not dependent on the copy number of the invader plasmid but depends on the origin type that is carried by the plasmid. The fact that, upon entering the cell, the plasmid is presumably present only in single-copy form makes any dependence on the copy number unlikely. While the two plasmids that trigger a defense reaction (pTA409 and pTA352) use an ORC-based mode of replication,26,27 plasmid pTA232 possesses the replication origin pHV2, which uses a distinct replication mode (presumably Rep-dependent28,44). Additional experiments will be needed to show whether the different modes of replication interfere sterically with the defense system (the origin of replication and the invader sequence are located directly next to each other in all three plasmids).
Materials and Methods
Strains
H. volcanii strains H26 (ΔpyrE2) and H11929 were grown aerobically at 45°C in Hv-YPC medium or in Hv-Ca medium (www.haloarchaea.com/resources/halohandbook/Halohandbook_2008_v7.pdf). E. coli strains DH5α (Invitrogen) and GM12145 were grown aerobically at 37°C in 2YT medium.46
Construction of invader plasmid mutants
The initial invader plasmid construct was generated based on the Haloferax shuttle vector pTA40927 (pHV1, pyrE2), including spacer 1 of the CRISPR locus P1 (P1.1) and the PAM sequence TTC (PAM3) or ACT (PAM9)15. To generate the mutations in the seed regions, an overlap-extension reaction was performed with Pfu polymerase (Fermentas, ThermoScientific) and different sets of oligonucleotides (Table S1). The reaction products were cloned into the EcoRV-digested pTA40947 vector, and the resulting plasmids were sequenced. Plasmids were passaged through E. coli GM121 cells (to avoid methylation) and were then introduced into Haloferax cells using the PEG method.29,48 The additional plasmids that were used for the invader tests were pTA35226 (pHV1, leuB) and pTA23229 (pHV2, leuB).
Transformation of H. volcanii
Plasmids were introduced into H. volcanii strain H2629 (ΔpyrE2) or H11929 (ΔpyrE2, ΔleuB, ΔtrpA). Transformants were selected on Hv-Ca plates without uracil. As a positive control, strain H26 or H119 was transformed with plasmid pTA409. Transformation rates were calculated as the number of uracil prototroph colonies, divided by the number of clones that could be grown in the absence of selection pressure. Each transformation reaction was conducted at least twice using independent preparations of plasmid. To confirm the identification of a functional invader sequence, H. volcanii cells were transformed at least three times with the plasmid-invader construct, using at least two different plasmid preparations. As has been observed in similar studies,13,15 it is difficult to accurately determine transformation rates: therefore, we defined only those sequences that led to at least a 100-fold reduction in transformation rates in this plasmid assay as a functional invader sequences for H. volcanii.
Isolation of the crRNA fraction and sequencing of cDNAs
Total RNA was isolated from exponentially growing H. volcanii H119 cells (OD: 0,8) using TriZol (Invitrogen, Life Technologies) and separated by 8% PAGE. The fraction of RNA ranging from 55–80 nucleotides length was eluted. Because crRNAs have been reported to contain 3′ phosphate and 5′ OH groups, the eluted RNA was treated with T4 polynucleotide kinase to generate 3′ OH groups49 and to obtain 5′ phosphate groups. For the generation of cDNA and for sequencing, the RNA was sent to LGC Genomics GmbH (Berlin). Shortly, 3 µg of gel-purified RNA was ligated with 20 pM of 454 LibA-Adaptor RNA oligo using T4 RNA ligase (Epicenter Biotechnologies). The RNA was reverse-transcribed using a 454 LibB-Adaptor-nonaN DNA oligo and AffinityScript Reverse Transcriptase (Takara Bio Inc.). The resulting first strand cDNAs were amplified by PCR, using the standard 454 Adaptor primers 454-F/454-R. The PCR products were sequenced using the Roche/454 GS FLX system with the Roche/454 Titanium chemistry according to the manufacturer’s instructions (454 Life Sciences). The sequencing was performed on 1/8th of a PTP.
Analysis of the sequencing data
Due to the internal priming of the 3′ oligonucleotides used in the sequencing protocol, we first extracted crRNA sequences from all of the read sequences. Extraction was performed by identifying the individual crRNA 5′ tags and cutting up to 35 nucleotides of the subsequent spacer sequence. The crRNA reads were extracted for each locus individually and then mapped to the genome using Segemehl, version 0.1.3.50 We used the sequenced genome for H. volcanii DS2, with the accession NC_013967.1 for the chromosome and CP001955.1 for the plasmid pHV4. The crRNA abundance profiles for each locus were visualized using the Integrative Genomics Viewer (IGV), version 2.0.3.51
Repeat structure predictions
The minimum free-energy structures of each repeat were determined using RNA fold from the Vienna Package, version 1.8.4,24 with the options -d2-noLP-p (Figs. S2 and S3). A single repeat representative was taken from one of the other 21 Haloarchaeal genomes in the CRISPRdb database.52 The consensus structural alignments were generated using the LocARNA web server.53 To determine the influence of the context sequence on the individual repeats, the entire repeat-spacer array was folded using RNAplfold, also from the Vienna Package, version 1.8.4,54 with the options -W 200-L 150-noLP (Figs. S2 and S3). The locality parameter settings for the window size (W) and the maximum base-pair span (L) were taken from Lange et al.55
NMR analyses
The 1D-1H-NMR-spectra were recorded on a Bruker Avance 600-MHz NMR-spectrometer equipped with a cryogenic probe, in a buffer containing 25 mM K2HPO4/KH2PO4, pH 6.5, 50 mM KCl and 10% (v/v) D2O, using a Jump-Return-Echo pulse sequence56 for water suppression at temperatures of 283 and 298 K.
Structural probing
The repeat RNA for P1 and P2 was obtained from Biomers (biomers.net). The RNA was labeled at the 5′ end and subjected to digestion with the RNases T1, V1 and S1 to identify single-stranded and double-stranded regions, according to the manufacturer’s instructions (Ambion, Invitrogen, Life Technologies).
Supplementary Material
Acknowledgments
We are grateful to Thorsten Allers (University of Nottingham) for the Haloferax vectors and genetic tools. We thank Elli Bruckbauer for her expert technical assistance. This work was funded by the German Research Council (Deutsche Forschungsgemeinschaft) through the research group FOR 1680 “Unravelling the prokaryotic immune system” (grant DFG MA1538/17-1 and BA2168/5-1). We would like to thank the state of Hessen for funding the Biomolecular Magnetic Resonance Center (BMRZ). We thank members of the research program for their helpful discussion.
Glossary
Abbreviations:
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cas
CRISPR associated
- crRNA
CRISPR RNA
- PAM
protospacer adjacent motif
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/24282
References
- 1.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 3.Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Review: Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem. 2011;2011:7. doi: 10.1515/BC.2011.042. [DOI] [PubMed] [Google Scholar]
- 4.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. 2011;45:273–97. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 5.Garrett RA, Vestergaard G, Shah SA. Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol. 2011;19:549–56. doi: 10.1016/j.tim.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Marchfelder A, Fischer S, Brendel J, Stoll B, Maier LK, Jäger D, et al. Small RNAs for defence and regulation in archaea. Extremophiles. 2012;16:685–96. doi: 10.1007/s00792-012-0469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 9.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet. 2012;46:311–39. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 11.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–76. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–40. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 13.Gudbergsdottir S, Deng L, Chen Z, Jensen JV, Jensen LR, She Q, et al. Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol. 2011;79:35–49. doi: 10.1111/j.1365-2958.2010.07452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108:10098–103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer S, Maier LK, Stoll B, Brendel J, Fischer E, Pfeiffer F, et al. An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J Biol Chem. 2012;287:33351–63. doi: 10.1074/jbc.M112.377002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–36. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 17.Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011;108:10092–7. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–56. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Randau L. RNA processing in the minimal organism Nanoarchaeum equitans. Genome Biol. 2012;13:R63. doi: 10.1186/gb-2012-13-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richter H, Zoephel J, Schermuly J, Maticzka D, Backofen R, Randau L. Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012;40:9887–96. doi: 10.1093/nar/gks737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofacker IL, Fontanta W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh Chem. 1994;125:167–88. doi: 10.1007/BF00818163. [DOI] [Google Scholar]
- 25.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norais C, Hawkins M, Hartman AL, Eisen JA, Myllykallio H, Allers T. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 2007;3:e77. doi: 10.1371/journal.pgen.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delmas S, Shunburne L, Ngo HP, Allers T. Mre11-Rad50 promotes rapid repair of DNA damage in the polyploid archaeon Haloferax volcanii by restraining homologous recombination. PLoS Genet. 2009;5:e1000552. doi: 10.1371/journal.pgen.1000552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charlebois RL, Lam WL, Cline SW, Doolittle WF. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc Natl Acad Sci USA. 1987;84:8530–4. doi: 10.1073/pnas.84.23.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allers T, Ngo HP, Mevarech M, Lloyd RG. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl Environ Microbiol. 2004;70:943–53. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartman AL, Norais C, Badger JH, Delmas S, Haldenby S, Madupu R, et al. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One. 2010;5:e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc Natl Acad Sci USA. 2011;108:21218–22. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–8. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–7. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 34.Juranek S, Eban T, Altuvia Y, Brown M, Morozov P, Tuschl T, et al. A genome-wide view of the expression and processing patterns of Thermus thermophilus HB8 CRISPR RNAs. RNA. 2012;18:783–94. doi: 10.1261/rna.031468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–9. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE) J Biol Chem. 2011;286:21643–56. doi: 10.1074/jbc.M111.238485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat Struct Mol Biol. 2011;18:688–92. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 38.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–64. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng L, Kenchappa CS, Peng X, She Q, Garrett RA. Modulation of CRISPR locus transcription by the repeat-binding protein Cbp1 in Sulfolobus. Nucleic Acids Res. 2012;40:2470–80. doi: 10.1093/nar/gkr1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O’Toole GA. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol. 2012;194:5728–38. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–61. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woods WG, Dyall-Smith ML. Construction and analysis of a recombination-deficient (radA) mutant of Haloferax volcanii. Mol Microbiol. 1997;23:791–7. doi: 10.1046/j.1365-2958.1997.2651626.x. [DOI] [PubMed] [Google Scholar]
- 45.Allers T, Barak S, Liddell S, Wardell K, Mevarech M. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl Environ Microbiol. 2010;76:1759–69. doi: 10.1128/AEM.02670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller JH. 1972. Experiments in Molecular Genetics Cold Spring Harbour, N. Y.: Cold Spring Harbour Laboratory Press. [Google Scholar]
- 47.Hölzle A, Fischer S, Heyer R, Schütz S, Zacharias M, Walther P, et al. Maturation of the 5S rRNA 5′ end is catalyzed in vitro by the endonuclease tRNase Z in the archaeon H. volcanii. RNA. 2008;14:928–37. doi: 10.1261/rna.933208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cline SW, Schalkwyk LC, Doolittle WF. Transformation of the archaebacterium Halobacterium volcanii with genomic DNA. J Bacteriol. 1989;171:4987–91. doi: 10.1128/jb.171.9.4987-4991.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schürer H, Lang K, Schuster J, Mörl M. A universal method to produce in vitro transcripts with homogeneous 3′ ends. Nucleic Acids Res. 2002;30:e56. doi: 10.1093/nar/gnf055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, et al. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput Biol. 2009;5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38(Web Server issue):W373-7. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernhart SH, Hofacker IL, Stadler PF. Local RNA base pairing probabilities in large sequences. Bioinformatics. 2006;22:614–5. doi: 10.1093/bioinformatics/btk014. [DOI] [PubMed] [Google Scholar]
- 55.Lange SJ, Maticzka D, Möhl M, Gagnon JN, Brown CM, Backofen R. Global or local? Predicting secondary structure and accessibility in mRNAs. Nucleic Acids Res. 2012;40:5215–26. doi: 10.1093/nar/gks181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fürtig B, Richter C, Wöhnert J, Schwalbe H. NMR spectroscopy of RNA. Chembiochem. 2003;4:936–62. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.